Abstract
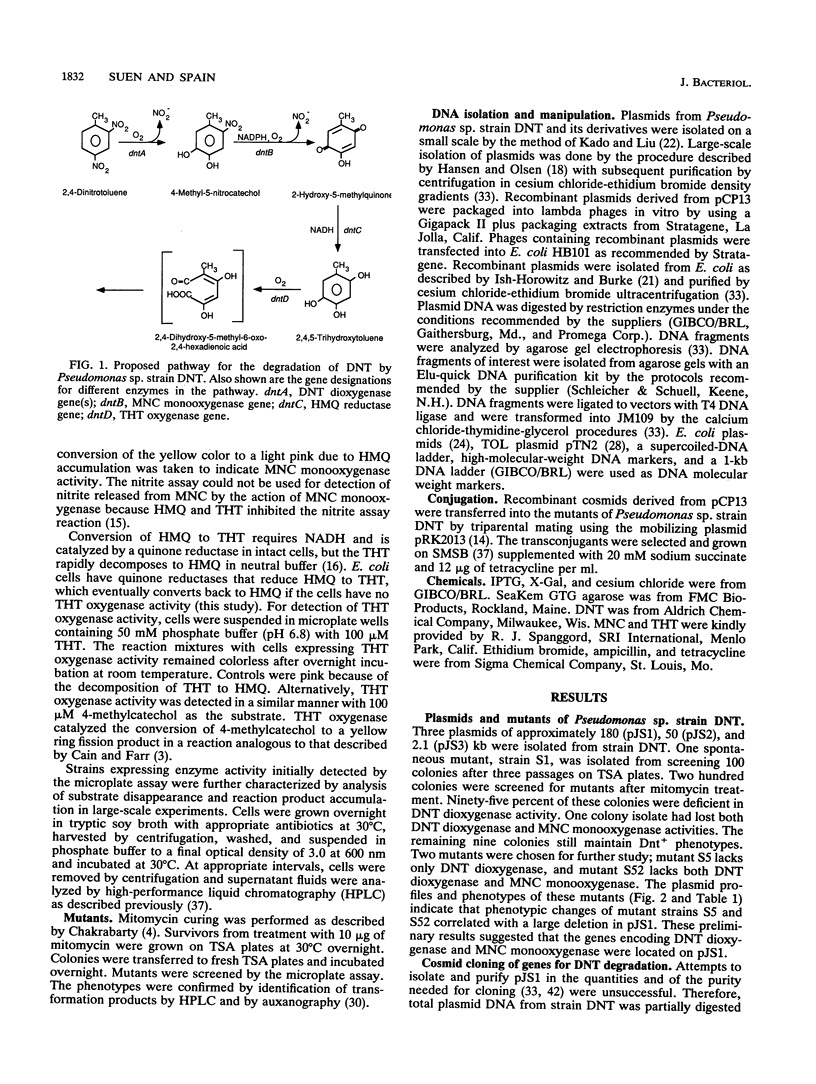
The degradation of 2,4-dinitrotoluene (DNT) by Pseudomonas sp. strain DNT is initiated by a dioxygenase attack to yield 4-methyl-5-nitrocatechol (MNC) and nitrite. Subsequent oxidation of MNC by a monooxygenase results in the removal of the second molecule of nitrite, and further enzymatic reactions lead to ring fission. Initial studies on the molecular basis of DNT degradation in this strain revealed the presence of three plasmids. Mitomycin-derived mutants deficient in either DNT dioxygenase only or DNT dioxygenase and MNC monooxygenase were isolated. Plasmid profiles of mutant strains suggested that the mutations resulted from deletions in the largest plasmid. Total plasmid DNA partially digested by EcoRI was cloned into a broad-host-range cosmid vector, pCP13. Recombinant clones containing genes encoding DNT dioxygenase, MNC monooxygenase, and 2,4,5-trihydroxytoluene oxygenase were characterized by identification of reaction products and the ability to complement mutants. Subcloning analysis suggests that the DNT dioxygenase is a multicomponent enzyme system and that the genes for the DNT pathway are organized in at least three different operons.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Cain R. B., Farr D. R. Metabolism of arylsulphonates by micro-organisms. Biochem J. 1968 Feb;106(4):859–877. doi: 10.1042/bj1060859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain R. B. Utilization of anthranilic and nitrobenzoic acids by Nocardia opaca and a flavobacterium. J Gen Microbiol. 1966 Feb;42(2):219–235. doi: 10.1099/00221287-42-2-219. [DOI] [PubMed] [Google Scholar]
- Chakrabarty A. M. Genetic basis of the biodegradation of salicylate in Pseudomonas. J Bacteriol. 1972 Nov;112(2):815–823. doi: 10.1128/jb.112.2.815-823.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A., Wubbolts M. G., Abril M. A., Ramos J. L. Nitroaromatics Are Substrates for the TOL Plasmid Upper-Pathway Enzymes. Appl Environ Microbiol. 1992 Jan;58(1):415–417. doi: 10.1128/aem.58.1.415-417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel O., Knackmuss H. J. Catabolism of 1,3-dinitrobenzene by Rhodococcus sp. QT-1. Arch Microbiol. 1991;157(1):76–79. doi: 10.1007/BF00245339. [DOI] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J Bacteriol. 1985 Jan;161(1):466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1985 Jan;161(1):85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D., Gibson D. T., Laborde A. L. Oxidation of naphthalene by a multicomponent enzyme system from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1982 Mar;149(3):948–954. doi: 10.1128/jb.149.3.948-954.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensley B. D., Ratzkin B. J., Osslund T. D., Simon M. J., Wackett L. P., Gibson D. T. Expression of naphthalene oxidation genes in Escherichia coli results in the biosynthesis of indigo. Science. 1983 Oct 14;222(4620):167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Haigler B. E., Spain J. C. Biotransformation of nitrobenzene by bacteria containing toluene degradative pathways. Appl Environ Microbiol. 1991 Nov;57(11):3156–3162. doi: 10.1128/aem.57.11.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Wubbolts M., Rose K., Leppik R. A., Timmis K. N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989 Sep;171(9):5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Thomson K., Anderson A. C. Identification of nitroso compounds from biotransformation of 2,4-dinitrotoluene. Appl Environ Microbiol. 1984 Jun;47(6):1295–1298. doi: 10.1128/aem.47.6.1295-1298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- McCormick N. G., Cornell J. H., Kaplan A. M. Identification of biotransformation products from 2,4-dinitrotoluene. Appl Environ Microbiol. 1978 May;35(5):945–948. doi: 10.1128/aem.35.5.945-948.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick N. G., Feeherry F. E., Levinson H. S. Microbial transformation of 2,4,6-trinitrotoluene and other nitroaromatic compounds. Appl Environ Microbiol. 1976 Jun;31(6):949–958. doi: 10.1128/aem.31.6.949-958.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa T., Hayashi E., Yokota T., Ebina Y., Nakazawa A. Isolation of TOL and RP4 recombinants by integrative suppression. J Bacteriol. 1978 Apr;134(1):270–277. doi: 10.1128/jb.134.1.270-277.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidle E. L., Shapiro M. K., Ornston L. N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J Bacteriol. 1987 Dec;169(12):5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert D. E., Butterworth B. E., Popp J. A. Dinitrotoluene: acute toxicity, oncogenicity, genotoxicity, and metabolism. Crit Rev Toxicol. 1984;13(3):217–234. doi: 10.3109/10408448409003373. [DOI] [PubMed] [Google Scholar]
- Robertson J. B., Spain J. C., Haddock J. D., Gibson D. T. Oxidation of nitrotoluenes by toluene dioxygenase: evidence for a monooxygenase reaction. Appl Environ Microbiol. 1992 Aug;58(8):2643–2648. doi: 10.1128/aem.58.8.2643-2648.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain J. C., Wyss O., Gibson D. T. Enzymatic oxidation of p-nitrophenol. Biochem Biophys Res Commun. 1979 May 28;88(2):634–641. doi: 10.1016/0006-291x(79)92095-3. [DOI] [PubMed] [Google Scholar]
- Spanggord R. J., Spain J. C., Nishino S. F., Mortelmans K. E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991 Nov;57(11):3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streber W. R., Timmis K. N., Zenk M. H. Analysis, cloning, and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987 Jul;169(7):2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M., Hayakawa T., Shaw J. P., Rekik M., Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991 Mar;173(5):1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli K., Brock B. J., Joshi D. K., Gold M. H. Degradation of 2,4-dinitrotoluene by the lignin-degrading fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1992 Jan;58(1):221–228. doi: 10.1128/aem.58.1.221-228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whited G. M., Gibson D. T. Separation and partial characterization of the enzymes of the toluene-4-monooxygenase catabolic pathway in Pseudomonas mendocina KR1. J Bacteriol. 1991 May;173(9):3017–3020. doi: 10.1128/jb.173.9.3017-3020.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yeh W. K., Gibson D. T., Liu T. N. Toluene dioxygenase: a multicomponent enzyme system. Biochem Biophys Res Commun. 1977 Sep 9;78(1):401–410. doi: 10.1016/0006-291x(77)91268-2. [DOI] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Karl M. R., Blatt L. M., Simon M. J., Winter R. B., Fausset P. R., Lu H. S., Harcourt A. A., Chen K. K. Cloning and characterization of a Pseudomonas mendocina KR1 gene cluster encoding toluene-4-monooxygenase. J Bacteriol. 1991 Sep;173(17):5315–5327. doi: 10.1128/jb.173.17.5315-5327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zylstra G. J., Gibson D. T. Toluene degradation by Pseudomonas putida F1. Nucleotide sequence of the todC1C2BADE genes and their expression in Escherichia coli. J Biol Chem. 1989 Sep 5;264(25):14940–14946. [PubMed] [Google Scholar]
- Zylstra G. J., McCombie W. R., Gibson D. T., Finette B. A. Toluene degradation by Pseudomonas putida F1: genetic organization of the tod operon. Appl Environ Microbiol. 1988 Jun;54(6):1498–1503. doi: 10.1128/aem.54.6.1498-1503.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer J. R., van Neerven A. R., de Vries E. J., de Vos W. M., Zehnder A. J. Cloning and characterization of plasmid-encoded genes for the degradation of 1,2-dichloro-, 1,4-dichloro-, and 1,2,4-trichlorobenzene of Pseudomonas sp. strain P51. J Bacteriol. 1991 Jan;173(1):6–15. doi: 10.1128/jb.173.1.6-15.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]




