Abstract
Our aim has been to understand the features of erbB receptor homo- and heterodimer assembly to develop approaches to disrupt receptor activation. We have developed a general approach to cause erbB receptor-specific trans inhibition of human neoplasia. The clonal progression of human astrocytomas to a more malignant phenotype often involves the amplification and overexpression of the epidermal growth factor receptor (EGFr) gene. We have selectively targeted the EGFr in human glioblastoma cells with kinase-deficient mutants of the erbB family derived from the ectodomain of the Neu oncogene that are able to form heterodimers with EGFr and inhibit EGFr-dependent phenotypes. In EGFr-positive U87MG human glioblastoma cells, expression of the Neu ectodomain inhibits EGF-, but not platelet-derived growth factor-, induced DNA synthesis; inhibits cell proliferation in the presence of EGF, but not platelet-derived growth factor; inhibits the ability of U87MG to form colonies in soft agar; and inhibits transforming efficiency in athymic mice. These studies establish that EGFr-mediated signal transduction is important in the maintenance of malignant glioma, and that trans receptor inhibition is a novel way to abrogate abnormal growth of these tumors. Neu ectodomains will be useful in determining the manner in which the EGFr contributes to glial tumorigenesis and in the design of pharmaceuticals that disable erbB family oncoproteins. In addition, these studies provide a rationale for the application of the Neu ectodomain in gene therapy approaches to human malignant glioma and, potentially, to other systemic epithelial malignancies expressing erbB family receptors.
Keywords: epidermal growth factor receptor, glioma/Neu
Amplification and/or alteration of the epidermal growth factor receptor (EGFr) gene is frequently observed in glial tumor progression (1, 2), particularly in glioblastoma, the most malignant glial tumor (3–8). A significant proportion of these tumors show EGFr amplification with or without gene alteration (2–4), and this has been correlated with a shorter interval to disease recurrence and poorer survival (8).
EGFr amplification can be associated with aberrant EGFr transcripts along with normal EGFr transcripts (1). Frequent amplification and subsequent structural alteration suggests the EGFr may be important for the maintenance of the phenotype of malignant glioma. A frequently observed EGFr mutant has been identified in a subset of human glioblastomas and results from an in-frame truncation of 801 bp (corresponding to exons 2–7) in the extracellular domain of the receptor (1, 2, 9–11), which is thought to result in constitutive kinase activation and may also affect the ligand-binding properties of the molecule (7, 12).
Observed mutations of EGFr in human epithelial malignancies consist of overexpression with or without amplification and, less commonly, of coding sequence alterations. Oncogenic transformation caused by mutants of EGFr appear to be tissue-specific and have been observed in erythroid leukemia, fibrosarcoma, angiosarcoma, and melanoma, as well as glioblastoma (13). Overexpression of the normal EGFr may cause oncogenic transformation in certain cases, probably in an EGF-dependent manner (13, 14). Transfection of high amounts of wild-type EGFr into NIH 3T3 cells results in ligand-dependent but incomplete transformation (15). Overexpression may cause altered cell-cycle regulation of the EGFr kinase and contribute to the transformed state, as has been observed for oncogenic p185neu (16).
Previous work has shown that U87MG human glioblastoma cells expressing the 801-bp ectodomain-deleted EGFr mutant seen in a subset of human glioblastoma tumor samples have increased oncogenicity in vivo but not in vitro when compared with U87MG cells expressing only endogenous EGFr (7). This feature may relate to an altered subcellular localization of the constitutively active mutant form (17). These data suggest a role for EGFr-mediated signaling in glial tumorigenesis. EGFr-expressing clones, which respond in a ligand-independent manner, may provide a growth advantage for glioblastoma cells in vivo (7). The mechanism by which EGFr may confer a growth advantage in vivo is unknown, but may include an effect on angiogenesis (7, 18).
Ligand binding of receptor tyrosine kinases induces receptor dimerization (19), transphosphorylation of receptor proteins, and phosphorylation of intracellular substrates, leading to cell growth or differentiation (20, 21). EGFr and Neu/c-erbB-2 (rat/human form) interact (22–24), and form an active, heterodimeric kinase complex both in vitro and in vivo (23, 25–27). Heterodimer formation has also been observed among other erbB receptors and may be a more general mechanism of receptor activation among other receptor tyrosine kinases (21, 28). Structurally altered receptor deletion mutants have been observed to function in a dominant–negative manner by suppressing the function of wild-type receptors, in particular the EGFr, flk-1 (receptor for vascular endothelial growth factor), and the insulin receptor, leading to phenotypic change (29–32).
The Neu ectodomain associates with EGFr and virtually eliminates high-affinity EGF binding sites in a fibroblast-derived background (33, 34). The unproductive kinase mutant heterodimer dramatically limits synergistic transformation and tumorigenicity of human tumor cells, establishing a trans receptor inhibitory property for human neoplasia. In this work, we demonstrate that transfer of the Neu ectodomain to EGFr-containing human glioblastoma cells inhibits the transformed phenotype, and show that this inhibition is mediated by specific inhibition of the EGFr. Cell growth and proliferation often revert to a quiescent normal level after introduction of the ectodomain-derived mutant Neu protein, indicating that nonproliferating cells would be unaffected by the expression of Neu ectodomains. Based on these studies, Neu ectodomains will facilitate study of the role played by the EGFr in glial tumorigenesis, and may be important in the design of pharmaceuticals that disable erbB family receptors and/or erbB signal transduction pathways. Finally, ectodomain-derived Neu constructs may be used for therapeutic gene delivery into human glioblastomas.
MATERIALS AND METHODS
Vector Construction.
The deletion mutants T691stop and N691stop were derived from pSV2Tneu and pSV2Nneu (35), respectively, by substitution of a stop codon for the normal codon Thr-691, as described (34).
A fragment containing the neor gene isolated from pSV2NEOr (36) was subcloned into pSV2T691stop and pSV2N691stop. The simian virus 40 early promoter fragment was then replaced by the human cytomegalovirus (CMV) promoter/enhancer from the plasmid p290 (37), resulting in pCMV/T691stop/neor and pCMV/N691stop/neor for eukaryotic expression. In addition, the full-length Neu cDNA [Nneu (35)] was subcloned into this expression vector to create pCMV/Nneu/neor.
Maintenance of Cells and Development of Stably Transfected Cell Lines.
The U87MG human glioblastoma cell line was obtained from Webster Cavenee (Ludwig Cancer Institute, San Diego). For stable cell transfections, 10 micrograms of either the pCMV/N691stop/neor, pCMV/T691stop/neor, or pCMV/Nneu/neor construct was transfected into U87MG cells via the lipofectamine reagent (GIBCO/BRL) under conditions determined by transfections using the pCMV-β (bacterial β-galactosidase) (CLONTECH) reporter construct. Optimal transfection efficiency was determined by chemiluminescence as detected by a luminometer (Tropix, Bedford, MA). After 2–3 weeks in selection media containing 0.8 mg/ml G418 sulfate (Geneticin, GIBCO/BRL), established clones were isolated and screened by flow cytometric analysis with anti-neu mAb 7.16.4 staining (34). All cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Bio-Whittaker) with 10% fetal bovine serum (FBS) (HyClone), 100 units penicillin, 50 μg/ml streptomycin, and 2 mM l-glutamine (GIBCO/BRL). All cultured cells were maintained at 37°C in 5% CO2/95% air. The medium for stably transfected cell lines was supplemented with 0.4 mg/ml G418 sulfate (Geneticin) for maintenance of transgene expression. Stably transfected cell lines were periodically checked by flow cytometric analysis with mAb 7.16.4 to document stable levels of transgene expression.
Metabolic Labeling of Cells Followed by Immunoprecipitation.
Full details have been described by our laboratory (33, 34, 38). Briefly, subconfluent cells (1 × 106) are seeded overnight onto 10-cm dishes in full-growth medium (10% FBS/DMEM). The following day, cells are starved in cysteine-free DMEM for 1 hr then pulsed with [35S]cysteine (50 μCi/ml; 1 Ci = 37 GBq; Amersham) for 15 hr in 3% dialyzed FBS/cysteine-free DMEM. Lysates are harvested after two washes in PBS using a PI/RIPA buffer (34). Immunoprecipitations are carried out on ice for 60 min and complexes are separated by binding to protein A-Sepharose prior to separation by 8% SDS/PAGE, drying, and exposure to film. mAb 7.16.4 against the Neu ectodomain has been described (33, 34). mAb 225 against the EGFr was provided by John Mendelsohn (Memorial Sloan–Kettering Cancer Center). A polyclonal antisera against all forms of platelet-derived growth factor receptor (PDGF-R) was obtained from Upstate Biotechnology (Lake Placid, NY) and a mAb against erbB3 was a gift from W. J. Gillick (Imperial Cancer Research Fund, London). Five micrograms of antibody was used for immunoprecipitating proteins from lysates recovered from 10-cm dishes.
Proliferation Assay as Measured by MTT [3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide] Incorporation.
Cell lines are plated in 96-well plates at 3,000 cells per well in 10% DMEM. After seeding, cells are starved and synchronized in either 0.05% or 0.5% DMEM × 48 hr before incubation with various concentrations of EGF or PDGF for 24–48 hr. MTT is given to the cells for 4 hr (39). Cells are lysed in 50% SDS/20% dimethyl sulfoxide and kept at 37°C overnight. Proliferation is assessed by taking optical density readings at 570 nm (39), using an ELISA reader. This reading is directly proportional to viable cell number. The number of cells used in this assay was determined to be within the linear range for this cell type.
In Vitro and in Vivo Tumorigenicity Assays.
Anchorage-independent growth was determined by assessing the colony-forming efficiency of cells suspended in soft agar. Cells (1–3 × 103) were suspended in a 1 ml top layer (0.18% agarose/10% FBS/DMEM) in 6-cm culture dishes containing a 3 ml cell-free feeder layer consisting of 0.25% agarose in DMEM supplemented with 10% FBS and 20 mM Hepes (pH 7.5). Colonies (>0.3 mm) were visualized and counted on day 21–28 for all cell lines after staining with p-iodonitrotetrazolium violet (1 mg/ml). Each cell line was examined in triplicate for three separate experiments.
NCr homozygous nude mice (6–8 weeks old) were purchased from the National Cancer Institute. Cells (1 × 106) were suspended in 0.1 ml of PBS and injected intradermally into the mid-dorsum of each animal. Parental U87MG cells were injected on one side of individual animals and stably transfected cell lines were injected on the contralateral side to make direct comparisons of growth within each animal. PBS alone was also injected into each animal as an additional control. Animals were maintained in accordance with the guidelines of the Committee on Animals of the University of Pennsylvania and those prepared by the Committee on Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources. Tumor growth was monitored twice weekly for 10–12 weeks. Tumor size was calculated by measuring tumor volume (length × width × thickness).
Measurements of DNA Synthesis.
DNA synthesis was assessed by incorporation of the thymidine analog 5-bromodeoxyuridine (BrdUrd) and its detection by indirect immunofluorescence (40). Subconfluent cells (1.25 × 105 per well) were seeded overnight onto coverslips in 6-well plates in full-growth media and then grown in serum-free media for 48 hr. Ligand at the indicated concentrations, 10% serum, or serum-free media was then added back in media containing BrdUrd for 24 hr. Cells were then fixed in 95% ethanol/5% acetic acid for 15 min at room temperature. Prior to antibody staining, cells were permeabilized in PBS containing 0.1% Tween for 2 min at room temperature. BrdUrd incorporation was detected by staining with a mouse monoclonal anti-BrdUrd antibody (Amersham) followed by a Texas red-coupled anti-mouse antibody (Amersham). The Texas red-coupled antibody was diluted 1:200 in PBS containing homologous cell extract and 0.1 mg of BSA per ml. Cells were counted under indirect immunofluorescence after being counterstained with Hoechst 33528 at 1:10,000 in PBS. The percentage of BrdUrd-positive cells was determined by comparing BrdUrd-positive cells to Hoechst-positive cells and counts were confirmed by an independent observer.
RESULTS
Characteristics of Stably Transfected U87MG Cells Expressing Kinase-Deficient Neu cDNAs.
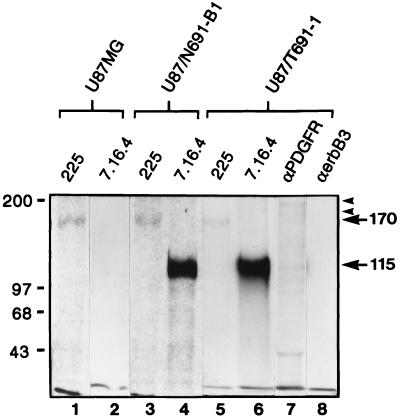
The presence of EGFr (Mr = 170 kDa) in U87MG cells was determined by flow cytometric analysis and by metabolic labeling with [35S]cysteine, followed by immunoprecipitation, and SDS/PAGE (Fig. 1). We have developed clonal U87MG-derived cell lines expressing either the T691stop (subclone U87/T691-1, expressing the truncated Neu protein containing the oncogenic rat Neu transmembrane point mutation) or N691stop (subclone U87/N691-B1, expressing the truncated Neu protein containing the protooncogenic Neu transmembrane region) form of kinase-deficient Neu. Expression of each ectodomain form results in a protein with a cytoplasmic domain deletion of 591 carboxyl-terminal amino acids, which includes the kinase domain and autophosphorylation sites. Both N691stop and T691stop Neu ectodomain proteins are capable of forming heterodimers with EGFr in U87MG cells (data not shown). The U87/T691-1 and U87/N691-B1 cell lines express maximal levels of the transgene relative to five other clonal cell lines expressing either form of truncated Neu we characterized.
Figure 1.
Comparison of expression levels of EGFr and truncated forms of Neu proteins in U87MG human glioblastoma cells after metabolic labeling. Subconfluent cells (1 × 106 cells per lane) were labeled with [35S]cysteine for 15 hr. Cells include U87MG parental human glioblastoma cells and U87MG cell lines transfected with either the N691stop (U87/N691-B1) or the T691stop form of Neu (U87/T691-1). Cell lysates were then immunoprecipitated with either anti-EGFr mAb 225 (lanes 1, 3, and 5); anti-neu mAb 7.16.4 (lanes 2, 4, and 6), which recognizes the Neu ectodomain; anti-PDGF-R polyclonal antisera (lane 7); or anti-erbB3 mAb (lane 8). Immune complexes were resolved and separated by 8% SDS/PAGE. Protein signals representing EGFr (170 kDa), truncated Neu proteins (either N691stop or T691stop form, each 115 kDa), PDGF-R (180 and 190 kDa forms), and human erbB3 were observed after autoradiography (3-day exposure).
Expression of the truncated Neu protein (Mr = 115 kDa) was confirmed by both flow cytometric analysis and metabolic labeling with [35S]cysteine in both U87/T691-1 and U87/N691-B1 (Fig. 1). There is no endogenous human p185c-erbB2 as determined by flow cytometric analysis in the U87MG cell line. Flow cytometric analysis revealed very low levels of erbB3 in these cells, which was beyond the resolution of metabolic labeling (Fig. 1, lane 8). Flow cytometric analysis revealed that the level of endogenous EGFr on all subclones of U87MG expressing the Neu ectodomain is approximately equal to that observed in parental U87MG. Varying the stoichiometry of the ectodomain-derived Neu peptide did not alter levels of endogenous EGFr in U87MG cells. Densitometric analysis after metabolic labeling with [35S]cysteine (Fig. 1) indicated that the relative ratio of protein precipitated with mAb 7.16.4 to that precipitated by mAb 225 was higher in U87/T691-1 cells than in U87/N691-B1 cells. The higher stoichiometric ratio of mutant Neu to EGFr in the U87/T691-1 subclone was supported by flow cytometric analysis.
Phenotypic Characteristics of U87MG-Derived Cell Lines Expressing the Neu Ectodomain.
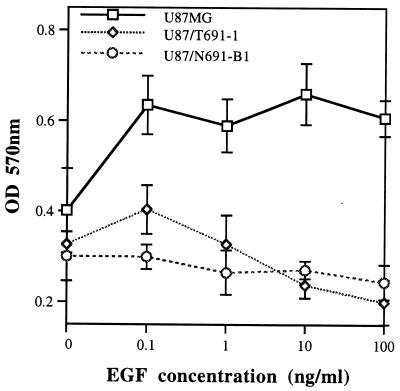
Abrogation of in vitro proliferation in response to EGF after serum starvation was observed in U87MG subclones stably transfected with either the N691stop or T691stop form (Fig. 2). Parental U87MG cells exhibited a 64.39% and 51.37% increase in proliferation to 10 ng/ml and 100 ng/ml EGF, respectively (Fig. 2). The U87/N691-B1 and U87/T691–1 cell lines did not exhibit an increase in proliferation at these EGF concentrations in this representative experiment. Transient transfections of either the N691stop or T691stop form of truncated Neu into U87MG cells also resulted in a blunted mitogenic response to EGF and in a 32–44% reduction in the ability to form morphologically transformed foci at 2 weeks relative to U87MG cells transfected with pCMV-β. Capacity for growth in low serum in these clones was also reduced. U87/N691-B1 cell numbers were reduced 63.2 ± 1.31% (mean ± SD) and U87/T691-1 cell numbers were reduced 73 ± 15.2% in three experiments of growth in reduced serum.
Figure 2.
Proliferation in response to EGF as determined by MTT incorporation. Three thousand cells of each cell line were seeded in a 96-well plate in 10% FBS/DMEM overnight before being starved in 0.05% DMEM for 48 hr. Recombinant EGF at the indicated concentrations was then added for 48 hr at which point MTT was added for 4 hr prior to cell lysis. The plates were then incubated at 37°C overnight before spectrophotometric analysis of viable cell number with an ELISA reader (OD570). Data points represent mean and standard deviation for four samples at each ligand concentration. Similar experiments were performed on six independent occasions. The number of cells used was determined to be within the linear range of this assay.
Inhibition of Anchorage-Independent Growth in Vitro.
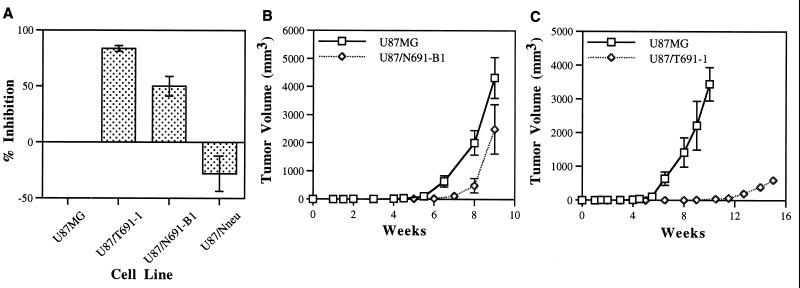
The ability of U87MG cells expressing mutant Neu proteins to form colonies in soft agar was dramatically reduced (Fig. 3A). The transforming efficiency of the U87/T691-1 clonal line was inhibited 83.7 ± 2.6% (mean ± SEM) and in U87/N691-B1 by 50 ± 8.7% in three independent experiments. A U87MG-derived cell line expressing full-length, protooncogenic Neu (U87/Nneu) used as a control was more transformed in these assays than parental U87MG. The EGFr+/p185c-neu-expressing line displayed a 27 ± 15.7% increase in transforming efficiency relative to the parental U87MG. This observation supports prior experiments in our laboratory indicating a synergistic role in both normal signaling and transformation between full-length Neu and EGFr in a fibroblast-derived context (22, 26).
Figure 3.
Growth inhibition of U87MG glioblastoma cells by truncated Neu ectodomains. (A) Anchorage-independent growth assay. Cells (1,000–3,000 of each cell line) were seeded in soft agar dishes and cultured for 21–28 days. Colonies were then visualized and counted after staining. Each experiment was performed in triplicate. The mean degree of inhibition and standard error (SEM) observed in three independent experiments is shown. The U87/Nneu cell line is a control cell line that expresses full-length normal p185neu and exhibits increased transforming efficiency in vitro in these experiments. (B) Comparison of tumor growth in athymic mice between U87MG and U87MG cells expressing the N691stop form of Neu (U87/N691-B1) and (C) between U87MG and U87MG cells expressing the T691stop form of Neu (U87/T691-1). 106 cells of each cell line were injected intradermally on day 0 and tumor volume was recorded weekly. U87MG cells were injected on one side and the transfected cell line was injected into the contralateral side of the same animal. Data represents the mean and standard deviation (SD) of tumors derived from each cell line (U87MG, n = 19; U87/N691-B1, n = 9; U87/T691–1, n = 9).
In Vivo Inhibition of Cell Lines Expressing Either the N691stop or T691stop Form of the Neu Ectodomain.
We compared growth of the glioblastoma clones after injection into athymic mice. Both U87/T691–1 and U87/N691-B1 were inhibited relative to U87MG parental cells, with U87/T691–1 exhibiting a profound degree of inhibition (Fig. 3 B and C). The U87/T691–1 transfected line did not form appreciable tumors until 12 weeks after implantation (Fig. 3C), which is beyond the period of time when some animals injected with parental U87MG cells had to be sacrificed due to excessive tumor burden. Many animals survived greater than 100 days with tumors < 0.5 cm3 after subcutaneous inoculation with U87/T691–1 cells. Additionally, more than 50% of the subcutaneous injections with this cell line failed to produce tumors of any measureable size. These data were reproduced when different subclones of both U87/N691 and U87/T691 were used in both the in vitro and in vivo transformation assays. The U87MG-derived cell line expressing full-length normal Neu (U87/Nneu), which exhibited increased tumorigenicity in vitro (Fig. 3A), displayed nearly identical growth kinetics to U87MG parental glioblastoma cells in vivo. This observation was further corroborated by analysis of in vivo growth of an additional U87MG subclone expressing full-length p185c-neu.
Specificity of Inhibition of EGFr Signaling by the Neu Ectodomain.
To address the specificity of inhibition mediated by the Neu ectodomain, we examined whether the more inhibited transfected line, U87/T691–1, was capable of responding in vitro to PDGF. A PDGF-R signaling pathway has been implicated in glial tumorigenesis (41). A polyclonal antibody reactive with all dimeric forms of PDGF-R precipitated predominant bands of 180 kDa and 190 kDa in the U87MG parental line (data not shown) and U87/T691–1 (Fig. 1, lane 7). These bands correspond to PDGF receptors observed in other human glioblastoma cell lines (42–44). The 180 kDa form represents the mature β-type receptor, which others have precipitated in the U87MG human glioblastoma cell line (43). We observed more significant proliferation in U87/T691–1 to PDGF-BB [binds all dimeric forms of the PDGF-R and binds β receptors with high affinity (43)] than to EGF (Table 1). We also observed more significant proliferation to PDGF-BB than to EGF in fibroblasts expressing EGFr, PDGF-R, and kinase-deficient Neu proteins.
Table 1.
Percent increase in proliferation in response to ligand
| Ligand conc., ng/ml | U87MG (parental)
|
U87/T691-1
|
||
|---|---|---|---|---|
| EGF | PDGF-BB | EGF | PDGF-BB | |
| 0 | — | — | — | — |
| 0.01 | 18.5 ± 0.6 | 35.8 ± 6.1 | 6.5 ± 5.1 | 30.0 ± 5.6 |
| 0.1 | 53.3 ± 3.9 | 47.3 ± 9.2 | 6.8 ± 4.5 | 40.8 ± 4.8 |
| 1.0 | 58.6 ± 4.9 | 43.5 ± 5.1 | 8.8 ± 4.5 | 47.1 ± 7.4 |
| 10.0 | 64.5 ± 7.8 | 43.5 ± 6.9 | 3.7 ± 2.2 | 45.9 ± 3.9 |
| 100 | 61.1 ± 6.2 | 59.9 ± 9.3 | 9.0 ± 5.4 | 53.2 ± 12.7 |
Proliferation in response to either EGF or PDGF-BB is shown between the U87MG cells and the U87/T691-1 cell line (expressing the T691stop truncated form of Neu). These data represent a compilation of six independent experiments (the response of U87/T691-1 to EGF represents data from eight separate experiments). Proliferation in response to ligand was determined by the incorporation of MTT under conditions described in Materials and Methods. The mean % increase above baseline (no ligand) ± SEM is shown. The measured proliferation was determined to be over a linear range for this cell line. conc., concentration.
To corroborate this observation with an additional measure, and to analyze, independently, S-phase events in response to ligand, we addressed the ability of PDGF-BB to initiate DNA synthesis in U87MG parental cells and in U87/T691–1 transfected cells as measured by BrdUrd incorporation (Table 2). PDGF-BB initiates DNA synthesis to a more significant degree than EGF in transfected U87/T691–1, and these cells respond nearly as well as U87MG to PDGF-BB after serum starvation. The response of Neu ectodomain-transfected cells to EGF is significantly blunted relative to nontransfected glioblastoma cells after serum starvation. These data suggest that the inhibition produced by the Neu ectodomain is primarily mediated via inhibition of EGFr-mediated signal transduction.
Table 2.
Percent BrdUrd incorporation
| Condition | Serum-free media | 10% serum | EGF, 10 ng/ml | PDGF-BB, 10 ng/ml | PDGF-BB, 50 ng/ml |
|---|---|---|---|---|---|
| U87MG | 19.4 ± 4.9 | 90.5 ± 7.8 | 45.5 ± 0.9 | 22.7 ± 4.4 | 36.9 ± 6.8 |
| U87/T691-1 | 15.4 ± 0.7 | 83.6 ± 0.4 | 17.3 ± 7.0 | 19.5 ± 6.2 | 30.5 ± 4.7 |
DNA synthesis in response to either EGF or PDGF-BB for U87MG cells and U87/T691-1 cells (expressing the T691stop truncated form of Neu) is shown. After starvation in serum-free media for 48 hr, cells were either placed back into serum-free media, 10% FBS DMEM (complete growth media), or ligand in serum-free media at the indicated concentration for 24 hr. BrdUrd incorporation was determined by immunohistochemistry and positive cells were counted as described in Materials and Methods. The data shown are mean ± SD for triplicate samples. Similar results were observed in three independent experiments. The results of this representative experiment were confirmed by an independent observer.
DISCUSSION
The heterodimerization between EGFr and either wild-type or mutant Neu proteins can be detected even in the absence of EGF, and is preferred thermodynamically over either form of homodimerization (33, 34). Recent evidence suggests that erbB-2 forms heterodimers under physiologic conditions with EGFr and erbB-3, increasing signaling in response to EGF and Neu differentiation factor (NDF or heregulin), respectively, and supporting the important paradigm that full-length Neu/c-erbB-2 potentiates growth factor signaling in trans (45).
The Neu ectodomain appears to be more effective than dominant–negative constructs of EGFr in binding to EGFr and causing trans inhibition of EGFr-mediated signal transduction. Homodimerization and codimerization between EGFr and cytoplasmic domain-deleted EGFr have been shown to be equally efficient but are notably EGF-dependent (30). Expression of the N691stop Neu mutant leads to elimination of the high-affinity class of EGFrs, an inhibition of EGF-induced DNA synthesis, less efficient EGF-induced internalization and down-regulation of EGFr, and reduced oncogenic potential in mutant Neu-EGFr coexpressing fibroblasts (33). These observations provided biochemical evidence that the Neu ectodomain is sufficient to associate with the EGFr and modulate EGFr-dependent phenotypes. Furthermore, the Neu ectodomain appears to associate preferentially with EGFr, rather than with other Neu monomers, again suggesting that kinase-deficient Neu may more efficiently disable EGFr function than cytoplasmic domain-deleted EGFr. There is an increase in tyrosine phosphorylation of EGFr in response to EGF in U87MG human glioblastoma cells and constitutively active, ligand-independent EGFr mutants increase the tumorigenicity of the U87MG cell line, showing that the EGFr signaling pathway is important for the transformed phenotype in these cells (7).
Neu ectodomain proteins are capable of forming heterodimers with EGFr in U87MG glioblastoma cells. Neu ectodomains inhibit EGF-induced DNA synthesis (Table 2), block the in vitro proliferation of EGFr-positive human glioblastoma cells to EGF (Fig. 2, Table 1), lower total cell numbers in culture in reduced serum, reduce the ability of glioblastoma cells to form morphologically transformed foci and grow in an anchorage-independent manner in soft agar (Fig. 3A), and reduce the ability of human glioblastoma cells to form tumors in athymic mice (Fig. 3 B and C).
The ectodomain forms of p185neu with (T691stop) and without (N691stop) the transmembrane mutation are both effective at causing trans inhibition in EGFr-positive U87MG human glioblastoma cells. The inhibitory effect is erbB family specific, while PDGF signaling is intact. U87/T691-1 cells undergo DNA synthesis and mitosis in response to PDGF-BB nearly to the same degree as parental U87MG cells over a range of ligand concentrations (Tables 1 and 2). In U87MG cells, EGF induces DNA synthesis to a greater degree, and is more mitogenic, than PDGF (Fig. 2, Tables 1 and 2), which has been observed by others in glioblastoma cells (46). In U87MG cells transfected with T691stop (U87/T691-1), PDGF induced S-phase events and M-phase events much more efficiently than EGF (Fig. 2, Tables 1 and 2). These results were corroborated with U87/N691-B1 and other subclones of U87MG expressing either N691stop or T691stop. Similar results were seen when the PDGF-AB form of PDGF was used as a ligand.
Because many of the same Src homology 2- and Src homology 3-containing substrates are posttranslationally modified after both EGFr and PDGF-R phosphorylation, one might have expected that the mitogenic response to PDGF in cells expressing Neu ectodomains would be blunted, even if it is unlikely that the Neu ectodomain can physically associate with PDGF-R. This was not observed and is in agreement with previous work from our laboratory, which showed that intracellular substrates were not significantly altered in fibroblasts as a result of expression of a full-length, kinase-deficient mutant of Neu (p185K757M, possessing an intact carboxyl terminus but a mutated ATP-binding site) capable of inhibiting the transformed phenotype (47). The PDGF receptor has been implicated in glial tumorigenesis (41), and dominant–negative mutants of PDGF-R have been shown to inhibit the phenotype of rat C6 glioma cells (48). However, it appears as if an EGFr-dependent signal transduction pathway(s), independent of signaling through the PDGF receptor, is important for the initiation and/or maintenance of the transformed phenotype of human glioblastoma.
An analysis of other subclones expressing Neu ectodomains supports a relationship between the expression level of kinase-deficient Neu and phenotypic modulation in U87MG cells. The minimum ratio of mutant Neu proteins to EGFr required to functionally disable EGFr is unknown in this cell type, but has been observed to be as low as 1:1 in certain fibroblast-derived lines (33). However, the differential expression of mutant Neu proteins in U87/T691–1 and U87/N691-B1 cells (Fig. 1) is unlikely to account fully for the differences in phenotypic inhibition observed between these two clones. The T691stop-expressing U87MG subclones, including U87/T691–1, exhibit an altered morphology, with cells appearing flat and having a significantly reduced tendency to form foci in culture. Although both the N691stop and T691stop mutants physically associate with EGFr, the T691stop Neu mutant has been observed to homodimerize more efficiently than N691stop (19, 33), and for this reason has been observed to form heterodimers with EGFr less efficiently than the N691stop mutant (X.Q. and M.I.G., unpublished data). The more significant inhibition of EGFr observed with the T691stop mutant suggests that this form is more effective in disabling EGFr in U87MG cells, and may indicate that the activated receptor complex is composed of oligomers, which are more efficiently inactivated by T691stop than N691stop. There is structural evidence to suggest that the activated ligand-binding domain of EGFr is composed of dimers, trimers, and larger multimers (49).
These studies provide direct evidence for a role of the EGFr in the maintenance of human malignant glioma. The Neu ectodomain can assemble efficiently with the EGFr in glioblastoma cells, and thereby disrupt EGFr-mediated phenotypes in a trans-dominant fashion. Glial tumors are also notable for alterations in cell-cycle regulatory proteins, in particular, U87MG cells lack p16 (50). However, these cells are still inhibited by disabling the erbB family of proteins by expression of the Neu ectodomain, which is very promising for the treatment of advanced human disease. We have recently shown that the T691stop form of Neu is able to form heterodimers with the constitutively active, ectodomain deletion mutant of EGFr found in a subset of high-grade gliomas (1, 2, 7, 9–11). Additionally, T691stop inhibits tumor formation in vivo of U87MG-derived cells coexpressing endogenous EGFr and mutant EGFrs (D.M.O. and M.I.G., unpublished data).
Because of the high affinity of the Neu ectodomain for the EGFr, the importance of this receptor in glial transformation, the absence of toxicity as a consequence of transfection into nonproliferating cells, and the specific nature of the inhibition of EGFr-mediated signal transduction, the Neu ectodomain appears to be a viable biologic construct for genetic therapies of human glioblastoma. The T691stop mutant Neu form may be the preferred form for use in glial-derived human tumors. Neu ectodomain polypeptides with and without the transmembrane mutation may be important in the design of biologic pharmaceuticals, which disable erbB family oncoproteins. The use of the Neu ectodomain could theoretically be extended to other tumor types expressing oncogenic erbB family receptor tyrosine kinases, including other neural tumors and, potentially, tumors derived from systemic epithelial tissues.
Acknowledgments
This work was supported by the American Association of Neurological Surgeons Research Foundation, by Grant IRG-135P from the American Cancer Society, and by the McCabe Fund of the University of Pennsylvania to D.M.O., and by grants from the National Cancer Institute, the Council for Tobacco Research, and the Astral Corporation to M.I.G.
ABBREVIATIONS
- EGFr
epidermal growth factor receptor
- PDGF
platelet-derived growth factor
- PDGF-R
PDGF receptor
References
- 1.Sugawa N, Ekstrand A J, James C D, Collins V P. Proc Natl Acad Sci USA. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ekstrand A J, Sugawa N, James C D, Collins V P. Proc Natl Acad Sci USA. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Libermann T A, Nusbaum H R, Razon N, Kris R, Lax I, Soreq H, Whittle N, Waterfield M D, Ullrich A, Schlessinger J. Nature (London) 1985;313:144–147. doi: 10.1038/313144a0. [DOI] [PubMed] [Google Scholar]
- 4.Wong A J, Bigner S H, Bigner D D, Kinzler K W, Hamilton S R, Vogelstein B. Proc Natl Acad Sci USA. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.James C D, Carlbom E, Dumanski J P, Hansen M, Nordenskjold M, Collins V P, Cavenee W K. Cancer Res. 1988;48:5546–5551. [PubMed] [Google Scholar]
- 6.Cavenee W K. Cancer. 1992;70:1788–1793. doi: 10.1002/1097-0142(19920915)70:4+<1788::aid-cncr2820701621>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 7.Nishikawa R, Ji X D, Harmon R C, Lazar C S, Gill G N, Cavenee W K, Huang H J. Proc Natl Acad Sci USA. 1994;91:7727–7731. doi: 10.1073/pnas.91.16.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schlegel J, Merdes A, Stumm G, Albert F K, Forsting M, Hynes N, Kiessling M. Int J Cancer. 1994;56:72–77. doi: 10.1002/ijc.2910560114. [DOI] [PubMed] [Google Scholar]
- 9.Malden L T, Novak U, Kaye A H, Burgess A W. Cancer Res. 1988;48:2711–2714. [PubMed] [Google Scholar]
- 10.Humphrey P A, Wong A J, Vogelstein B, Zalutsky M R, Fuller G N, Archer G E, Friedman H S, Kwatra M M, Bigner S H, Bigner D D. Proc Natl Acad Sci USA. 1990;87:4207–4211. doi: 10.1073/pnas.87.11.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong A J, Ruppert J M, Bigner S H, Grzeschik C H, Humphrey P A, Bigner D S, Vogelstein B. Proc Natl Acad Sci USA. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callaghan T, Antczak M, Flickinger T, Raines M, Myers M, Kung H J. Oncogene. 1993;8:2939–2948. [PubMed] [Google Scholar]
- 13.Carter T H, Kung H J. Crit Rev Oncogen. 1994;5:389–428. doi: 10.1615/critrevoncog.v5.i4.40. [DOI] [PubMed] [Google Scholar]
- 14.Haley J D, Hsuan J J, Waterfield M D. Oncogene. 1989;4:273–283. [PubMed] [Google Scholar]
- 15.Yamazaki H, Ohba Y, Tamaoki N, Shibuya M. Jpn J Cancer Res. 1990;81:773–779. doi: 10.1111/j.1349-7006.1990.tb02644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiyokawa N, Yan D H, Brown M E, Hung M C. Proc Natl Acad Sci USA. 1995;92:1092–1096. doi: 10.1073/pnas.92.4.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ekstrand A J, Liu J, He J, Hamid M L, Longo N, Collins V P, James C D. Oncogene. 1995;10:1455–1460. [PubMed] [Google Scholar]
- 18.Goldman C K, Kim J, Wong W L, King V, Brock T, Gillespie G Y. Mol Biol Cell. 1993;4:121–133. doi: 10.1091/mbc.4.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiner D B, Liu J, Cohen J A, Williams W V, Greene M I. Nature (London) 1989;339:230–231. doi: 10.1038/339230a0. [DOI] [PubMed] [Google Scholar]
- 20.Ullrich A, Schlessinger J. Cell. 1990;61:203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- 21.Heldin C H. Cell. 1995;80:213–223. doi: 10.1016/0092-8674(95)90404-2. [DOI] [PubMed] [Google Scholar]
- 22.Kokai Y, Myers J N, Wada T, Brown V I, LeVea C M, Davis J G, Dobashi K, Greene M I. Cell. 1989;58:287–292. doi: 10.1016/0092-8674(89)90843-x. [DOI] [PubMed] [Google Scholar]
- 23.Wada T, Qian X, Greene M I. Cell. 1990;61:1339–1347. doi: 10.1016/0092-8674(90)90697-d. [DOI] [PubMed] [Google Scholar]
- 24.Dougall W C, Qian X, Greene M I. J Cell Biochem. 1993;53:61–73. doi: 10.1002/jcb.240530108. [DOI] [PubMed] [Google Scholar]
- 25.Goldman R, Ben-Levy R, Peles E, Yarden Y. Biochemistry. 1990;29:11024–11028. doi: 10.1021/bi00502a002. [DOI] [PubMed] [Google Scholar]
- 26.Qian X L, Decker S J, Greene M I. Proc Natl Acad Sci USA. 1992;89:1330–1334. doi: 10.1073/pnas.89.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spivak-Kroizman T, Rotin D, Pinchasi D, Ullrich A, Schlessinger J, Lax I. J Biol Chem. 1992;267:8056–8063. [PubMed] [Google Scholar]
- 28.Carraway K L, Cantley L C. Cell. 1994;78:5–8. doi: 10.1016/0092-8674(94)90564-9. [DOI] [PubMed] [Google Scholar]
- 29.Honnegger A M, Schmidt A, Ullrich A, Schlessinger J. Mol Cell Biol. 1990;10:4035–4044. doi: 10.1128/mcb.10.8.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashles O, Yarden Y, Fischer R, Ullrich A, Schlessinger J. Mol Cell Biol. 1991;11:1454–1463. doi: 10.1128/mcb.11.3.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou O K, Dull T J, Russell D S, Lebwohl G D, Ullrich L A, Rosen D. J Biol Chem. 1987;262:1842–1847. [PubMed] [Google Scholar]
- 32.Millauer B, Shawver L K, Plate K H, Risau W, Ullrich A. Nature (London) 1994;367:576–579. doi: 10.1038/367576a0. [DOI] [PubMed] [Google Scholar]
- 33.Qian X L, Dougall W C, Hellman M E, Greene M I. Oncogene. 1994;9:1507–1514. [PubMed] [Google Scholar]
- 34.Qian X L, Dougall W C, Hellman M, Greene M I. Proc Natl Acad Sci USA. 1994;91:1500–1504. doi: 10.1073/pnas.91.4.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bargmann C I, Hung M-C, Weinberg R A. Nature (London) 1986;319:226–230. doi: 10.1038/319226a0. [DOI] [PubMed] [Google Scholar]
- 36.Southern P J, Berg P. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 37.Reed J C, Cuddy M, Haldar S, Croce C, Nowell P, Makover D, Bradley K. Proc Natl Acad Sci USA. 1990;87:3660–3664. doi: 10.1073/pnas.87.10.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian X L, Dougall W C, Fei Z, Greene M I. Oncogene. 1995;10:211–219. [PubMed] [Google Scholar]
- 39.Hansen M B, Nielson S E, Berg K. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 40.Kupperman E, Wen W, Meinkoth J L. Mol Cell Biol. 1993;13:4477–4484. doi: 10.1128/mcb.13.8.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermanson M, Funa K, Hartman M, Claesson-Welsh L, Heldin C H, Westermark B, Nister M. Cancer Res. 1992;52:3213–3219. [PubMed] [Google Scholar]
- 42.Claesson-Welsh L, Hammacher A, Westermark B, Heldin C H, Nister M. J Biol Chem. 1989;264:1742–1747. [PubMed] [Google Scholar]
- 43.Nister M, Claesson-Welsh L, Eriksson A, Heldin C H, Westermark B. J Biol Chem. 1991;266:16755–16763. [PubMed] [Google Scholar]
- 44.Pelicci G, Lanfrancone L, Salcini A E, Romano A, Mele S, Grazia Borrello M, Segatto O, Di Fiore P P, Pelicci P G. Oncogene. 1995;11:899–907. [PubMed] [Google Scholar]
- 45.Karungaran D, Tzahar E, Beerli R R, Chen X, Graus-Porta D, Ratzkin B J, Seger R, Hynes N E, Yarden Y. EMBO J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen P H, Ness G O, Engerbraaten O, Bjerkvig R, Lillehaug J R, Laerum O D. Int J Cancer. 1994;56:255–261. doi: 10.1002/ijc.2910560219. [DOI] [PubMed] [Google Scholar]
- 47.Dougall W C, Qian X, Miller M J, Greene M I. DNA Cell Biol. 1996;15:31–40. doi: 10.1089/dna.1996.15.31. [DOI] [PubMed] [Google Scholar]
- 48.Strawn L M, Mann E, Elliger S S, Chu L M, Germain L L, Niederfellner G, Ullrich A, Shawver L K. J Biol Chem. 1994;269:21215–21222. [PubMed] [Google Scholar]
- 49.Lax I, Mitra A K, Ravara C, Hurwitz D R, Rubinstein M, Ullrich A, Stroud R M, Schlessinger J. J Biol Chem. 1991;266:13828–13833. [PubMed] [Google Scholar]
- 50.He J, Olson J J, James C D. Cancer Res. 1995;55:4833–4836. [PubMed] [Google Scholar]