Abstract
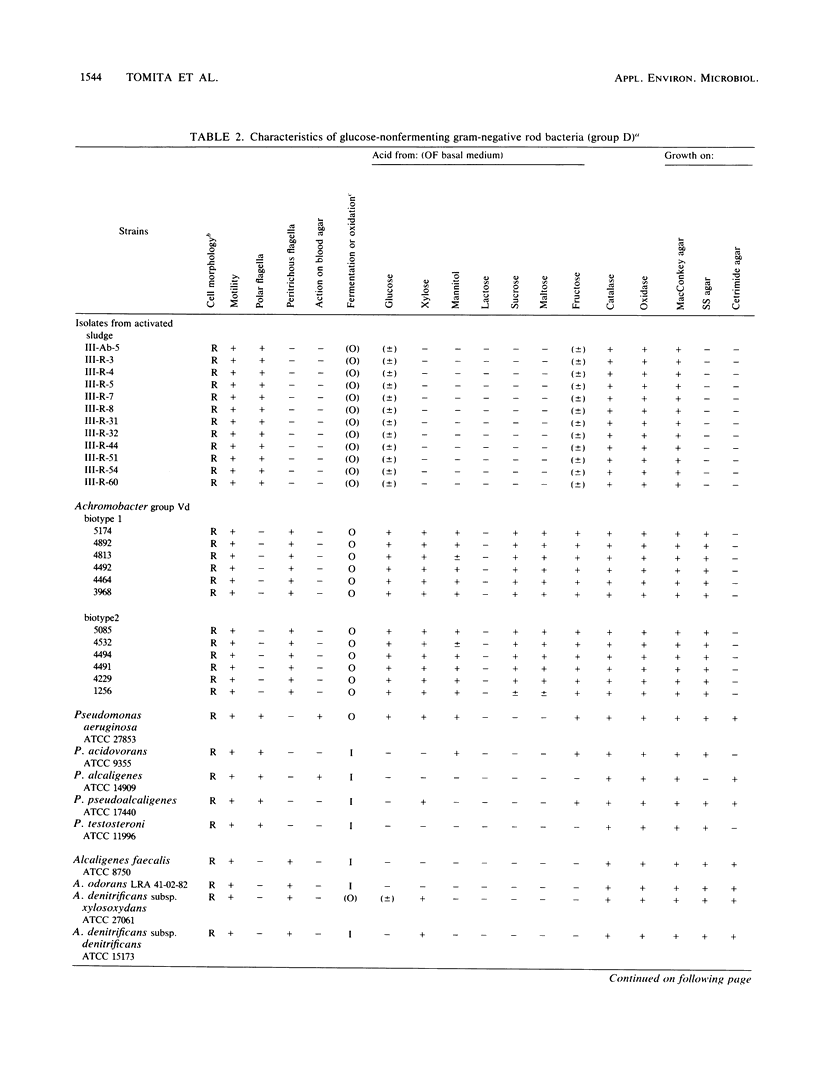
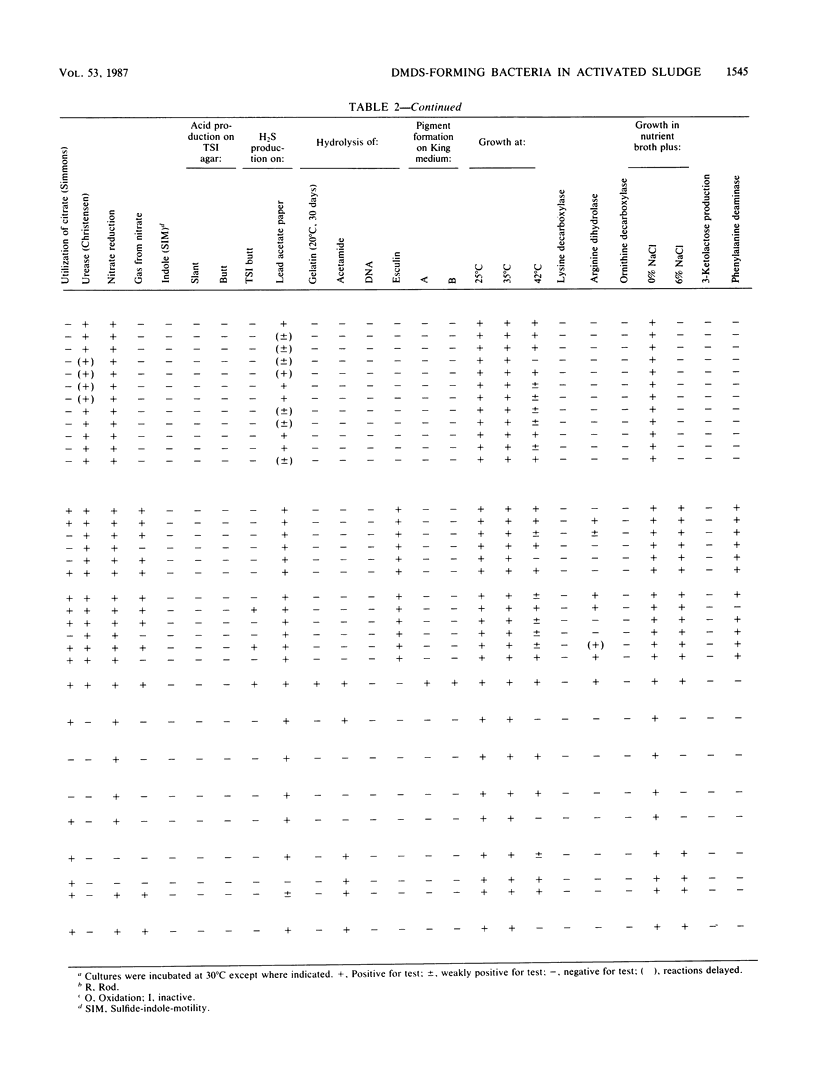
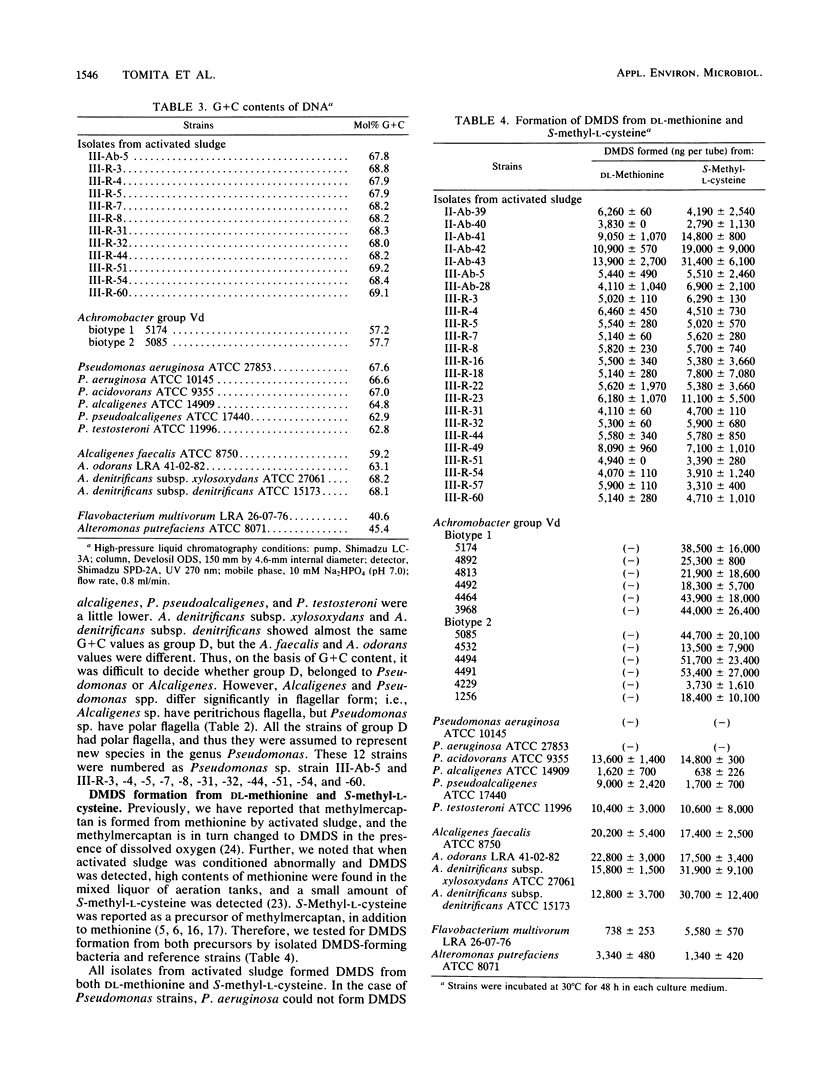
Twenty-four strains with high dimethyl disulfide (DMDS)-forming ability were isolated from activated sludge and identified to the genus level. These bacteria were classified into four groups (A, B, C, and D) by the API ZYM System (API System S.A., Montalieu, France). Group A (three strains) was identified as genus Lactobacillus by the API 20B System, by the method of Cowan and Steel, and by production of lactic acid as confirmed by gas-liquid chromatography. Group B (eight strains) was identified as genus Corynebacterium by API 20B and the Cowan and Steel method. Group C (one strain) was suggested to belong to genus Corynebacterium by the API 20B System. Group D (12 strains) was identified as genus Pseudomonas or Alcaligenes by the API 20B System, as genus Alcaligenes by the Cowan and Steel method, and as Achromobacter group Vd by the API 20NE System. However, on the basis of guanine-plus-cytosine contents in DNA and form of flagella, these strains were identified as genus Pseudomonas. Formation of DMDS from DL-methionine and S-methyl-L-cysteine was tested. DMDS-forming bacteria isolated from activated sludge formed DMDS from both precursors. In genus Pseudomonas, P. aeruginosa could not form DMDS from either precursor, but P. acidovorans, P. alcaligenes, P. pseudoalcaligenes, and P. testosteroni formed DMDS. In genus Alcaligenes, A. denitrificans subsp. xylosoxydans, A. denitrificans subsp. denitrificans, A. faecalis, and A. odorans formed DMDS from both precursors. Achromobacter group Vd formed DMDS from S-methyl-L-cysteine, but could not from DL-methionine.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chester B., Cooper L. H. Achromobacter species (CDC group Vd): morphological and biochemical characterization. J Clin Microbiol. 1979 Mar;9(3):425–436. doi: 10.1128/jcm.9.3.425-436.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi G. L. Characterization of nonfermentative nonfastidious gram negative bacteria encountered in medical bacteriology. J Appl Bacteriol. 1971 Sep;34(3):623–644. doi: 10.1111/j.1365-2672.1971.tb02326.x. [DOI] [PubMed] [Google Scholar]
- Hayward N. J., Jeavons T. H., Nicholson A. J., Thornton A. G. Methyl mercaptan and dimethyl disulfide production from methionine by Proteus species detected by head-space gas-liquid chromatography. J Clin Microbiol. 1977 Sep;6(3):187–194. doi: 10.1128/jcm.6.3.187-194.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Nakamura T., Eguchi Y. Purification and characterization of methioninase from Pseudomonas putida. J Biochem. 1976 Jun;79(6):1263–1272. doi: 10.1093/oxfordjournals.jbchem.a131180. [DOI] [PubMed] [Google Scholar]
- Kadota H., Ishida Y. Production of volatile sulfur compounds by microorganisms. Annu Rev Microbiol. 1972;26:127–138. doi: 10.1146/annurev.mi.26.100172.001015. [DOI] [PubMed] [Google Scholar]
- Kreis W., Hession C. Isolation and purification of L-methionine-alpha-deamino-gamma-mercaptomethane-lyase (L-methioninase) from Clostridium sporogenes. Cancer Res. 1973 Aug;33(8):1862–1865. [PubMed] [Google Scholar]
- LEIFSON E. Staining, shape and arrangement of bacterial flagella. J Bacteriol. 1951 Oct;62(4):377–389. doi: 10.1128/jb.62.4.377-389.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A., 3rd, Scanlan R. A., Lee J. S., Libbey L. M. Volatile compounds produced in sterile fish muscle (Sebastes melanops) by Pseudomonas putrefaciens, Pseudomonas fluorescens, and an Achromobacter species. Appl Microbiol. 1973 Jul;26(1):18–21. doi: 10.1128/am.26.1.18-21.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore D. P., Thompson J. F. Methionine biosynthesis from S-methylcysteine by thiomethyl transfer in Neurospora. Biochem Biophys Res Commun. 1967 Aug 7;28(3):474–479. doi: 10.1016/0006-291x(67)90336-1. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Starkey R. L. Dissimilation of methionine by fungi. J Bacteriol. 1969 Aug;99(2):544–551. doi: 10.1128/jb.99.2.544-551.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Segal W., Starkey R. L. Microbial decomposition of methionine and identity of the resulting sulfur products. J Bacteriol. 1969 Jun;98(3):908–913. doi: 10.1128/jb.98.3.908-913.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]


