Abstract
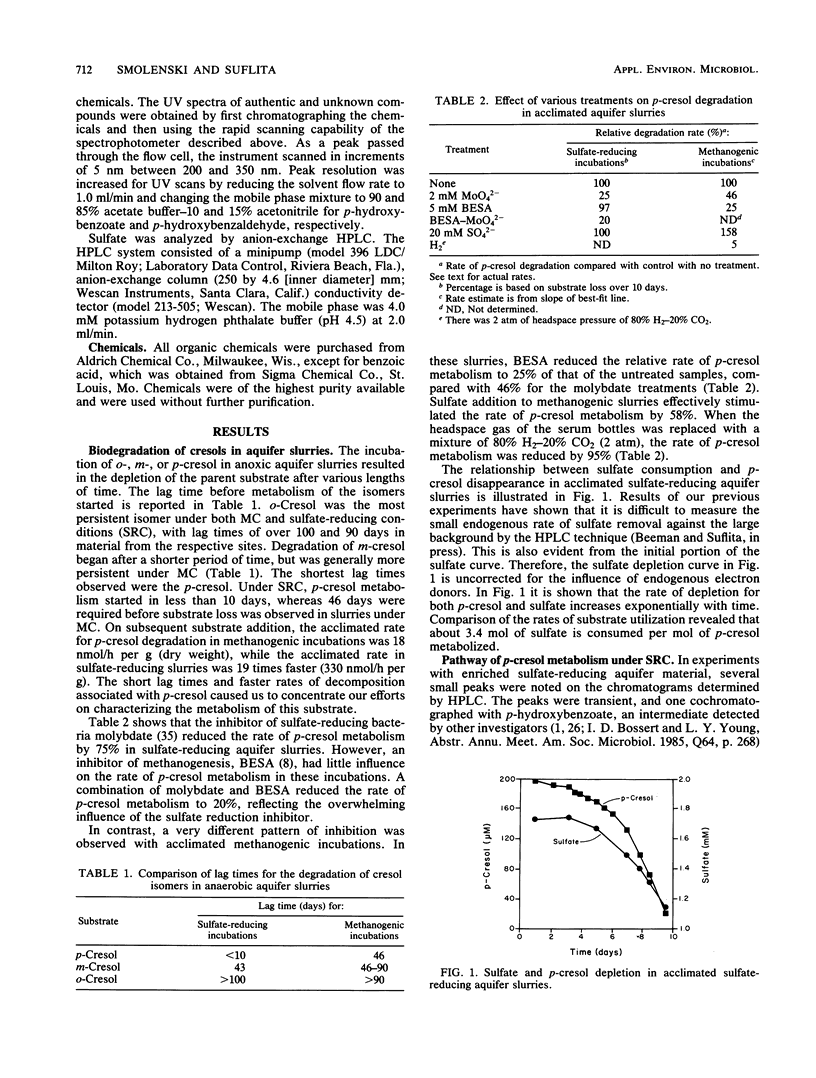
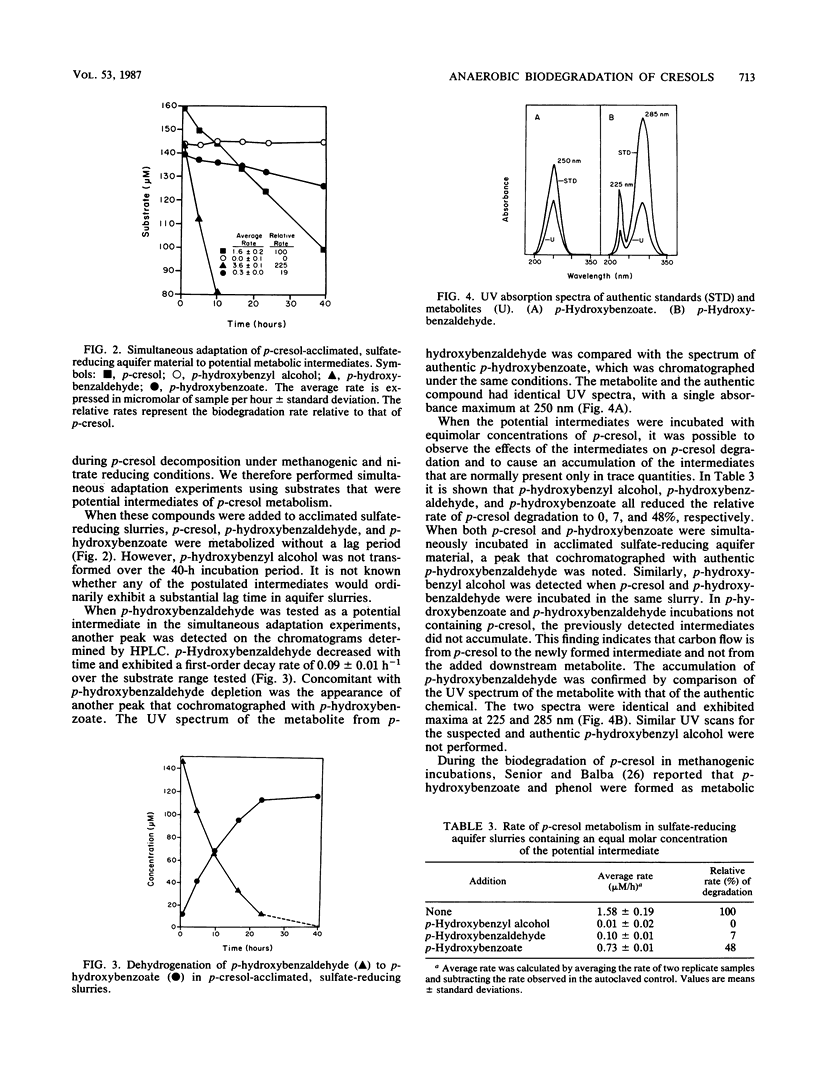
The biodegradation of o-, m-, and p-cresol was examined in material obtained from a shallow anaerobic alluvial sand aquifer. The cresol isomers were preferentially metabolized, with p-cresol being the most easily degraded. m-Cresol was more persistent than the para-isomer, and o-cresol persisted for over 90 days. Biodegradation of cresol isomers was favored under sulfate-reducing conditions (SRC) compared with that under methanogenic conditions (MC). Slurries that were acclimated to p-cresol metabolism transformed this substrate at 18 and 330 nmol/h per g (dry weight) for MC and SRC, respectively. Inhibition of electron flow to sulfate reduction with 2.0 mM molybdate reduced p-cresol metabolism in incubations containing sulfate. When methanogenesis was blocked with 5 mM bromoethanesulfonic acid in incubations lacking sulfate, p-cresol catabolism was retarded. Under SRC 3.4 mol of sulfate was consumed per mol of p-cresol metabolized. The addition of sulfate to methanogenic incubations stimulated p-cresol degradation. Simultaneous adaptation studies in combination with spectrophotometric and chromatographic analysis of metabolites indicated that p-cresol was oxidized under SRC to p-hydroxybenzoate via the corresponding alcohol and aldehyde. This series of reactions was inhibited under sulfate-limited or aerobic conditions. Therefore, the primary catabolic event for p-cresol decomposition under SRC appears to involve the hydroxylation of the aryl methyl group.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bossert I. D., Young L. Y. Anaerobic oxidation of p-cresol by a denitrifying bacterium. Appl Environ Microbiol. 1986 Nov;52(5):1117–1122. doi: 10.1128/aem.52.5.1117-1122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd S. A., Shelton D. R., Berry D., Tiedje J. M. Anaerobic biodegradation of phenolic compounds in digested sludge. Appl Environ Microbiol. 1983 Jul;46(1):50–54. doi: 10.1128/aem.46.1.50-54.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant M. P., Campbell L. L., Reddy C. A., Crabill M. R. Growth of desulfovibrio in lactate or ethanol media low in sulfate in association with H2-utilizing methanogenic bacteria. Appl Environ Microbiol. 1977 May;33(5):1162–1169. doi: 10.1128/aem.33.5.1162-1169.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAGLEY S., PATEL M. D. Oxidation of p-cresol and related compounds by a Pseudomonas. Biochem J. 1957 Jun;66(2):227–233. doi: 10.1042/bj0660227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson S. A., Suflita J. M. Extrapolation of biodegradation results to groundwater aquifers: reductive dehalogenation of aromatic compounds. Appl Environ Microbiol. 1986 Oct;52(4):681–688. doi: 10.1128/aem.52.4.681-688.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Romesser J. A., Wolfe R. S. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry. 1978 Jun 13;17(12):2374–2377. doi: 10.1021/bi00605a019. [DOI] [PubMed] [Google Scholar]
- Healy J. B., Young L. Y. Anaerobic biodegradation of eleven aromatic compounds to methane. Appl Environ Microbiol. 1979 Jul;38(1):84–89. doi: 10.1128/aem.38.1.84-89.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J. Incorporation of [18O]water in the formation of p-hydroxybenzyl alcohol by the p-cresol methylhydroxylase from Pseudomonas putida. Biochem J. 1978 Oct 1;175(1):345–347. doi: 10.1042/bj1750345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J., Taylor D. G. The purification and properties of p-cresol-(acceptor) oxidoreductase (hydroxylating), a flavocytochrome from Pseudomonas putida. Biochem J. 1977 Oct 1;167(1):155–162. doi: 10.1042/bj1670155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper D. J. The hydroxylation of P-cresol and its conversion to P-hydroxybenzaldehyde in Pseudomonas putida. Biochem Biophys Res Commun. 1976 Mar 22;69(2):462–468. doi: 10.1016/0006-291x(76)90544-1. [DOI] [PubMed] [Google Scholar]
- Keat M. J., Hopper D. J. P-cresol and 3,5-xylenol methylhydroxylases in Pseudomonas putida N.C.I.B. 9896. Biochem J. 1978 Nov 1;175(2):649–658. doi: 10.1042/bj1750649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keat M. J., Hopper D. J. The aromatic alcohol dehydrogenases in Pseudomonas putida N.C.I.B. 9869 grown on 3,5-xylenol and p-cresol. Biochem J. 1978 Nov 1;175(2):659–667. doi: 10.1042/bj1750659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R., Dwyer D. F., Klug M. J. Kinetic analysis of competition between sulfate reducers and methanogens for hydrogen in sediments. Appl Environ Microbiol. 1982 Jun;43(6):1373–1379. doi: 10.1128/aem.43.6.1373-1379.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney M. J., Bryant M. P. Anaerobic Degradation of Lactate by Syntrophic Associations of Methanosarcina barkeri and Desulfovibrio Species and Effect of H(2) on Acetate Degradation. Appl Environ Microbiol. 1981 Feb;41(2):346–354. doi: 10.1128/aem.41.2.346-354.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton D. R., Tiedje J. M. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic Acid. Appl Environ Microbiol. 1984 Oct;48(4):840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y. Simultaneous Adaptation: A New Technique for the Study of Metabolic Pathways. J Bacteriol. 1947 Sep;54(3):339–348. doi: 10.1128/jb.54.3.339-348.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suflita J. M., Robinson J. A., Tiedje J. M. Kinetics of microbial dehalogenation of haloaromatic substrates in methanogenic environments. Appl Environ Microbiol. 1983 May;45(5):1466–1473. doi: 10.1128/aem.45.5.1466-1473.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. F., Campbell W. L., Chinoy I. Anaerobic degradation of the benzene nucleus by a facultatively anaerobic microorganism. J Bacteriol. 1970 May;102(2):430–437. doi: 10.1128/jb.102.2.430-437.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]


