Abstract
Whether, and to what extent, lineage restriction contributes to the organization of the mammalian brain remains unclear. Here we address this issue by examining the distribution of clonally related cells in chimeric mice generated by injecting genetically tagged embryonic stem (ES) cells into blastocyst embryos. Our examination of postnatal chimeric brains revealed that the vast majority of labeled ES cell descendents were confined within a different subset of brain regions in each animal. Moreover, the deployment of labeled cells in different brain regions was distinctive. The pattern of ordered and binomial colonization suggested that early diversified founder cells may constrain the fates of their descendants through a restriction of dispersion. In addition, the symmetrical distribution of ES cell descendants suggests that bilaterally corresponding structures may arise from a common set of progenitor cells. Finally, clones of cells formed a continuous band within the deep strata of the neocortex. This later finding in conjunction with the radial distribution of clones in remaining layers observed in previous studies indicates that the cerebral neocortex may derive from two groups of founder cells, which is consistent with the hypothesis of dual phylogenetic origins of the mammalian cerebral cortex.
The mammalian central nervous system comprises an array of functionally and structurally distinct regions. The developmental events that generate the regional diversity of the mammalian brain remain to be clarified (1–4). Relevant mechanisms may include a combination of lineage restriction and environmental induction. A regionalized distribution of homologs of homeobox and other putative regulatory genes was found in the vertebrate nervous system (reviewed in ref. 5), and their ectopic expression can induce transformation of rhombomere identity (6) whereas null mutations lead to an absence of neural segments (7). These findings strongly suggest that a hierarchical combination of regulatory genes is involved in the specification of regional identities of the mammalian brain.
In contrast, it is far less clear whether developmental outcomes of cells are predisposed by their ancestors (1). Recent transplantation studies demonstrated that early embryonic cells retained the capacity to switch their phenotypic fates in accordance with the host sites (8, 9). One explanation for these findings is that the mechanism of lineage restriction in the nervous system, unlike the immune system, is not an irreversible modification of the progenitor genome (10). Alternatively, it might be argued from these observations that the lineage origin bears little or no influence on the developmental outcome of cells. Transplantation experiments test only the commitment state of a cell, so lineage studies are required to test the existence of lineage restriction in an unperturbed situation.
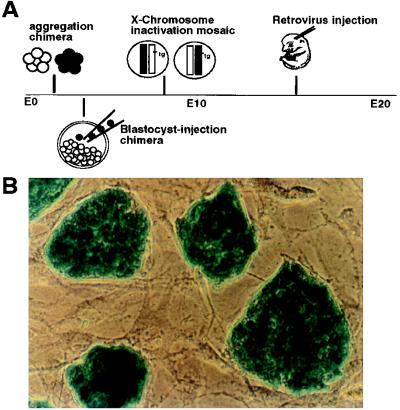
The intrauterine location of the mammalian embryo poses a challenge to lineage studies. Although short term lineage tracing can be conducted by injecting marker dye into mouse embryos cultured in vitro (11), labeling clones by genetic methods, such as chimeras, mosaics, or retroviral vectors, are nevertheless required for long term lineage tracing (12–18; summarized in Fig. 1A). Each of these methods labels a different number and size of clones depending on its unique timing and mechanism. The clonal allocation pattern is subsequently deduced from analysis of the number and position of labeled cells. In a previous aggregation chimera study, it was concluded that the cerebellar Purkinje cells are derived from a small number of progenitors, implicating a role of lineage restriction in the determination of cell fates (12). However, this conclusion was challenged with regard to whether the embryonic founder cells for the entire somatic tissues were interpreted as progenitors of an individual cell type in that study (refs. 19 and 20 but see 21).
Figure 1.
(A) Schematic illustration of the major approaches for retrospective clonal analysis in the mouse. These include: making aggregated embryos between genetically distinct strains at the 4- to 8-cell stage (12, 13); injecting genetically tagged ES cells into blastocyst embryos (14); taking advantage of random X chromosome inactivation of transgene in female mice (15); and infecting embryos with replication-defected retrovirus (16–18). The timing of labeling clones in each method also is indicated in perspective. In the present study, D3 ES cells were first transfected and selected for stable expression of the lacZ gene and then used for injection into mouse blastocyst embryos. As shown in B, colonies of undifferentiated ES cells (actin 69) display homogeneous expression of the lacZ gene on top of a fibroblast feeder cell layer in vitro.
In the present study, we injected genetically tagged embryonic stem (ES) cells into blastocyst embryos and subsequently examined the distribution of labeled and therefore clonally related cells in the chimeric mouse brain. This method labels fewer cells than the aggregation–chimera method (14) and retains the merit of marking a clone before the gastrulation stage of development. We reasoned that the distribution of ES cell descendants should be haphazard if clones were randomly dispersed, with specification solely dependent on their fortuitous positions. Conversely, if embryonic progenitor cells were diversified early and gave rise to descendants of a stereotyped fate, an ordered and distinct allocation pattern would be obtained. Thus, whether there is lineage restriction of cell fates can be tested directly by the method adopted in the present study.
MATERIALS AND METHODS
Generation of Blastocyst–Injection Chimeras.
ES–D3 cells (107) in 0.7 ml of medium were transfected with a 25-μg mixture (6:1 ratio) of a 4.9-kb linearized lacZ gene expression cassette and a 1.5-kb neomycin resistance cassette by a gene pulser (25 mF, 0.32 kV; Bio-Rad). The lacZ gene (3.9 kb) was driven by a 277-bp chicken β-actin promoter taken from the pBA-neo vector (22). Procedures for maintaining ES cells at an undifferentiated state and selection for neomycin (G418)-resistant clones have been reported (23). After a 10-day drug selection, 72 colonies were picked and expanded. Half of the cells in each colony was frozen–preserved, and the other half was passaged in the medium without G418 for 2 additional weeks to test the expression stability of the transgene. Four colonies were found to exhibit homogeneously high expression of the lacZ gene. The injection of ES–D3 cells into C57BL/6J blastocyst embryos to generate black–agouti chimeras was identical to the procedure of homologous recombination techniques (23). All 11 chimeras examined in this study were derived from one lacZ-expressing clone (actin 69). Another clone was used for injection but failed to produce chimeras.
Histology and Plotting of Cellular Locations.
Ten-day- or 5-month-old mice were anesthetized and transcardically perfused with 4% paraformaldehyde and 0.2% glutaraldehyde. Mouse brains were frozen, serially sectioned, and collected onto salinated slides before staining by 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) histochemistry (24) at 37°C overnight. After X-Gal histochemical staining, sections were either counterstained by 0.1% neutral red or immunostained for glial fibrillary acidic protein to assist the identification of the location and the cell types of positively stained cells.
PCR of Histologic Specimens.
Procedures modified from Greer et al. (25) were used to detect the existence of genome-incorporated lacZ genes from paraformaldehyde/glutaraldehyde-fixed tissue sections. After cover slides were removed from xylene, tissue specimens were gradually rehydrated. Tissue samples 30 μm thick and ≈400 μm in diameter were carefully picked up using #26 syringe needles under a dissection microscope. Each tissue sample was put into a microcentrifuge tube containing 50 μl of digestion solution (50 mM Tris, pH 8.5/1 mM EDTA/0.5% Tween 20/0.4 mg/ml proteinase K) and was incubated at 55°C for 14 h. Proteinase K was then inactivated by heating the tubes at 94°C for 8 min; 50 μl of autoclaved, distilled water was then added to each tube to increase the volume. An equal volume of phenol/chloroform/isoamyl alcohol (50%/48%/2%) was added for the first extraction, and then an equal volume of chloroform/isoamyl alcohol (24:1) was added for the second extraction. The final extract was then ethanol-precipitated in a PCR Eppendorf tube. After centrifugation and evaporation of excess ethanol, the reaction was set up in the final volume of 50-μl mixtures containing 10 mM Tris, pH 8.3/50 mM KCl/1.25 mM MgCl2 (using 10× PCR buffer from Perkin–Elmer), a 50-μM 2′-deoxynucleoside 5′-triphosphate mixture (Boehringer Mannheim), and 0.1 μM each of oligonucleotide primers of lacZ 5, lacZ 3, SMGA5, and SMGA3. The primer set of lacZ 5 (5′-CGAATCTCTATCGTGCGGTG-3′) and lacZ 3 (5′-TCGTCTGCTCATCCATGACC-3′) amplifies a 198-bp fragment of the bacterial β-galactosidase (lacZ) gene. The inclusion of the primer sets of SMGA 5 (5′-GCCTTGGCTCCCAGCACCAT-3′) and SMGA 3 (5′-CCAAGACCCAGCAACTCCTC-3′) was aimed to amplify a 109-bp fragment of the endogenous smooth muscle γ-enteric actin gene (26) as an internal control of successful harvest of genomic DNA. The PCR Eppendorf tubes were heated to 94°C for 4 min, cooled to 60°C for 1 min, and then kept at 72°C while 1.25 units of Taq polymerase (Perkin–Elmer) was added. The PCR reaction was cycled at 94°C for 1 min, 60°C for 1 min, and 72°C for 2 min for 30 times and then at 72°C for 5 min for the final extension. For every set of PCRs, several tubes containing all reaction mixtures except DNA were added to exclude the possibility of false-positives due to cross contamination. Data of the whole set of PCRs were discarded once false-positive amplification was observed, which occurred very rarely when the PCR cycle was set to 30. An aliquot of the PCR product was run on a 3% agarose gel (Boehringer Mannheim) containing ethidium bromide for visualization of the DNA bands. In our experience, phenol extraction produces much cleaner PCR products, and the sensitivity of PCR detection decreases considerably when the amplification target sequence is longer than 200 bp, as has been reported (25).
RESULTS
Restricted and Distinct Allocation of ES Cell Descendants.
Cells from one G418-resistant and lacZ-expressing clone (Fig. 1B) were injected into C57BL/6J blastocyst embryos to generate chimeric mice. Approximately one-seventh of the mouse pups in a litter were coat–color chimeras. The extent of coat–color chimerism ranged from 10 to 60%. Brains of 10-day- or 5-month-old chimeras were serially sectioned and stained by X-Gal histochemistry (24) to locate the clonally related lacZ-positive (+) cells. The number of lacZ+ cells found in the brain correlated with the degree of coat–color chimerism (27). In addition to the nervous system, lacZ+ cells also were found in a variety of organs in accordance with the pluripotentiality of the ES cells and the ubiquitous expression of the actin promoter.
In any given brain region, lacZ+ cells always were intermingled with unstained and therefore clonally unrelated cells, indicating the absence of selective aggregation between the same strained cells. Nevertheless, the vast majority of lacZ+ cells was confined in a multitude of differently shaped patches, and they were arranged differently in each brain region (Fig. 2A). In general, lacZ+ cells formed small clusters in regions comprised of nuclei and were distributed in a laminar fashion in laminated structures. The areas of laminated distribution of lacZ+ cells included the cerebral cortex (Fig. 2A), the superior colliculus (Fig. 2B), and the dentate gyrus (Fig. 2C). Of interest, the pattern of laminar distribution of clonally related cells in the dentate gyrus also was observed in two independent studies of interspecies, aggregation chimeras (13, 28).
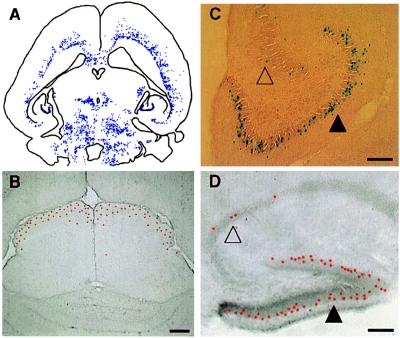
Figure 2.
(A) The general distribution pattern is illustrated here by camera lucida plotting of labeled cells in chimera 4. Although complicated, the distribution pattern is not haphazard. Three distinguished features of distribution are illustrated in B, C, and D. First (B), labeled cells tend to occupy certain layers in laminated brain regions. For example, labeled cells (red—pseudocolored to highlight the position of cells at low magnification) are restricted to the outermost superficial gray layer in the superior colliculus in chimera 5. Second (C), the deployment of labeled cells in deferent brain regions is distinct. As shown here, labeled cells were distributed evenly along the outer margin of the dentate gyrus (filled triangle) but were scattered discontinuously in the nearby Ammon’s horn (unfilled triangle; chimera 4). Third (D), the same region-specific deployment of labeled cells can be found in other animals, such as the hippocampal region of chimera 1 shown here. (Bar = 100 μm.)
The deployment of labeled cells in different brain regions was distinct. For example, lacZ+ cells were continuously allocated in the dentate gyrus in contrast to their scattered and clustered distribution in the nearby Ammon’s horn of the hippocampus (Fig. 2C). This regionally distinct allocation of labeled cells was not accidental; the same pattern also was found in other animals (Fig. 2D). In addition, when lacZ+ cells were found in one brain region, they tended to populate the whole structure in a highly consistent pattern. For example, the distribution patterns of lacZ+ cells shown in Fig. 2 B–D were consistently found throughout the entire superior colliculus and the hippocampal formation. The main exception to the continuous distribution was the cerebellum, where the allocation of lacZ+ cells extended across several folia but did not enter the entire cerebellar hemisphere (Fig. 3). However, although there was a virtual absence of lacZ+ cells in the cerebellum of one chimeric mouse (Fig. 3A), there was a large population of labeled cells in another animal (Fig. 3B). Such large differences in the number of labeled cells and their ordered alignment strongly suggested a binomial nature of the colonization of ES cell descendants in individual brain regions.
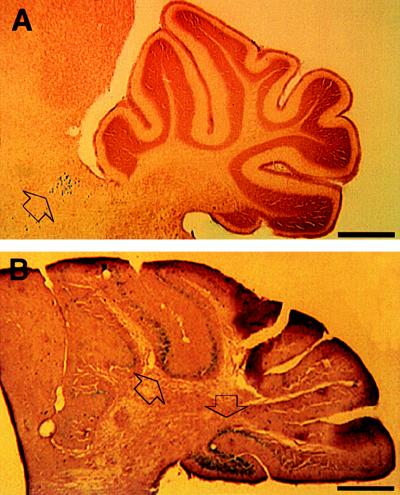
Figure 3.
(A) Labeled cells were found in the brainstem (arrowhead) in chimera 1 but not in the cerebellum. (B) On the contrary, a densely packed population of ES cell descendants (arrowhead) was allocated in the cerebellum across several folia in chimera 11. Such a great magnitude of change of presence of labeled cells and their ordered alignment indicates a binomial colonization of ES cell descendants in a brain region. (Bar = 500 μm.)
ES Cells Preferentially Colonized Certain Brain Regions.
It is known that the distribution of different strains of cells is not completely random in chimeric animals. This unequal representation in different tissues is described as site-specific selection (19) or chimeric imbalance (29). To evaluate its impact on clonal allocation, we compared the distribution sites of labeled cells in 11 chimera mice derived from injection of the same ES cell clone (actin 69) to standardize the genetic background.
The comparison revealed that lacZ+ cells were found consistently in brain regions such as the superior colliculus, never were observed in some structures like the basal ganglion, and occasionally were located in regions such as the cerebellum and the habenular nucleus (Table 1). Taken together, a different density of lacZ+ cells was found in a different subset of brain regions in each animal. Thus, despite a certain degree of preferential incorporation, descendants of injected ES cells did not colonize the same set of brain regions in each animal. Three male chimeras of ≈50% coat–color chimerism were crossed with C57BL/6 females, but germline transmission was not achieved. Therefore, the possibility that the reporter gene might be inactivated in some brain regions could not be completely excluded.
Table 1.
Distribution profile of lacZ+ cells in 11 chimeric mice
| Brain region | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Olfactory nucleus | +/+ | +/+ | −/− | ++/++ | −/− | ++/++ | ND | −/− | +/+ | +/+ | −/− |
| Cerebral cortex* | +/+ | −/− | +/+ | ++/++ | ++/++ | +/+ | ++/++ | −/− | −/− | −/− | −/− |
| Corpus striatum | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− | −/− |
| Cornus ammonis | +/+ | +/+ | −/− | +/+ | +/+ | +/+ | ++/++ | −/− | +/− | +/− | −/− |
| Dentate gyrus | ++/++ | −/− | −/− | +++/+++ | ++/++ | +/+ | ++/++ | +/− | +/− | −/− | +/+ |
| Amygdaloid nucleus | +/+ | +/+ | +/+ | ++/++ | −/− | +/+ | ND | −/− | ++/++ | ++/++ | ++/++ |
| Septal area | +/+ | −/− | −/− | +/+ | +/+ | +/+ | +/+ | −/− | ++/++ | +/+ | +/+ |
| Habenular nucleus | −/− | −/− | −/− | −/− | ++/++ | −/− | +/+ | +/− | ++/++ | +/+ | +/+ |
| Dorsal thalamus* | +/+ | +/+ | +/+ | ++/++ | ++/++ | +/+ | ++/++ | +/− | −/− | +/+ | ++/++ |
| Hypothalamus* | +/+ | +/+ | +/+ | ++/++ | ++/++ | +/+ | ND | −/− | ++/++ | ++/++ | ++/++ |
| Superior colliculus* | +/+ | ND | +/+ | ++/++ | +++/+++ | +/+ | +++/+++ | +/− | ++/++ | ++/++ | ++/++ |
| Inferior colliculus* | +/+ | ND | −/− | +/+ | −/− | ++/++ | +/+ | −/− | −/− | −/− | ND |
| Cerebellum* | −/− | −/− | −/− | −/− | ++/++ | −/− | ++/++ | −/− | +++/+++ | ++/− | +++/+++ |
Results presented as right side/left side.
+, Densities of lacZ+ cells were designated after a comparison between different animals; ND, not determined.
lacZ+ cells were distributed in a restricted portion of the region.
ES Cell Descendants Colonize Bilaterally Corresponding Structures.
One salient feature of the distribution of lacZ+ cells is that bilaterally corresponding structures are typically both colonized in a concerted manner (Table 1). This bilaterally symmetric allocation was not only observed in animals of a high degree of chimerism or in preferentially incorporated regions (Fig. 4 A and B). However, it should be pointed out that the number of lacZ+ cells between bilateral structures was not always equivalent, and in rare occasions, a unilateral allocation of lacZ+ cells was nevertheless observed (Fig. 4C). Similar to our observations, both bilaterally symmetric genotype compositions (27, 29, 30) and occasionally unilateral allocations (21) were reported in previous aggregation chimera studies. We propose a lineage-based mechanism to explain such bilateral symmetry (Fig. 4D).
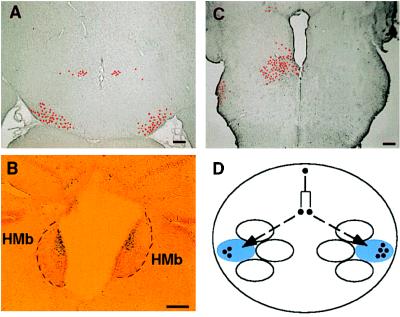
Figure 4.
Bilaterally corresponding structures are typically both populated by ES cell descendants. (A) Although only a small number of labeled cells (pseudocolored to highlight the position of cells at low magnification) was found in this section, the cells were allocated symmetrically in the diencephalon (chimera 4). (B) The medial habenular nucleus (HMb) was not a preferentially incorporated structure, but, when labeled cells were located on one side, they were symmetrically localized on the opposite side of the habenular nucleus (chimera 9). (C) Nevertheless, a unilateral allocation of labeled cells was observed occasionally (chimera 8). (D) One possible mechanism to generate such bilaterally symmetric allocations is that cells of the same ancestry tend to incorporate into bilaterally corresponding structures. The sizes of bilaterally sibling clones, as indicated in the figure, are not necessarily expanded to the same extent. (Bar = 100 μm.)
Clonal Allocation Restricted to the Deep Cortical Strata.
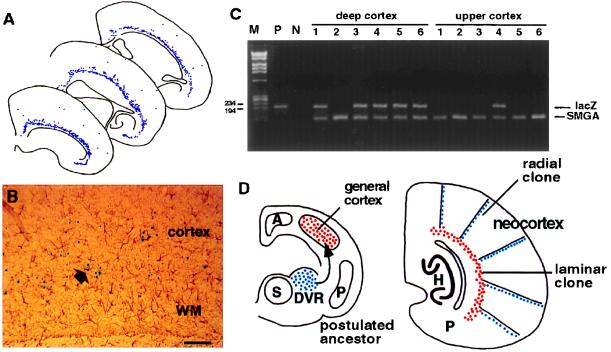
We found a continuous clonal allocation restricted to the deep layers of the entire cerebral cortex in 6 out of 11 chimeras (Fig. 5A). The granular and supragranular layers were not greatly labeled in any animal. Most of the lacZ+ cells were large cells, presumably pyramidal neurons situated within layers 5 and 6. Only occasional cells were immunoreactive with the glial fibrillary acidic protein antibody (Fig. 5B). Therefore, the labeled cells were not glial cells embedded in the white matter beneath the cerebral cortex. Although a small number of lacZ+ cells were scattered in the upper cortical layers, they were not deployed in any discernible alignment. The laminar distribution pattern was unexpected because previous studies had demonstrated that the majority of clonally related neurons was stacked up in a columnar fashion in the cortical plate (2, 3).
Figure 5.
(A) Camera lucida plotting of ES cell descendants from three sections of chimera 5 indicates a continuous laminar allocation restricted to the deep layers (V and VI) of the entire cerebral cortex. Labeled cells in the upper cortical plate were much scarcer and more sporadically distributed. (B) Double labeling with antibody against glial fibrillary acidic protein indicated that astrocytes (arrowhead) were only a small percentage of this group of lacZ+ cells located above the white matter (WM). (C) Tissue blocks of the deep and the upper cerebral cortices were picked individually from histological specimens and processed for PCR detection of the lacZ gene (200-bp signal). The endogenous γ-enteric actin gene (SMGA, 100-bp signal) also was amplified to ascertain the successful harvest of DNA from histologic specimens. Results of one representative set of samples are shown here. In a larger series of samples (n = 43), 72% of the deep cortical samples and 11.6% of the upper cortical samples were lacZ-positive. The correlation between X-Gal histochemical expression and the presence of the lacZ gene was statistically significant (P < 0.005 in χ2 analysis; df = 1). (D) Comparative studies indicated that the three-layered general cortex expanded and propelled the archicortex (A: hippocampus, H) and the paleocortex (P: piriform cortex, P) toward the medial wall of telencephalon during evolution. It also has been postulated that neuroblasts of the dorsal ventricular ridge (DVR) located in the dorsal striatum (S) migrated and incorporated into the general cortex in ancestral mammals to result in the modern six-layered isocortex (4). One possible explanation of the coexistence of radial and laminar clones in the mouse cerebral cortex is that these two phylogenetically distinct populations of cells remain ontogenetically segregated and are allocated differently. (D is modeled after ref. 4). (Bar = 100 μm.)
We first examined whether the reporter gene was present throughout the cortical plate but was only expressed in the deep laminae. Germline transmission was not obtained from these chimeras, so we compared the PCR detection of the lacZ gene with the X-Gal staining pattern between the upper and lower cortical layers. Small tissue blocks ≈400 μm in diameter and 30 μm thick were picked up from the histologic specimen. Genomic DNA samples were then prepared and subjected to PCR analysis of the existence of the lacZ gene (25). In 43 samples from the deep cortex, 31 gave positive signals of the lacZ gene (72%) whereas only 5 out of 43 samples from the upper cortex displayed the lacZ gene signal (11.6%) (Fig. 5C). There is a strong correlation between the existence of the lacZ gene and its expression, as detected by the X-Gal histochemistry (P < 0.005 in χ2 analysis; df = 1). Therefore, the presence of a large number of undetected ES cell descendants in the upper cortical plate is not favored by these data.
Deep cortical layers contain early generated cells (31), so we next considered whether ES cell descendants contributed to early generated neurons but were replaced by host cells in producing late generated cells of the upper cortical layers. Such a temporal change of genotype compositions, described as chimeric drift (32), was observed in aggregation chimeras before. However, such a scenario is unlikely because late generated cells in the superficial gray layer of the superior colliculus, the dentate gyrus, and in the internal granular layer of the cerebellum all were heavily populated by lacZ+ cells in our chimeras (Figs. 2 B and C, 3B). Therefore, we concluded that the observed distribution of lacZ+ cells in the deep cortical layers represented a genuine clonal allocation pattern (Fig. 5D, see below).
DISCUSSION
Methodological Considerations.
Lineage studies of mammals have relied on retrospective clonal analyses such as aggregation chimeras, X-inactivation mosaics, or retroviral lineage tracing (Fig. 1A). Each of these methods has its own merit and provides a unique angle to the issue of clonal allocation. The examination of clonally related cells using blastocyst–injection chimeras, adopted in the present study, offers several advantages. First, larger clones are obtained by this method than by retroviral gene transfer, and these larger clones allow better assessment of the predominant mode of distribution of the clonal cohorts (16–18). Second, the chimera method is not limited by the accessibility of mouse embryos for injection of retroviral vectors, and it permits clonal analysis of both early and late generated brain structures. Third, the blastocyst–injection chimera labels fewer cells than the aggregation chimera and thus assists the unveiling of subtle features of clonal allocation.
The injection–chimera method also has certain limitations. For example, unless it is assessed in germline-transmitted offspring, the full expression spectrum of the reporter gene cannot be ascertained without endogenous markers. In addition, the distribution of injected ES cell descendants may be biased toward a subset of brain regions, as is revealed in the comparison of their distribution sites (Table 1). The preferential incorporation (chimeric imbalance) may be due to the difference in strain backgrounds (13, 28, 29) or imprinted genes (33). In any event, it is possible that only a subset of the clonal allocation patterns is revealed in a particular strain combination of chimeras. Nevertheless, the blastocyst–injection chimera provide unique information to complement our understanding of the mammalian embryogenesis obtained by other methods.
Clonal Restriction.
The main goal of the present study was to examine whether there is a lineage restriction of cell fates in mammalian brains. The analysis showed that the deployment of ES cell descendants in different brain regions was both distinctive and specific (Fig. 2). Moreover, ES cell descendants were distributed in individual brain regions in a binomial fashion (Fig. 3; Table 1). Therefore, the distribution pattern indicates that different brain regions are populated by descendants of a small number of progenitor cells. The very existence of specific progenitor cells for different brain regions implies that cell fates are predisposed by their lineage origins.
The mechanism of lineage constraint of cell fates cannot be addressed by clonal analysis alone. In view of recent transplantation studies revealing a certain degree of indeterminacy of early embryonic cells (8, 9), the mechanism of lineage constraint is likely to be a restriction of cell mingling (34, 35) rather than an irreversible modification of the progenitor genome (10). Therefore, positionally constrained clonal cohorts exhibit similar fates because they are exposed to the same set of inductive stimuli but retain the capacity to switch to a new fate in a heterologous environment (8, 9).
Bilateral Symmetry.
We found that ES cell descendants tended to colonize bilaterally corresponding regions in a concerted fashion (Table 1). Comparable observations also were reported in aggregation chimeras in which bilaterally similar genotype compositions were noticed in the spinal cord (27), the retinas (29), and the cerebellum (30). It is important to note that an occasionally unilateral allocation of labeled cells was observed in both the present and previous aggregation chimera studies (21), which is characteristically different from the uniformly symmetric expression of transgenes. The most parsimonious explanation of the observed pattern is that labeled cells in bilaterally corresponding structures are derived from a common pool of ancestors that splits into two sibling clones at an early stage of development (Fig. 4D). In other words, the bilateral similarity of cell fates (analogy) is predicted by the similarity of ancestry (homology) in the development of the mammalian brain.
Bilaterally symmetric allocations of sibling clones also have been observed in the nematode (36), the amphibian (37), and the fish brain (38). Therefore, bilaterally concerted clonal allocation may be an evolutionary conserved strategy to generate the structural symmetry of the nervous system. The inequality of the number of labeled cells between bilateral structures indicates that the two sibling clones might not expand to the same extent in the vertebrate nervous system. Finally, bilateral allocation may be more liable in the vertebrate than in the invertebrate. Thus, similar to our observation in mammals, unilateral allocation of clonal descendants also was observed occasionally in the zebrafish (38).
Composite Clonal Allocation in the Neocortex.
The predominant mode of clonal allocation in the murine cerebral cortex appears to be radial, as indicated in X chromosome inactivation mosaic animals (15; reviewed in ref. 3). Thus, the continuous clonal allocation restricted to the deep layers of the entire cerebral cortex is both striking and unexpected (Fig. 5A). These clones are mostly neurons rather than glial cells of the white matter (Fig. 5B). Furthermore, the possibility of a deep layer-restricted expression of the reporter gene was not supported by our PCR analysis (Fig. 5C). Neither did we notice a tendency of ES cell descendants to contribute only to early generated cells. Thus, the deep laminar distribution of labeled cells appears to represent a genuine clonal allocation compartment. This conclusion is consistent with previous studies indicating separate sets of progenitor cells for the superficial and deep cortical layers (28, 39, 40). Of interest, it has been noticed that the deep cortical layer VI was exempt from the radial columnar patterns in the X chromosome inactivation mosaics (15). Thus, clonal allocations in the cerebral cortex appear to be mixed with a core of clones restricted to deep strata of the cortex overlaid by an array of radial clones situated in the more superficial layers (Fig. 5D). A certain degree of infiltration of the radial clones into the layer VI (the multiform layer) is nevertheless compatible with the scenario. It is unclear why the clones in our chimera mice were restricted to deep cortical strata whereas another blastocyst–injection chimera study revealed predominantly radial clones in the upper strata (14). One possibility is due to different preferential incorporation sites of injected stem cells in the two chimera studies of different strain–combinations.
The observation of superficially located radial and deeper located laminar clones within the cerebral cortex suggests a possibility that two distinct pools of progenitor cells contribute to cortical formation. The root of dual ontogenetic origins of cortical cells may lie in the phylogenetic history of the mammalian cerebral cortex. The comparative anatomical studies suggest that the three-layered lateral pallium, or general cortex, of amphibians, birds, and reptiles may be homologous to the six-layered mammalian neocortex (41). During the evolution, the expansion of the lateral pallium caused displacement of the archicortex (hippocampal formation) and the paleocortex (piriform cortex) toward the medial wall of the telencephalon (Fig. 5D). Simultaneously, the lateral pallium was transformed from a simple three-layered structure into a more complex six-layered mammalian cortex. The cellular events underlying this cytoarchitectonic elaboration are unknown. One attractive hypothesis is that, during evolution, the neuroblasts of dorsal ventricular ridge in ancient reptiles and birds have migrated into the lateral pallium and given rise to the six-layered neocortex (4). According to this hypothesis, the mammalian cerebral cortex is formed by the contribution from two distinct populations of cells. Our results further suggest that these two phylogenetically discrete populations may remain segregated during the ontogeny and form separate laminar and radial clones of the mammalian cerebral cortex.
Acknowledgments
We thank Drs. M. Jacobson, H. Karten, D. Goldowitz, and K. Herrup for their critical reading of, and suggestions on, an early version of this manuscript; Dr. R. Levenson for advice on construction of the transgene marker; J. Elsemore, C. Hughes, and D. Butkis for their excellent technical assistance; and Dr. H. Komuro for his useful discussions throughout the project. This study was supported by Public Health Service Grant NS14841 (P.R.).
ABBREVIATIONS
- ES
embryonic stem
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
Footnotes
Present address: Alexion Pharmaceuticals, New Haven, CT 06511.
References
- 1.Jacobson M. Developmental Neurobiology. 3rd Ed. New York: Plenum Press; 1991. [Google Scholar]
- 2.Rakic P. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 3.Rakic P. Proc Natl Acad Sci USA. 1995;92:11323–11327. doi: 10.1073/pnas.92.25.11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nauta W, Karten H. In: The Neurosciences: Second Study Program. Schmitt F, editor. New York: Rockefeller Univ. Press; 1970. pp. 7–26. [Google Scholar]
- 5.Puelles L, Rubenstein J. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 6.Marsahll H, Nonchev S, Sham M, Muchaamore I, Lumsden A, Krumlauf R. Nature (London) 1992;360:737–741. doi: 10.1038/360737a0. [DOI] [PubMed] [Google Scholar]
- 7.Thomas K, Capecchi M. Nature (London) 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- 8.Renfranz P, Cunningham M, McKay R. Cell. 1991;66:713–729. doi: 10.1016/0092-8674(91)90116-g. [DOI] [PubMed] [Google Scholar]
- 9.Fishell G. Development. 1995;121:803–812. doi: 10.1242/dev.121.3.803. [DOI] [PubMed] [Google Scholar]
- 10.Hozumi N, Tonegawa S. Proc Natl Acad Sci USA. 1976;73:3628–3632. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawson K, Meneses J, Pedersen R. Development. 1991;113:891–911. doi: 10.1242/dev.113.3.891. [DOI] [PubMed] [Google Scholar]
- 12.Wetts R, Herrup K. J Neurosci. 1982;2:1494–1498. doi: 10.1523/JNEUROSCI.02-10-01494.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldowitz D. Neuron. 1989;3:705–713. doi: 10.1016/0896-6273(89)90239-0. [DOI] [PubMed] [Google Scholar]
- 14.Nakatsuji N, Kadokawa Y, Suemori H. Dev Growth Differ. 1991;33:571–578. doi: 10.1111/j.1440-169X.1991.00571.x. [DOI] [PubMed] [Google Scholar]
- 15.Tan S-S, Breen S. Nature (London) 1993;362:638–640. doi: 10.1038/362638a0. [DOI] [PubMed] [Google Scholar]
- 16.Luskin M, Pearlman A, Sanes J. Neuron. 1988;1:635–647. doi: 10.1016/0896-6273(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 17.Walsh C, Cepko C. Science. 1992;255:434–440. doi: 10.1126/science.1734520. [DOI] [PubMed] [Google Scholar]
- 18.Kornack D, Rakic P. Neuron. 1995;15:311–321. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 19.Soriano P, Jaenisch R. Cell. 1986;46:19–29. doi: 10.1016/0092-8674(86)90856-1. [DOI] [PubMed] [Google Scholar]
- 20.Jennings C. Trends Neurosci. 1988;11:46–49. doi: 10.1016/0166-2236(88)90161-0. [DOI] [PubMed] [Google Scholar]
- 21.Vogel M, Herrup K. Dev Biol. 1993;156:49–68. doi: 10.1006/dbio.1993.1058. [DOI] [PubMed] [Google Scholar]
- 22.Guild B, Finer M, Housman D, Mulligan R. J Virology. 1988;62:3795–3801. doi: 10.1128/jvi.62.10.3795-3801.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elliott E, Drake J, Amigorena S, Elsemore J, Webster P, Mellman I, Flavell R. J Exp Med. 1994;681:681–694. doi: 10.1084/jem.179.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanes J, Rubenstein J, Nicolas J-F. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greer C, Wheeler C, Manos M. PCR Methods Appl. 1994;3:S113–S122. doi: 10.1101/gr.3.6.s113. [DOI] [PubMed] [Google Scholar]
- 26.Szucsik J, Lessard J. Genomics. 1995;28:154–162. doi: 10.1006/geno.1995.1126. [DOI] [PubMed] [Google Scholar]
- 27.Musci T, Mullen R. Dev Biol. 1992;152:133–144. doi: 10.1016/0012-1606(92)90163-b. [DOI] [PubMed] [Google Scholar]
- 28.Fishell G, Rossant J, van der Kooy D. Dev Biol. 1990;141:70–83. doi: 10.1016/0012-1606(90)90102-o. [DOI] [PubMed] [Google Scholar]
- 29.Williams R, Goldowitz D. Proc Natl Acad Sci USA. 1992;89:1184–1188. doi: 10.1073/pnas.89.4.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herrup K, Wetts R, Diglio T. J Neurogenetics. 1984;1:275–288. doi: 10.3109/01677068409107092. [DOI] [PubMed] [Google Scholar]
- 31.Angevine J, Sidman R. Nature (London) 1961;192:766–768. doi: 10.1038/192766b0. [DOI] [PubMed] [Google Scholar]
- 32.Gearhart J, Oster-Granite M. J Hered. 1981;72:3–5. doi: 10.1093/oxfordjournals.jhered.a109421. [DOI] [PubMed] [Google Scholar]
- 33.Keverne E, Fundele R, Narasimha M, Barton S, Surani M. Dev Brain Res. 1996;92:91–100. doi: 10.1016/0165-3806(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 34.Fishell G, Mason C, Hatten M. Nature (London) 1994;362:636–638. doi: 10.1038/362636a0. [DOI] [PubMed] [Google Scholar]
- 35.Sheard P, Jacobson M. Science. 1984;236:851–854. doi: 10.1126/science.3576203. [DOI] [PubMed] [Google Scholar]
- 36.Sulston J. In: The Nematode Caenorhabditis elegans. Wood W, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 123–156. [Google Scholar]
- 37.Jacobson M. J Neurosci. 1984;3:1019–1038. doi: 10.1523/JNEUROSCI.03-05-01019.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimmel C, Warga R, Kane D. Development. 1994;120:265–276. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- 39.Crandall J, Herrup K. Exp Neurol. 1990;109:131–139. doi: 10.1016/s0014-4886(05)80014-7. [DOI] [PubMed] [Google Scholar]
- 40.Krushel L, Johnston J, Fishell G, Tibshirani R, van der Kooy D. Neuroscience. 1993;53:1035–1047. doi: 10.1016/0306-4522(93)90487-z. [DOI] [PubMed] [Google Scholar]
- 41.Northcutt G. Ann Rev Neurosci. 1981;4:301–350. doi: 10.1146/annurev.ne.04.030181.001505. [DOI] [PubMed] [Google Scholar]