Abstract
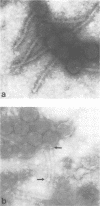
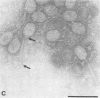
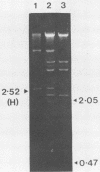
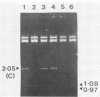
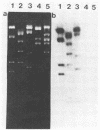
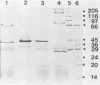
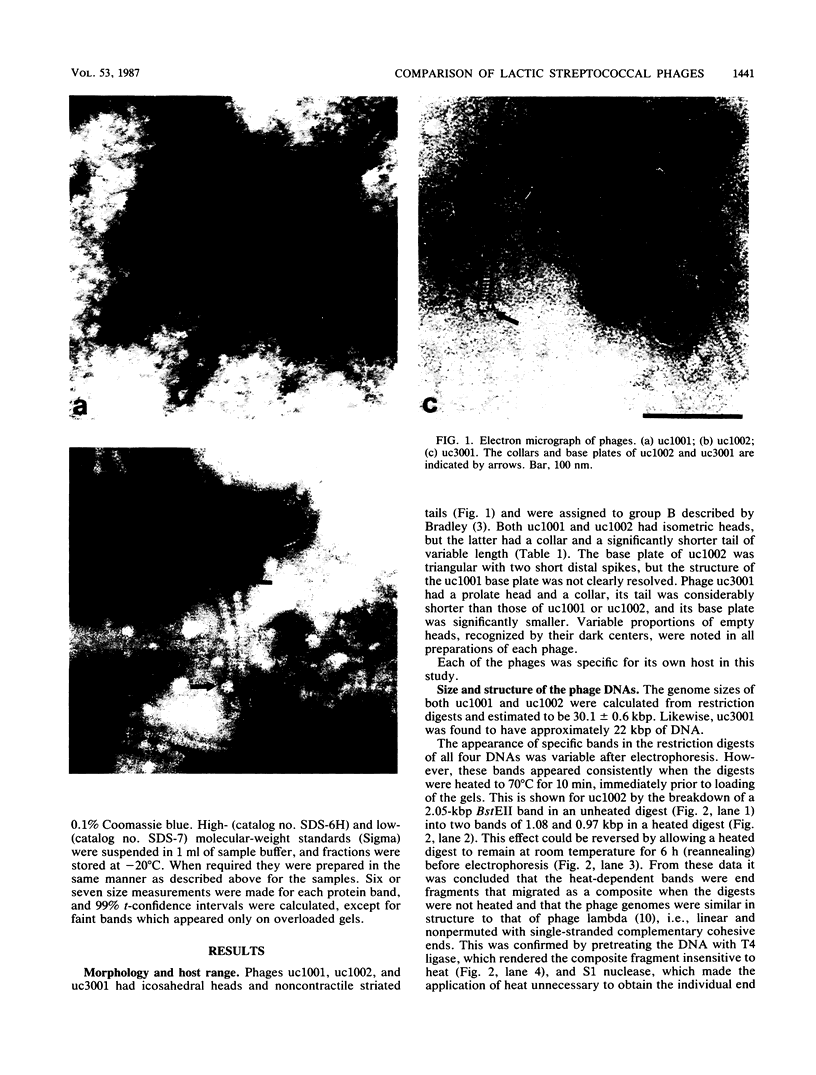
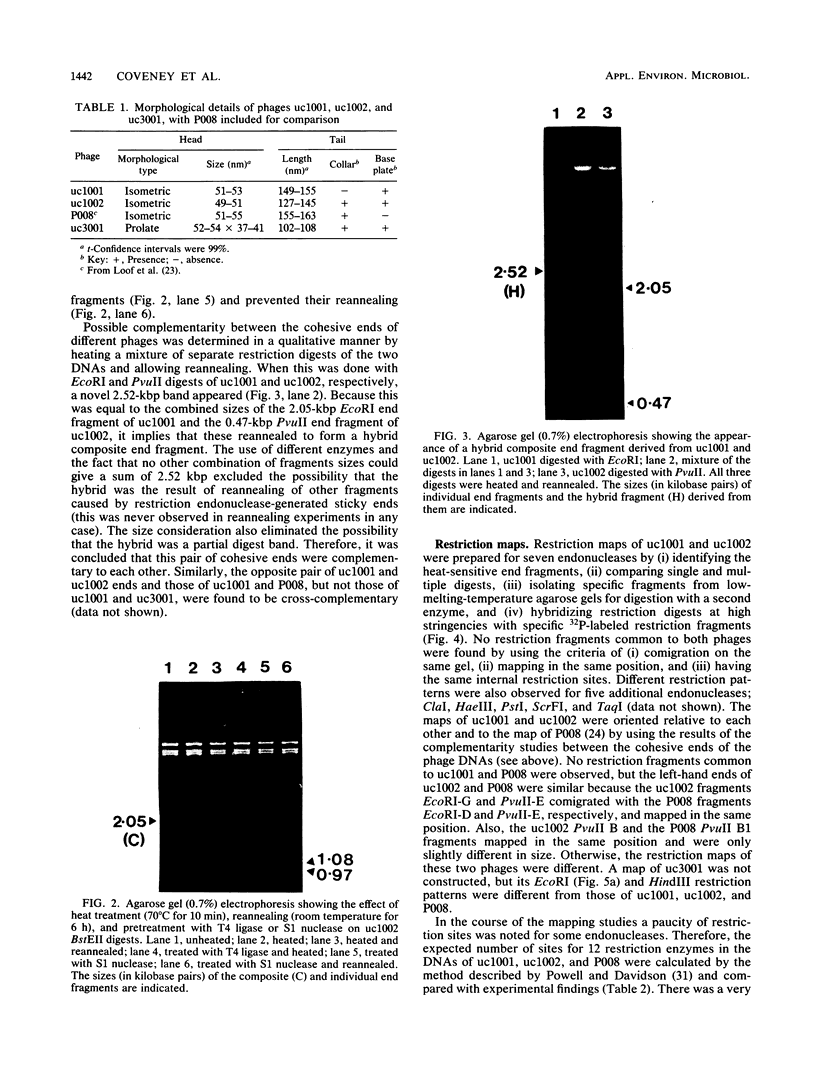
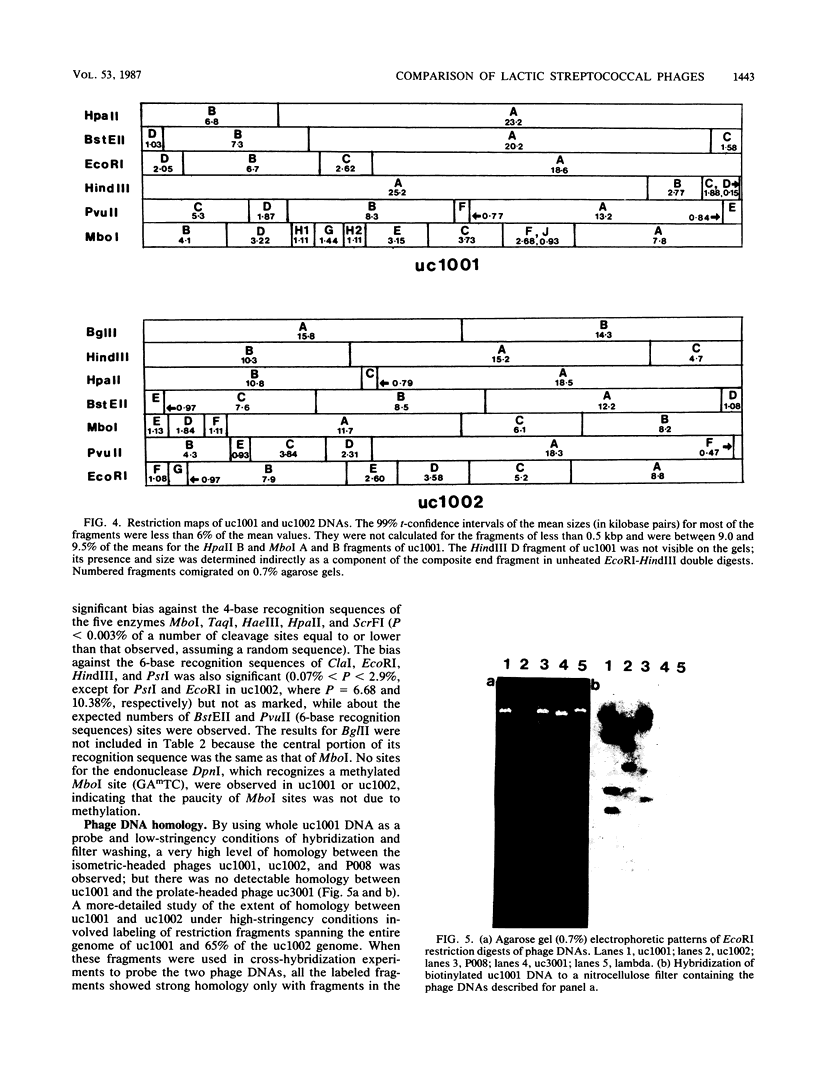
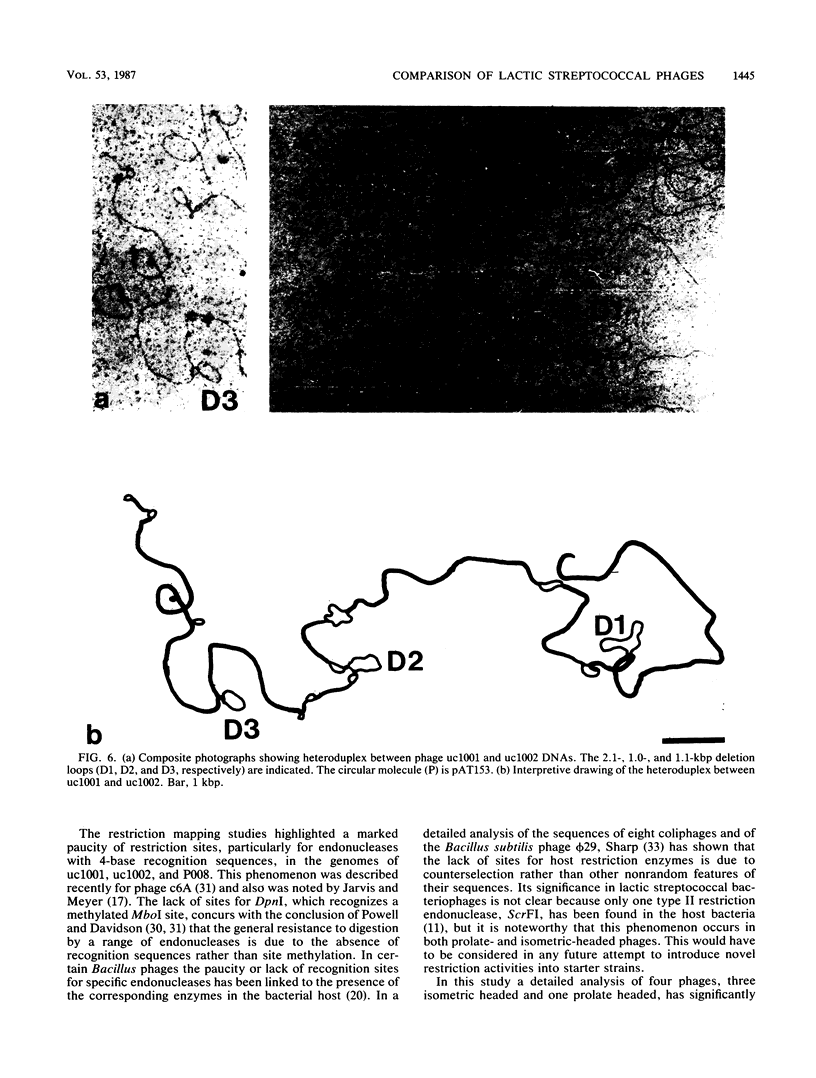
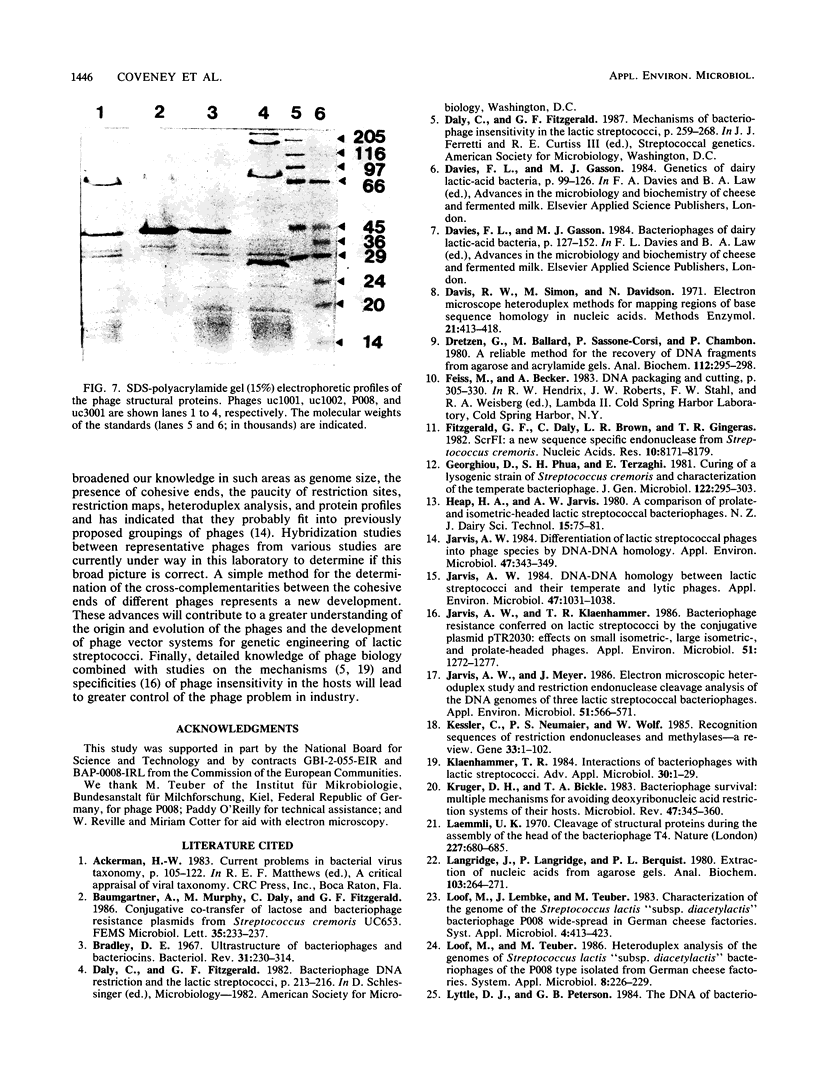
Bacteriophages uc1001 and uc1002, which are lytic for Streptococcus cremoris UC501 and UC502, respectively, were characterized in detail. Comparisons were made with a previously characterized phage, P008, which is lytic for Streptococcus lactis subsp. diacetylactis F7/2, and uc3001, which is a lytic phage for S. cremoris UC503. Phages uc1001 and uc1002 had small isometric heads (diameters, 52 and 50 nm, respectively) and noncontractile tails (lengths, 152 and 136 nm, respectively), and uc1002 also had a collar. Both had 30.1 ± 0.6 kilobase pairs (kbp) of DNA with cross-complementary cohesive ends. Restriction endonuclease maps made with seven endonucleases showed no common fragments. Despite this there was a very high level of homology between uc1001 and uc1002, and results of cross-hybridization experiments showed that the organization of both phage genomes was similar. Heteroduplex analysis confirmed this and quantified the level of homology at 83%. The regions of nonhomology comprised 2.1−, 1.1−, and 1.0-kbp deletion loops and 13 smaller loops and bubbles. The sodium dodecyl sulfate-polyacrylamide gel electrophoretic structural protein profiles were related, with a major band of about 40,000 molecular weight and minor bands of 35,000 and 34,000 molecular weight in common. There were also differences, however, in that uc1001 had a second major band of 68,000 molecular weight and two extra minor bands. Except for the restriction maps, which were strain specific, phages uc1001, uc1002, and P008 were closely related by all the criteria listed above. Their DNAs also showed a very significant bias against the cleavage sites of 9 of 11 restriction endonucleases. Phage uc3001 was unrelated to uc1001, uc1002, or P008 in that it had a prolate head (53 by 39 nm) and a shorter tail (105 nm), contained approximately 22 kbp of DNA, had unrelated cohesive ends, showed no DNA homology with the isometric-headed phages, and displayed a very different structural protein profile.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradley D. E. Ultrastructure of bacteriophage and bacteriocins. Bacteriol Rev. 1967 Dec;31(4):230–314. doi: 10.1128/br.31.4.230-314.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Fitzgerlad G. F., Daly C., Brown L. R., Gingeras T. R. ScrFI: a new sequence-specific endonuclease from Streptococcus cremoris. Nucleic Acids Res. 1982 Dec 20;10(24):8171–8179. doi: 10.1093/nar/10.24.8171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Meyer J. Electron microscopic heteroduplex study and restriction endonuclease cleavage analysis of the DNA genomes of three lactic streptococcal bacteriophages. Appl Environ Microbiol. 1986 Mar;51(3):566–571. doi: 10.1128/aem.51.3.566-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Neumaier P. S., Wolf W. Recognition sequences of restriction endonucleases and methylases--a review. Gene. 1985;33(1):1–102. doi: 10.1016/0378-1119(85)90119-2. [DOI] [PubMed] [Google Scholar]
- Krüger D. H., Bickle T. A. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol Rev. 1983 Sep;47(3):345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langridge J., Langridge P., Bergquist P. L. Extraction of nucleic acids from agarose gels. Anal Biochem. 1980 Apr;103(2):264–271. doi: 10.1016/0003-2697(80)90266-3. [DOI] [PubMed] [Google Scholar]
- Lyttle D. J., Petersen G. B. The DNA of bacteriophage 643: isolation and properties of the DNA of a bacteriophage infecting lactic Streptococci. Virology. 1984 Mar;133(2):403–415. doi: 10.1016/0042-6822(84)90406-9. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Tobian J. A., Jones K. R., Evans R. P., Clewell D. B. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene. 1982 Oct;19(3):345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- Mata M., Trautwetter A., Luthaud G., Ritzenthaler P. Thirteen Virulent and Temperate Bacteriophages of Lactobacillus bulgaricus and Lactobacillus lactis Belong to a Single DNA Homology Group. Appl Environ Microbiol. 1986 Oct;52(4):812–818. doi: 10.1128/aem.52.4.812-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Characterization of streptococcal bacteriophage c6A. J Gen Virol. 1985 Dec;66(Pt 12):2737–2741. doi: 10.1099/0022-1317-66-12-2737. [DOI] [PubMed] [Google Scholar]
- Powell I. B., Davidson B. E. Resistance to In Vitro Restriction of DNA from Lactic Streptococcal Bacteriophage c6A. Appl Environ Microbiol. 1986 Jun;51(6):1358–1360. doi: 10.1128/aem.51.6.1358-1360.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxelin M. L., Nurmiaho-Lassila E. L., Meriläinen V. T., Forsén R. I. Ultrastructure and Host Specificity of Bacteriophages of Streptococcus cremoris, Streptococcus lactis subsp. diacetylactis, and Leuconostoc cremoris from Finnish Fermented Milk "Viili". Appl Environ Microbiol. 1986 Oct;52(4):771–777. doi: 10.1128/aem.52.4.771-777.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986 Jan;3(1):75–83. doi: 10.1093/oxfordjournals.molbev.a040377. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Teuber M., Lembke J. The bacteriophages of lactic acid bacteria with emphasis on genetic aspects of group N lactic streptococci. Antonie Van Leeuwenhoek. 1983 Sep;49(3):283–295. doi: 10.1007/BF00399504. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]