Abstract
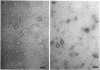
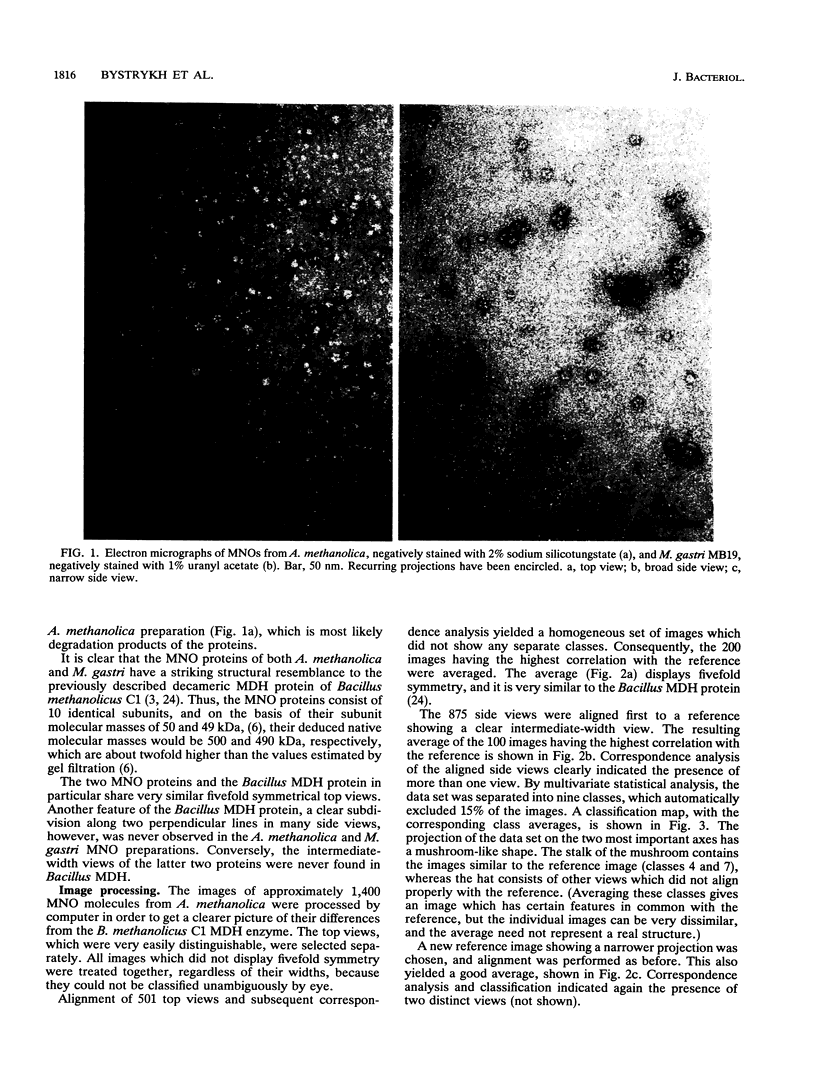
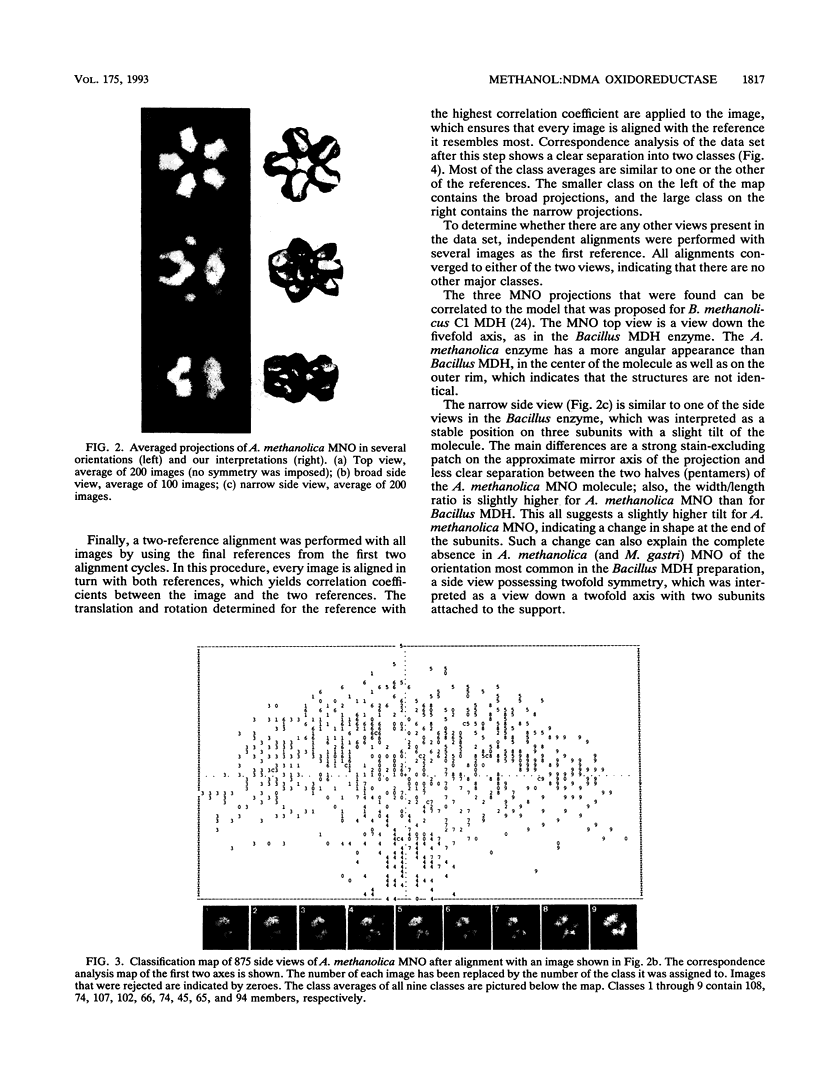
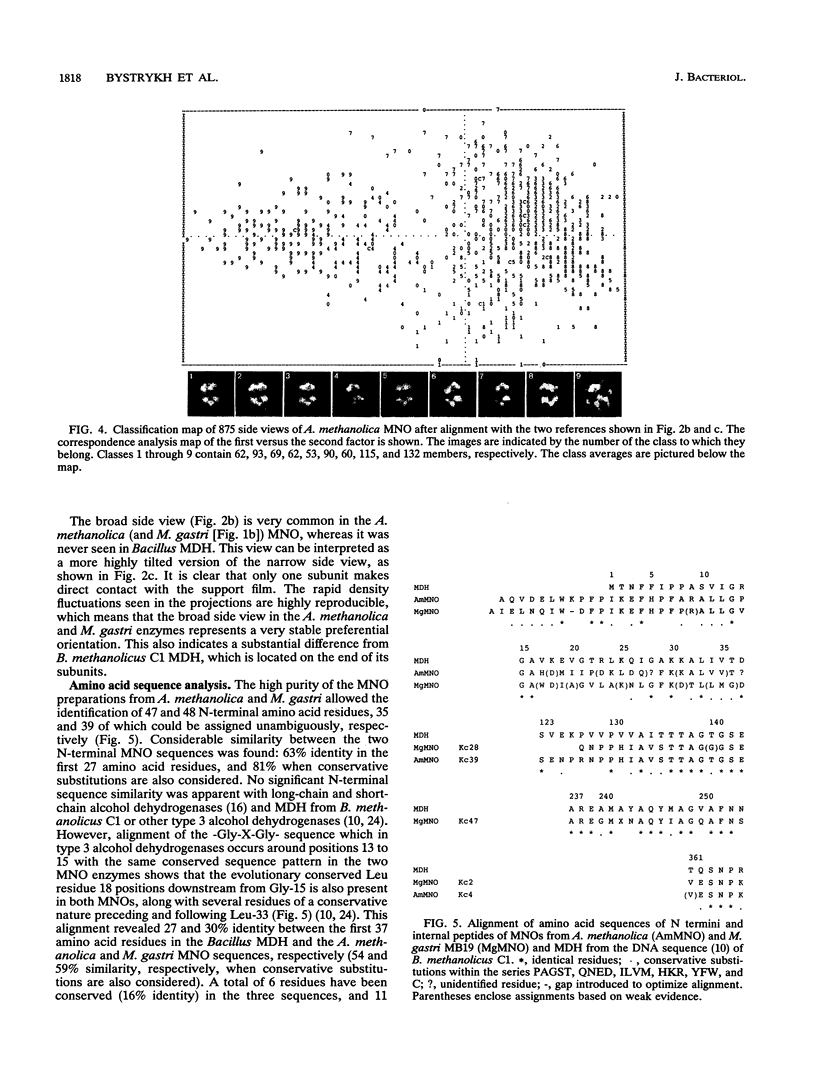
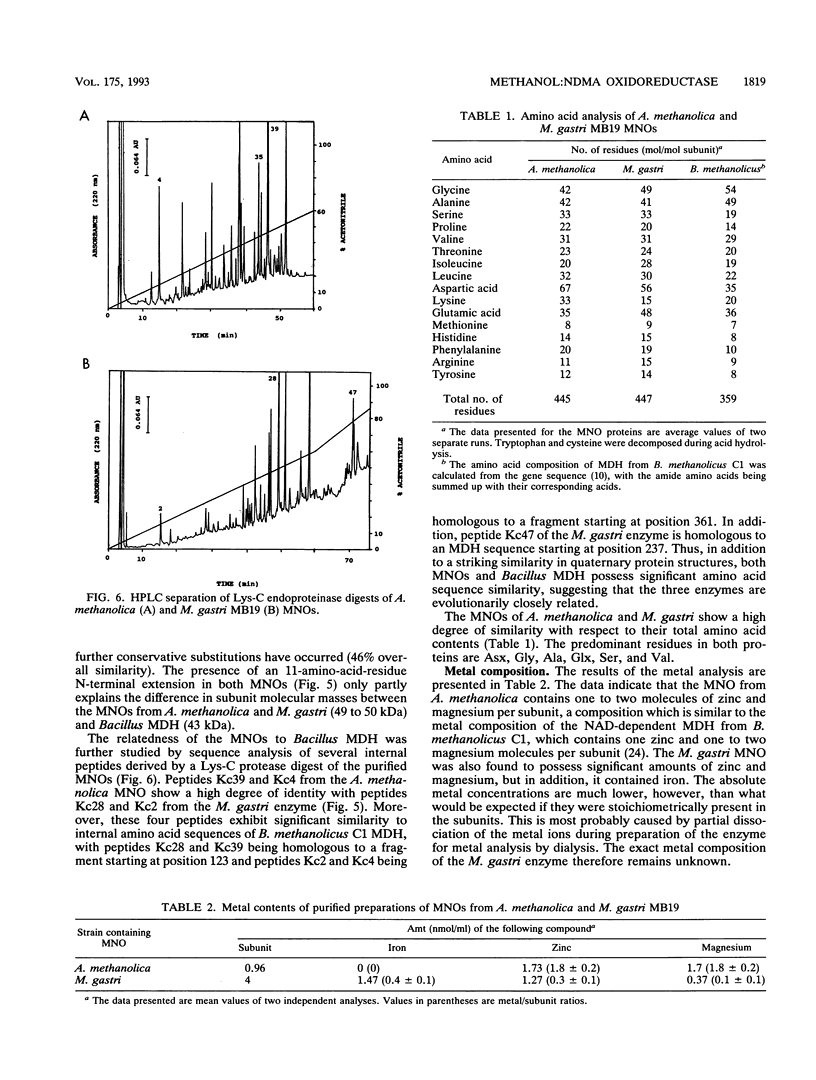
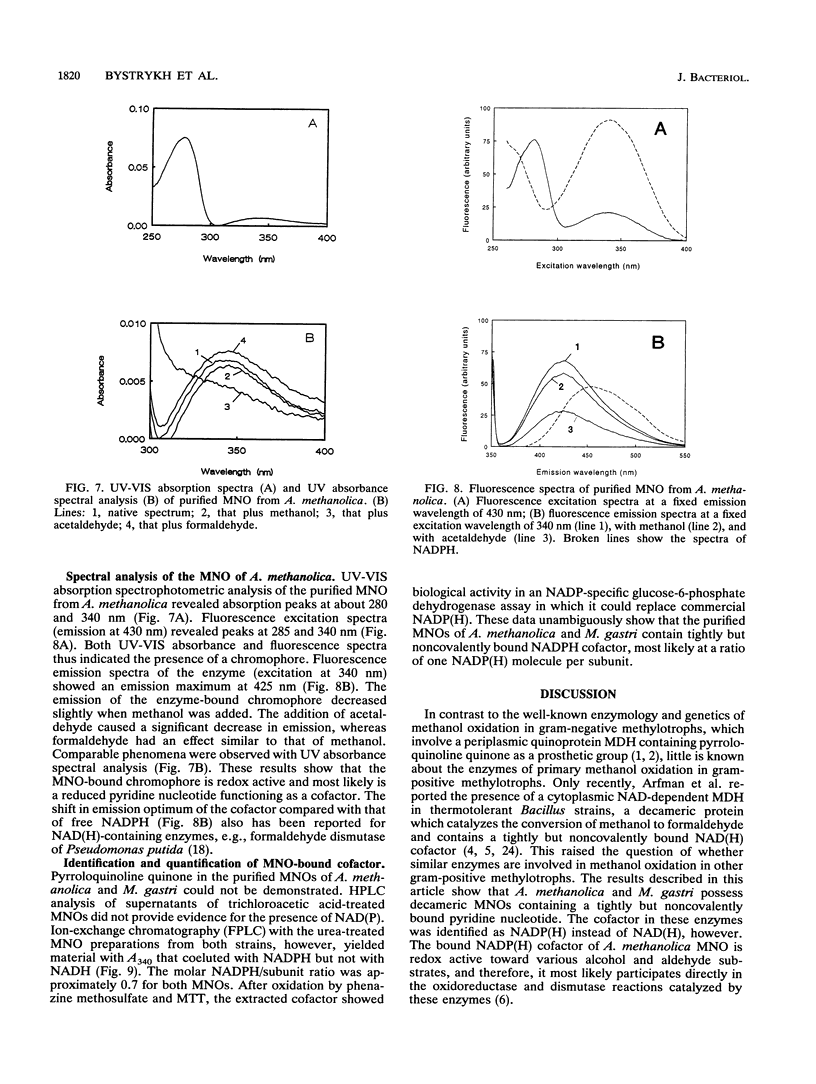
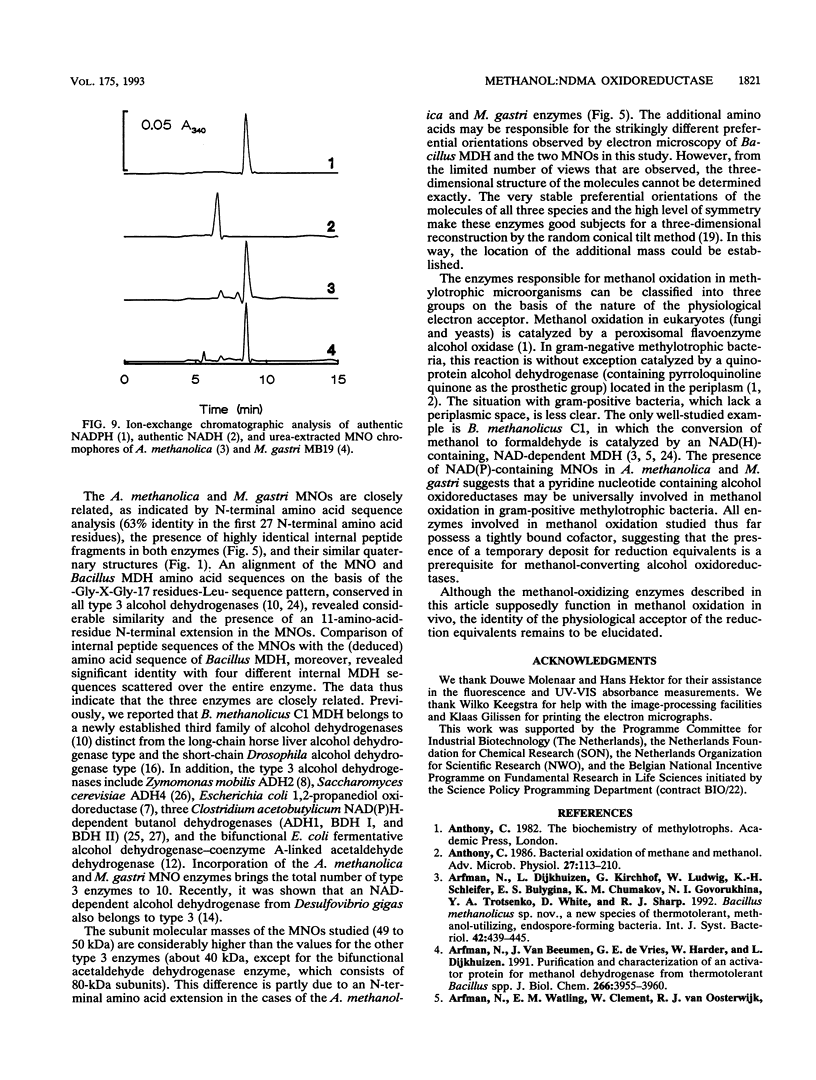
The quaternary protein structure of two methanol:N,N'-dimethyl-4-nitrosoaniline (NDMA) oxidoreductases purified from Amycolatopsis methanolica and Mycobacterium gastri MB19 was analyzed by electron microscopy and image processing. The enzymes are decameric proteins (displaying fivefold symmetry) with estimated molecular masses of 490 to 500 kDa based on their subunit molecular masses of 49 to 50 kDa. Both methanol:NDMA oxidoreductases possess a tightly but noncovalently bound NADP(H) cofactor at an NADPH-to-subunit molar ratio of 0.7. These cofactors are redox active toward alcohol and aldehyde substrates. Both enzymes contain significant amounts of Zn2+ and Mg2+ ions. The primary amino acid sequences of the A. methanolica and M. gastri MB19 methanol:NDMA oxidoreductases share a high degree of identity, as indicated by N-terminal sequence analysis (63% identity among the first 27 N-terminal amino acids), internal peptide sequence analysis, and overall amino acid composition. The amino acid sequence analysis also revealed significant similarity to a decameric methanol dehydrogenase of Bacillus methanolicus C1.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anthony C. Bacterial oxidation of methane and methanol. Adv Microb Physiol. 1986;27:113–210. doi: 10.1016/s0065-2911(08)60305-7. [DOI] [PubMed] [Google Scholar]
- Arfman N., Dijkhuizen L., Kirchhof G., Ludwig W., Schleifer K. H., Bulygina E. S., Chumakov K. M., Govorukhina N. I., Trotsenko Y. A., White D. Bacillus methanolicus sp. nov., a new species of thermotolerant, methanol-utilizing, endospore-forming bacteria. Int J Syst Bacteriol. 1992 Jul;42(3):439–445. doi: 10.1099/00207713-42-3-439. [DOI] [PubMed] [Google Scholar]
- Arfman N., Van Beeumen J., De Vries G. E., Harder W., Dijkhuizen L. Purification and characterization of an activator protein for methanol dehydrogenase from thermotolerant Bacillus spp. J Biol Chem. 1991 Feb 25;266(6):3955–3960. [PubMed] [Google Scholar]
- Arfman N., Watling E. M., Clement W., van Oosterwijk R. J., de Vries G. E., Harder W., Attwood M. M., Dijkhuizen L. Methanol metabolism in thermotolerant methylotrophic Bacillus strains involving a novel catabolic NAD-dependent methanol dehydrogenase as a key enzyme. Arch Microbiol. 1989;152(3):280–288. doi: 10.1007/BF00409664. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Similarity of Escherichia coli propanediol oxidoreductase (fucO product) and an unusual alcohol dehydrogenase from Zymomonas mobilis and Saccharomyces cerevisiae. J Bacteriol. 1989 Jul;171(7):3754–3759. doi: 10.1128/jb.171.7.3754-3759.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J., van Heel M. Correspondence analysis of aligned images of biological particles. J Mol Biol. 1982 Oct 15;161(1):134–137. doi: 10.1016/0022-2836(82)90282-0. [DOI] [PubMed] [Google Scholar]
- Goodlove P. E., Cunningham P. R., Parker J., Clark D. P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989 Dec 21;85(1):209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- Groen B. W., van Kleef M. A., Duine J. A. Quinohaemoprotein alcohol dehydrogenase apoenzyme from Pseudomonas testosteroni. Biochem J. 1986 Mar 15;234(3):611–615. doi: 10.1042/bj2340611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Analysis of phenylthiohydantoins by ultrasensitive gradient high-performance liquid chromatography. Methods Enzymol. 1983;91:486–493. doi: 10.1016/s0076-6879(83)91045-5. [DOI] [PubMed] [Google Scholar]
- Jörnvall H., Persson M., Jeffery J. Alcohol and polyol dehydrogenases are both divided into two protein types, and structural properties cross-relate the different enzyme activities within each type. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4226–4230. doi: 10.1073/pnas.78.7.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Kato N., Yamagami T., Shimao M., Sakazawa C. Formaldehyde dismutase, a novel NAD-binding oxidoreductase from Pseudomonas putida F61. Eur J Biochem. 1986 Apr 1;156(1):59–64. doi: 10.1111/j.1432-1033.1986.tb09548.x. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. J Electron Microsc Tech. 1988 Aug;9(4):359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Vonck J., Arfman N., De Vries G. E., Van Beeumen J., Van Bruggen E. F., Dijkhuizen L. Electron microscopic analysis and biochemical characterization of a novel methanol dehydrogenase from the thermotolerant Bacillus sp. C1. J Biol Chem. 1991 Feb 25;266(6):3949–3954. [PubMed] [Google Scholar]
- Walter K. A., Bennett G. N., Papoutsakis E. T. Molecular characterization of two Clostridium acetobutylicum ATCC 824 butanol dehydrogenase isozyme genes. J Bacteriol. 1992 Nov;174(22):7149–7158. doi: 10.1128/jb.174.22.7149-7158.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson V. M., Paquin C. E. Homology of Saccharomyces cerevisiae ADH4 to an iron-activated alcohol dehydrogenase from Zymomonas mobilis. Mol Gen Genet. 1987 Sep;209(2):374–381. doi: 10.1007/BF00329668. [DOI] [PubMed] [Google Scholar]
- Youngleson J. S., Jones W. A., Jones D. T., Woods D. R. Molecular analysis and nucleotide sequence of the adh1 gene encoding an NADPH-dependent butanol dehydrogenase in the Gram-positive anaerobe Clostridium acetobutylicum. Gene. 1989 May 30;78(2):355–364. doi: 10.1016/0378-1119(89)90238-2. [DOI] [PubMed] [Google Scholar]
- de Boer L., Dijkhuizen L., Grobben G., Goodfellow M., Stackebrandt E., Parlett J. H., Whitehead D., Witt D. Amycolatopsis methanolica sp. nov., a facultatively methylotrophic actinomycete. Int J Syst Bacteriol. 1990 Apr;40(2):194–204. doi: 10.1099/00207713-40-2-194. [DOI] [PubMed] [Google Scholar]
- de Vries G. E., Arfman N., Terpstra P., Dijkhuizen L. Cloning, expression, and sequence analysis of the Bacillus methanolicus C1 methanol dehydrogenase gene. J Bacteriol. 1992 Aug;174(16):5346–5353. doi: 10.1128/jb.174.16.5346-5353.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M., Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6(2):187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- van Heel M. Multivariate statistical classification of noisy images (randomly oriented biological macromolecules). Ultramicroscopy. 1984;13(1-2):165–183. doi: 10.1016/0304-3991(84)90066-4. [DOI] [PubMed] [Google Scholar]
- van Heel M., Stöffler-Meilicke M. Characteristic views of E. coli and B. stearothermophilus 30S ribosomal subunits in the electron microscope. EMBO J. 1985 Sep;4(9):2389–2395. doi: 10.1002/j.1460-2075.1985.tb03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]