Abstract
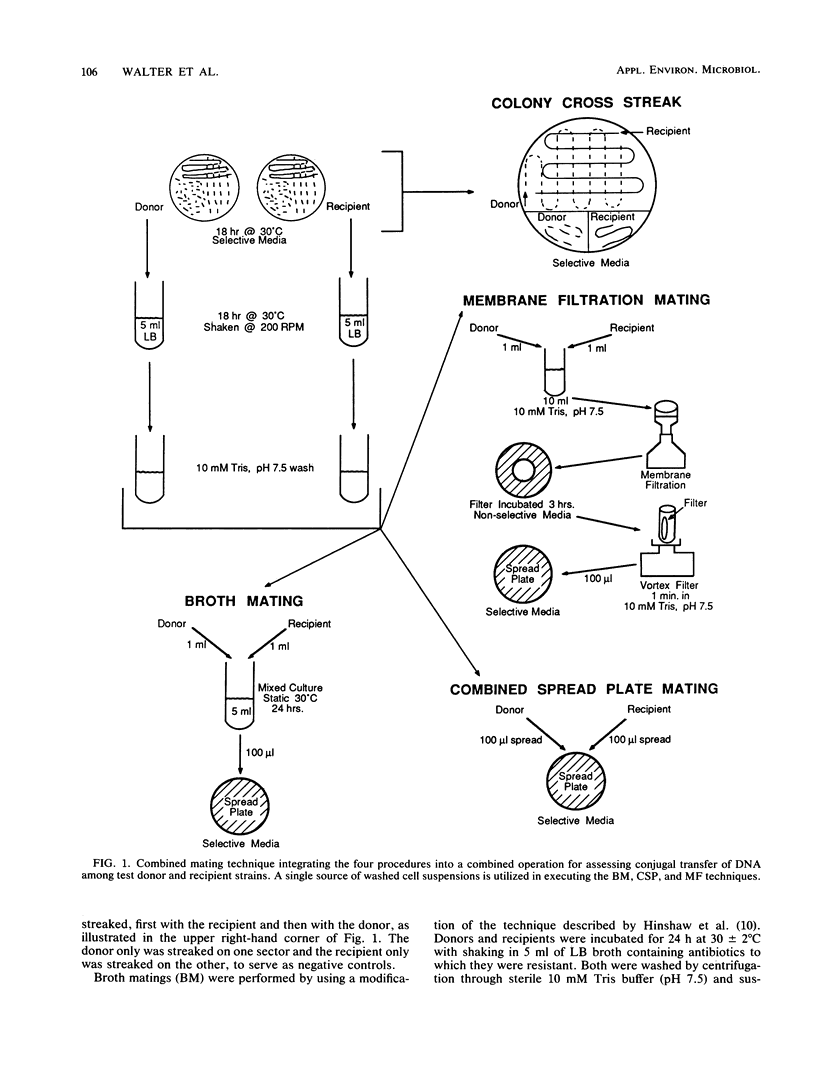
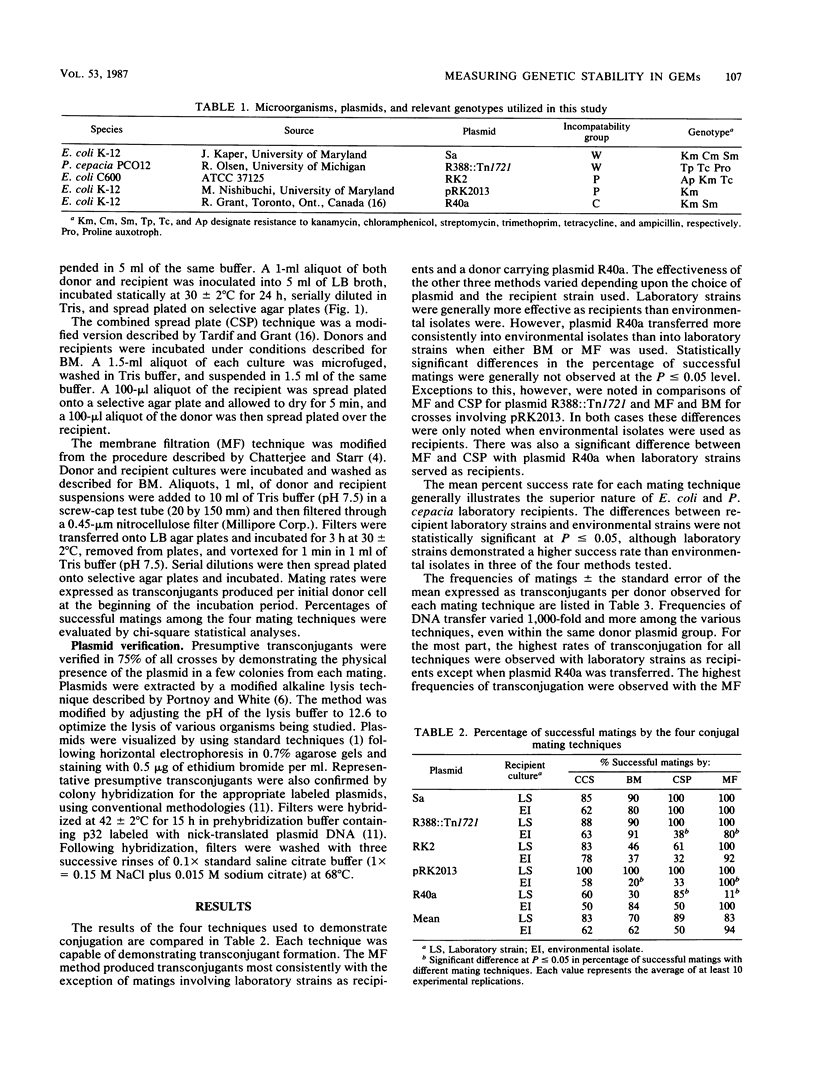
Four commonly used conjugation techniques, colony cross streak (CCS), broth mating (BM), combined spread plate (CSP), and membrane filtration (MF), were compared with each other regarding reliability, sensitivity, and complexity in evaluating the transfer of conjugative plasmids. Five plasmids representing several incompatibility groups plus a variety of laboratory and environmental isolates were used as mating pairs. The suitability of each method was evaluated for use in a routine assessment of the genetic stability of genetically engineered microorganisms. By the CSP and MF techniques with laboratory strains such as Escherichia coli and Pseudomonas species as recipients, transconjugants were usually produced in 100% of the mating trials. However, when environmental strains isolated from plants and soil were used as recipients, transconjugants were detected in 100% of some crosses and in as little as 30% in other crosses depending on the plasmid and recipient used. In general, differences in the percentage of successful matings between the CSP and MF techniques compared with the BM and CCS techniques were not statistically significant at the P less than or equal to 0.05 level. Occasionally, certain mating pairs consistently produced transconjugants by CCS or BM but not by CSP or MF. Since any single conjugation mating technique is not completely reliable in detecting transconjugants, we have developed a combined mating technique which integrates the CCS, CSP, BM, and MF methods as a single procedure to assess the mobility of plasmid DNA of genetically engineered microorganisms.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy P. S., Schulke J. W., Frazier L. M., Seidler R. J. Characterization of aquatic bacteria and cloning of genes specifying partial degradation of 2,4-dichlorophenoxyacetic acid. Appl Environ Microbiol. 1985 May;49(5):1237–1245. doi: 10.1128/aem.49.5.1237-1245.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E. S. The ecology of transferable drug resistance in the enterobacteria. Annu Rev Microbiol. 1968;22:131–180. doi: 10.1146/annurev.mi.22.100168.001023. [DOI] [PubMed] [Google Scholar]
- Chatterjee A. K., Starr M. P. Gene transmission among strains of Erwinia amylovora. J Bacteriol. 1973 Dec;116(3):1100–1106. doi: 10.1128/jb.116.3.1100-1106.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J Gen Microbiol. 1972 Sep;72(2):349–355. doi: 10.1099/00221287-72-2-349. [DOI] [PubMed] [Google Scholar]
- Davies J., Smith D. I. Plasmid-determined resistance to antimicrobial agents. Annu Rev Microbiol. 1978;32:469–518. doi: 10.1146/annurev.mi.32.100178.002345. [DOI] [PubMed] [Google Scholar]
- Helinski D. R. Plasmid determined resistance to antibiotics: molecular properties of R factors. Annu Rev Microbiol. 1973;27:437–470. doi: 10.1146/annurev.mi.27.100173.002253. [DOI] [PubMed] [Google Scholar]
- Hinshaw V., Punch J., Allison M. J., Dalton H. P. Frequency of R factor-mediated multiple drug resistance in Klebsiella and Aerobacter. Appl Microbiol. 1969 Feb;17(2):214–218. doi: 10.1128/am.17.2.214-218.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer R., Figurski D., Helinski D. R. Molecular vehicle properties of the broad host range plasmid RK2. Science. 1975 Dec 19;190(4220):1226–1228. doi: 10.1126/science.1060178. [DOI] [PubMed] [Google Scholar]
- Milewski E. A. Field testing of microorganisms modified by recombinant DNA techniques: applications, issues, and development of "points to consider" document. Recomb DNA Tech Bull. 1985 Sep;8(3):102–108. [PubMed] [Google Scholar]
- Rissler J. F. Research needs for biotic environmental effects of genetically-engineered microorganisms. Recomb DNA Tech Bull. 1984 Mar;7(1):20–30. [PubMed] [Google Scholar]
- Sinclair J. L., Alexander M. Role of resistance to starvation in bacterial survival in sewage and lake water. Appl Environ Microbiol. 1984 Aug;48(2):410–415. doi: 10.1128/aem.48.2.410-415.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif G., Grant R. B. Transfer of plasmids from Escherichia coli to Pseudomonas aeruginosa: characterization of a Pseudomonas aeruginosa mutant with enhanced recipient ability for enterobacterial plasmids. Antimicrob Agents Chemother. 1983 Aug;24(2):201–208. doi: 10.1128/aac.24.2.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATANABE T., FUKASAWA T. Episome-mediated transfer of drug resistance in Enterobacteriaceae. I. Transfer of resistance factors by conjugation. J Bacteriol. 1961 May;81:669–678. doi: 10.1128/jb.81.5.669-678.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]


