Abstract
Translational quality control is monitored at several steps, including substrate selection by aminoacyl-tRNA synthetases (aaRSs), and discrimination of aminoacyl-tRNAs by elongation factor Tu (EF-Tu) and the ribosome. Phenylalanyl-tRNA synthetase (PheRS) misactivates Tyr but is able to correct the mistake using a proofreading activity named editing. Previously we found that overproduction of editing-defective PheRS resulted in Tyr incorporation at Phe-encoded positions in vivo, although the misreading efficiency could not be estimated. This raised the question as to whether or not EF-Tu and the ribosome provide further proofreading mechanisms to prevent mistranslation of Phe codons by Tyr. Here we show that, after evading editing by PheRS, Tyr-tRNAPhe is recognized by EF-Tu as efficiently as the cognate Phe-tRNAPhe. Kinetic decoding studies using full-length Tyr-tRNAPhe and Phe-tRNAPhe, as well as a poly(U)-directed polyTyr/polyPhe synthesis assay, indicate that the ribosome lacks discrimination between Tyr-tRNAPhe and Phe-tRNAPhe. Taken together, these data suggest that PheRS editing is the major proofreading step that prevents infiltration of Tyr into Phe codons during translation.
Keywords: editing, proofreading, protein synthesis, quality control, translation, tRNA
INTRODUCTION
As a central process of biology, translation utilizes a conserved machinery throughout the three domains of life. Passage of genetic information from mRNA to protein can be separated into several steps, with transfer RNAs (tRNA) playing an adaptor role to pair amino acids with their cognate codons during translation. Amino acids are first attached to their cognate tRNAs by aminoacyl-tRNA synthetases (aaRSs); the resulting aminoacyl-tRNAs (aa-tRNAs) are subsequently delivered by elongation factors (EF-Tu in bacteria and EF-1α in archaea and eukarya) to ribosomes, where codon–anticodon recognition programs incorporation of amino acids into the elongating peptides. It is critical for organisms to maintain translational accuracy, as several studies have suggested that increased translational error rates can slow growth in bacteria (Nangle et al. 2002; Roy et al. 2004; Bacher et al. 2005) and cause neurodegeneration in mammals (Lee et al. 2006). To ensure quality control during translation the cell utilizes several strategies, including aaRS editing, EF-Tu discrimination, and ribosomal selection (Ibba and Söll 1999; Dale and Uhlenbeck 2005a).
aaRSs usually selectively activate their cognate amino acids during aminoacylation. However, errors in aminoacylation can occur when synthetases encounter noncognate amino acids or analogs that are structurally similar to their cognate substrates. For example, phenylalanyl-tRNA synthetase (PheRS) misactivates tyrosine at a rate significantly higher than the overall translational error rate (Roy et al. 2005). To maintain accuracy during translation, PheRS uses an editing domain that hydrolyzes misacylated tRNAPhe (Lin et al. 1984; Roy et al. 2004, 2005). Similar activities are found in many other aaRSs (Nureki et al. 1998; Silvian et al. 1999; Beuning and Musier-Forsyth 2000; Dock-Bregeon et al. 2000, 2004; Fukai et al. 2000; Ahel et al. 2003; Beebe et al. 2003, 2004; Lincecum et al. 2003; Wong et al. 2003; Korencic et al. 2004), suggesting that editing is widely used to proofread mistakes made in aminoacylation. Such editing activities are critical for the cell. It has been reported that a partial loss of alanyl-tRNA synthetase (AlaRS) editing function in mice results in protein misfolding and neurodegeneration (Lee et al. 2006), demonstrating the significance of maintaining a full editing activity.
EF-Tu was long considered to bind all aa-tRNAs with roughly equal efficiencies, the only exceptions being fMet-tRNAfMet, Ser/Sec-tRNAsec, Glu-tRNAGln, and Asp-tRNAAsn (Stanzel et al. 1994; Becker and Kern 1998; Dale and Uhlenbeck 2005a; Ambrogelly et al. 2007; Roy et al. 2007). The weak affinity of EF-Tu for Glu-tRNAGln and Asp-tRNAAsn prevents genetic ambiguity that would result from misincorporating Glu and Asp at Gln and Asn codons, respectively, while fMet-tRNAfMet and Sec-tRNASec instead bind specialized translation factors (Ambrogelly et al. 2007). Recently, Uhlenbeck and colleagues found that EF-Tu displays selectivity for both the amino acid and the tRNA body (LaRiviere et al. 2001; Asahara and Uhlenbeck 2002, 2005; Dale et al. 2004). It was proposed that, since EF-Tu binds amino acids with various affinities, their corresponding tRNAs have evolved to compensate for the differences in affinity thermodynamically, so that EF-Tu binds all cognate aa-tRNAs uniformly (Dale and Uhlenbeck 2005a). In contrast to cognate aa-tRNAs, EF-Tu binds their noncognate counterparts with a wide range of affinities, which may lead to reduced incorporation efficiencies for some mischarged amino acids.
The last step at which translational fidelity can be monitored is ribosomal decoding. Proper matching of codons and anticodons is a prerequisite for efficient decoding, and the ribosome utilizes both thermodynamic and kinetic discrimination mechanisms to reject aa-tRNAs with near-cognate or noncognate anticodons (Gromadski and Rodnina 2004a; Cochella and Green 2005). It is less clear, however, whether ribosomes are able to discriminate against misacylated tRNAs with cognate codons. Misacylated tRNAs have been shown to be utilized in translation of peptides in vitro and in vivo (Chapeville et al. 1962; Döring et al. 2001; Nangle et al. 2002; Bacher et al. 2005; Lee et al. 2006; Wang et al. 2006; Xie and Schultz 2006), although the decoding rates for these species are unknown. Several lines of evidence suggest that the ribosome may discriminate the side chains of amino acids. In a study by Bhuta and colleagues several amino acids were ligated to the C–A dinucleotide. The resulting aminoacyl-dinucleotides (C–A-aa) displayed a broad range of affinities for the ribosomal A site in a peptidyl transferase assay (Bhuta et al. 1981). Translation inhibition experiments using various puromycin derivatives also indicated that the ribosomal A site specifically recognizes the amino acid side chains of aa-tRNAs (Starck et al. 2003). Conversely, nonenzymatic binding studies using full-length aa-tRNAs showed that misacylated and correctly acylated tRNAs bind to the ribosomal A site with similar affinities (Fahlman et al. 2004; Dale and Uhlenbeck 2005b). To provide some insights into the discrepancy, we investigated the effect on decoding kinetics of full-length misacylated tRNAs, as part of a broader study of the quality control mechanisms that prevent misincorporation of Tyr at Phe codons. We found that EF-Tu efficiently recognizes Tyr-tRNAPhe synthesized by an editing-defective PheRS, and that the ribosome does not discriminate Tyr-tRNAPhe from the cognate Phe-tRNAPhe, as revealed by a fast-kinetic decoding experiment and poly(U)-directed polyPhe or polyTyr synthesis assays. Together, our data suggest that PheRS editing is the major proofreading step that prevents infiltration of Tyr into Phe codons during translation.
RESULTS AND DISCUSSION
Misacylation of tRNAPhe by editing-defective PheRSs
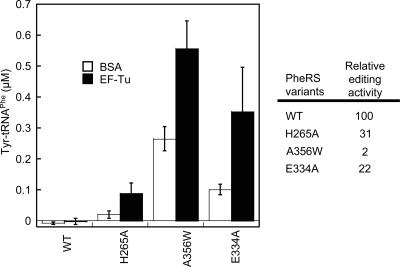
Previously we have shown that Escherichia coli PheRS possesses an editing activity against Tyr (Roy et al. 2004). PheRS editing site residues are mainly involved in substrate binding, and mutations of the conserved residues resulted in, at best, only a few fold decrease in editing (Ling et al. 2007). A question that arose is why the editing site residues are so conserved even though they do not contribute significantly to activity. Traditionally, editing experiments are performed in the absence of nonsynthetase cellular factors such as EF-Tu. Nevertheless, it has been shown that EF-Tu significantly increases the steady-state levels of cognate and misacylated tRNAs (Wolfson and Uhlenbeck 2002; Lee et al. 2006), which prompted us to further investigate the mischarging activities of PheRS variants in the presence and absence of EF-Tu. The wild-type (WT) PheRS did not produce accumulated Tyr-tRNAPhe irrespective of EF-Tu addition (Fig. 1). As the WT and editing defective PheRSs display similar aminoacylation activities (Ling et al. 2007), it is likely that the WT PheRS hydrolyzes Tyr-tRNAPhe before it is trapped by EF-Tu. In contrast, editing-defective PheRS variants displayed significantly increased tyrosylation levels in the presence of EF-Tu, likely through EF-Tu protection of synthesized Tyr-tRNAPhe from hydrolysis. The βH265A variant, which has a threefold reduced post-transfer editing activity, misacylated tRNAPhe very weakly in the absence of EF-Tu. Addition of EF-Tu dramatically increased the Tyr-tRNAPhe level, suggesting a critical role of βH265 for editing in vivo. It appears that the PheRS editing site has evolved to maintain a minimal editing efficiency while preventing “misediting” of the cognate Phe (Ling et al. 2007). Current views on the roles of aaRS editing site residues may need to be revisited in the context of in vivo conditions, as it now seems likely that aaRS editing sites have been fine-tuned to avoid uptake of misacylated tRNAs by elongation factors. This is consistent with the observation that a trans-editing factor, YbaK, associates with ProRS to compete with EF-Tu for mischarged Cys-tRNAPro (An and Musier-Forsyth 2005).
FIGURE 1.
Tyrosylation of E. coli tRNAPhe (5 μM) by PheRS variants (0.5 μM each) in the presence (filled bars) and absence (open bars) of EF-Tu (5 μM). The Y-axis represents the end levels of Tyr-tRNAPhe synthesized after 6 min. PheRS variants contain either a wild-type editing site (WT) or mutations in the β subunit that reduce the editing activity (Ling et al. 2007). Relative post-transfer editing activities determined previously are shown in the figure. Data sets are the average of three independent experiments.
EF-Tu efficiently recognizes Tyr-tRNAPhe
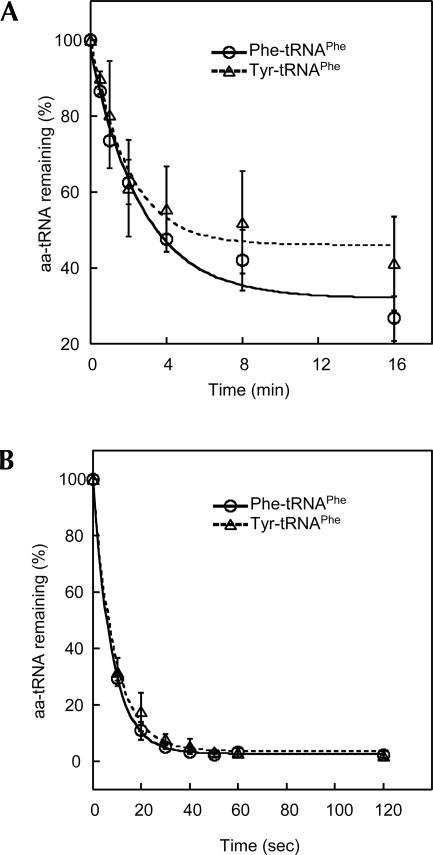
EF-Tu increased the tyrosylation level of tRNAPhe by PheRS, suggesting that Tyr-tRNAPhe might be protected by EF-Tu. To directly address this possibility, we tested the dissociation rates of Tyr-tRNAPhe and Phe-tRNAPhe from E. coli EF-Tu using an RNase A protection assay as previously described (LaRiviere et al. 2001; Asahara and Uhlenbeck 2002). The k off rates of Tyr-tRNAPhe and Phe-tRNAPhe were very similar at 2°C (Fig. 2A), comparable to values for cognate aa-tRNAs determined using Thermus thermophilus EF-Tu (LaRiviere et al. 2001; Dale et al. 2004). The k off values of both Tyr-tRNAPhe and Phe-tRNAPhe were increased by ∼30-fold when the reaction temperature was raised to 37°C (Fig. 2B), consistent with previous observations (Vorstenbosch et al. 2000). It has been predicted that some misacylated tRNAs bind EF-Tu either too tightly or too weakly to allow efficient delivery to the ribosome (LaRiviere et al. 2001; Asahara and Uhlenbeck 2005). However, many noncognate aa-tRNAs bind to EF-Tu with affinities similar to their cognate counterparts (Asahara and Uhlenbeck 2005). The data collected here show that E. coli EF-Tu does not discriminate Tyr-tRNAPhe from Phe-tRNAPhe. A cocrystal structure of EF-Tu complexed with Phe-tRNAPhe and a GTP analog revealed that the Phe side chain is stacked by a His residue (Nissen et al. 1995). Thus, it is likely that Tyr and Phe interact with the amino acid binding site of EF-Tu in the same manner, and that the presence of the p-hydroxyl group in Tyr does not hinder EF-Tu binding.
FIGURE 2.
Determination of k off rates of Phe- and Tyr-tRNAPhe from EF-Tu. The dissociation rates are (A) 0.21 ± 0.06 min−1 (Phe-tRNAPhe), 0.29 ± 0.19 min−1 (Tyr-tRNAPhe) at 2°C and (B) 7.7 ± 0.2 min−1 (Phe-tRNAPhe) and 6.8 ± 1.2 min−1 (Tyr-tRNAPhe) at 37°C. Data sets are the average of three independent experiments.
The ribosome does not discriminate Tyr-tRNAPhe from Phe-tRNAPhe
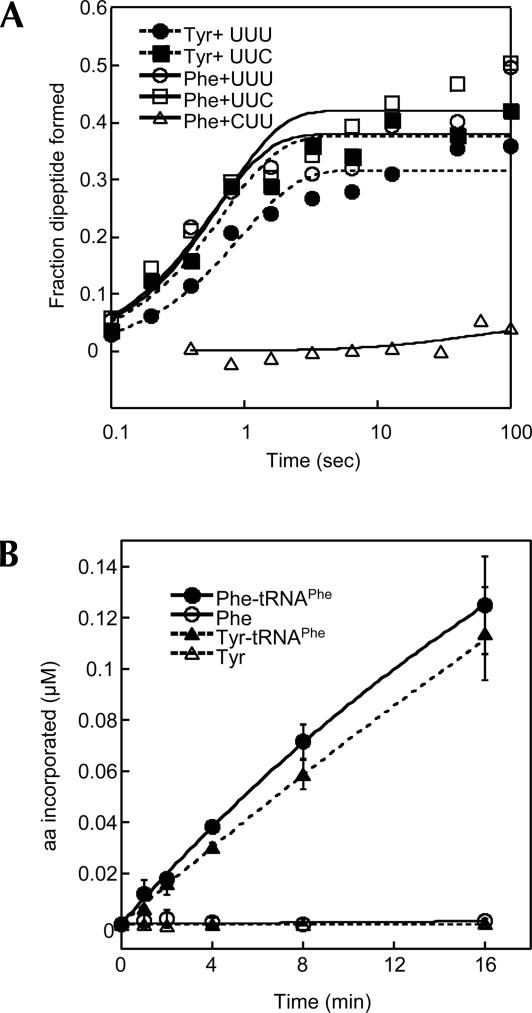
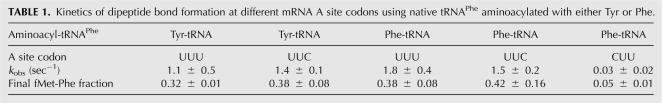
In a previously established in vivo assay using an amber suppressor tRNAPhe CUA, we observed that Tyr was misincorparated into UAG amber codons upon expression of an editing-defective PheRS (Roy et al. 2004), suggesting that the ribosome is able to use Tyr-tRNAPhe CUA as a substrate. What remained unknown, however, was how efficiently the ribosome recognizes the mischarged Tyr-tRNAPhe. To test decoding efficiency, we prepared 70S ribosomal initiation complexes with formyl-[35S]-Met-tRNAfMet in the P site and codon UUU, UUC, or CUU in the A site. Next, EF-Tu bound Tyr-tRNAPhe or Phe-tRNAPhe was added to the initiation complexes in a quenched-flow apparatus and the formation of dipeptides monitored. Fully modified native tRNAPhe was used to exclude potential impacts of modifications on decoding. The near-cognate CUU codon was decoded very poorly by Phe-tRNAPhe, while Tyr-tRNAPhe read UUU and UUC Phe codons as efficiently as Phe-tRNAPhe (Fig. 3A; Table 1), suggesting that the ribosomal A site recognizes Tyr-tRNAPhe and Phe-tRNAPhe equally well. To further test if the ribosome might discriminate Tyr-tRNAPhe at other steps in elongation, we performed poly(U)-directed polyTyr and polyPhe synthesis. The synthesis rates of polyTyr and polyPhe were almost identical (Fig. 3B), confirming that the ribosome lacks discrimination between Tyr and Phe side chains in the context of tRNAPhe. Previous studies showed that the ribosomal A site has different affinities for various aminoacyl-dinucleotide and puromycin derivatives (Bhuta et al. 1981; Starck et al. 2003), while nonenzymatic binding experiments suggested that the A site binds all tested misacylated tRNAs and cognate aa-tRNAs uniformly (Fahlman et al. 2004; Dale and Uhlenbeck 2005b). Upon codon–anticodon recognition, the ribosome undergoes a series of conformational changes (Pape et al. 1998, 1999, 2000; Ogle et al. 2002; Blanchard et al. 2004; Gromadski and Rodnina 2004a,b; Cochella and Green 2005). It is possible that, in nonenzymatic binding experiments, some rate-limiting induced-fit steps cannot be detected (Dale and Uhlenbeck 2005a). Alternatively, the aminoacyl-dinucleotide and puromycin derivatives may not bind to the ribosomal A site exactly as full-length aa-tRNAs do, as it has been suggested that the ribosomal active site adopts different conformations upon binding of puromycin derivatives or full-length aa-tRNAs (Youngman et al. 2004; Schmeing et al. 2005). The kinetic experiments demonstrate that Tyr-tRNAPhe is not discriminated by the ribosome; whether or not this conclusion is valid for other misacylated tRNAs now requires further systematic kinetic analyses.
FIGURE 3.
Decoding of Phe codons by Tyr-tRNAPhe and Phe-tRNAPhe. (A) fMet-Tyr and fMet-Phe dipeptide formation upon decoding of cognate (UUU and UUC) and near-cognate (CUU) Phe codons. Reactions contain 100 nM ribosomal initiation complex and 50 nM aa-tRNAPhe. The Y-axis shows the fraction of aa-tRNAPhe converted to dipeptide. Solid symbols, Tyr-tRNAPhe; open symbols, Phe-tRNAPhe. (B) Poly(U)-directed polyPhe and polyTyr synthesis. The reaction mixture contains 200 nM 70S ribosomes, 1 μM EF-G, 5 μM EF-Tu, 0.3 μg/μL poly(U) mRNA, and 1 μM [14C] Phe-tRNAPhe or [3H] Tyr-tRNAPhe synthesized with native E. coli tRNAPhe. As controls, 1 μM [14C] Phe or [3H] Tyr are added instead of [14C] Phe-tRNAPhe or [3H] Tyr-tRNAPhe. Data sets are the average of three independent experiments.
TABLE 1.
Kinetics of dipeptide bond formation at different mRNA A site codons using native tRNAPhe aminoacylated with either Tyr or Phe.
The editing activity of PheRS is essential for translational quality control
Our data demonstrated that, once Tyr-tRNAPhe evades editing by PheRS, incorporation of Tyr at Phe codons is inevitable. EF-Tu and the ribosome, which are downstream from aa-tRNA synthesis, do not provide further proofreading mechanisms to avoid Tyr misincorporation. This is distinct from some naturally misacylated tRNAs, such as Glu-tRNAGln and Asp-tRNAAsn, which are discriminated by EF-Tu (Stanzel et al. 1994; Becker and Kern 1998). Uhlenbeck and coworkers proposed that, since EF-Tu and possibly the ribosome display selectivity for different amino acids, their cognate tRNAs might have evolved to compensate for the binding affinities thermodynamically, so that all cognate aa-tRNAs can be translated uniformly (Dale and Uhlenbeck 2005a). Our findings support this hypothesis, as Tyr-tRNAPhe accumulation is normally prevented by the PheRS editing activity and as a result there is no selective pressure to drive further evolution of tRNAPhe. This hypothesis also explains why unnatural amino acids are usually efficiently incorporated into proteins in vivo, as the necessary orthogonal aaRSs do not exist in nature.
MATERIALS AND METHODS
Strains, plasmids, site-directed mutagenesis, and general methods
E. coli JM109/pKECA-Tu producing His6-tagged E. coli EF-Tu was a gift from B. Kraal (Leiden University, Leiden, The Netherlands). E. coli strain XL1-Blue/pQE31-FRS expressing the WT E. coli PheRS was a gift from D.A. Tirrell (California Institute of Technology). PheRS variants were previously obtained by site-directed mutagenesis in our laboratory (Roy et al. 2004). E. coli tRNAPhe and mRNA transcripts were prepared using in vitro T7 RNA polymerase runoff transcription as described (Roy et al. 2004). Native E. coli tRNAPhe, tRNAfMet, and poly(U) were purchased from Sigma-Aldrich.
Aminoacylation
Tyrosylation experiments were performed at 37°C as described (Roy et al. 2004) in the presence of 0.1 M Na-HEPES pH 7.2, 30 mM KCl, 10 mM MgCl2, 2 mM ATP, 5 μM E. coli tRNAPhe transcript, 50 μM [3H] Tyr (464 cpm/pmol), and 0.5 μM PheRS, with or without 5 μM E. coli EF-Tu. EF-Tu was activated in 50 mM Tris-HCl, 1 mM DTT, 68 mM KCl, 6.7 mM MgCl2, 2.5 mM phosphoenolpyruvate (PEP), 0.5 mM GTP, and 30 μg/mL pyruvate kinase (PK) at 37°C for 20 min before use.
Determination of k off values for EF-Tu
E. coli EF-Tu was activated in 50 mM Na-HEPES pH 7.2, 5 mM DTT, 150 mM NH4Cl, 20 mM MgCl2, 3 mM PEP, 2 mM GTP, and 30 μg/mL PK at 37°C for 20 min. A final concentration of 0.5 μM [14C] Phe-tRNAPhe or [3H] Tyr-tRNAPhe were added to 10 μM activated EF-Tu and incubated on ice for 5 min. RNase A was added to the reaction mixture to a final concentration of 100 μg/mL and aliquots were taken at each time point, spotted on 3 MM discs presoaked with 5% trichloric acid (TCA), washed, dried, and scintillation counted.
fMet-Phe and fMet-Tyr dipeptide formation
70S ribosomes from E. coli strain MRE600 were prepared as described (Fredrick and Noller 2002). Decoding experiments were performed in buffer A (50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, 3.5 mM MgCl2, 0.5 mM spermidine, 8 mM putrescine, and 2 mM DTT), which mimics in vivo conditions and is highly accurate for decoding. Initiation complexes were prepared by mixing 2 μM 70S ribosomes, 4 μM mRNA, 3 μM [35S] fMet-tRNAfMet , and 3 μM each of IF1, IF2, and IF3 in buffer B (50 mM Tris-HCl pH 7.5, 70 mM NH4Cl, 30 mM KCl, and 7 mM MgCl2). The reaction mix was incubated at 37°C for 20 min and purified through a Sephacryl S200 (Sigma-Aldrich) column. The initiation complexes were then diluted to a final concentration of 0.2 μM in buffer A. E. coli ternary complex mixture contained 0.1 μM native E. coli Phe-tRNAPhe or Tyr-tRNAPhe and 5 μM activated E. coli EF-Tu. The quench–flow experiment was done by rapidly mixing 15 μL initiation complex and 15 μL ternary complex at 25°C, followed by quenching with 1 M KOH at desired time points on a KinTek quench–flow machine. Reaction products were incubated at 37°C for 30 min to hydrolyze peptidyl- and aminoacyl-tRNAs, and separated on glass-back silica TLC plates (AnalTech). The solvent used was 3:1:1 (V/V/V) butanol:acetic acid:water containing 50 mM β-mercaptoethanol. Spots were visualized by phosphorimaging and quantified using ImageQuant.
Poly(U)-directed polyPhe and polyTyr synthesis
The reaction mixture contained 5 mM Tris-HCl pH 7.5, 5 mM MgAc2, 20 mM NH4Cl, 1 mM ATP, 30 μM GTP, 30 mM β-mercaptoethanol, 1 mM DTT, 5 mM PEP, 20 μg/mL PK, 200 nM 70S ribosomes, 1 μM E. coli EF-G, 5 μM E. coli EF-Tu, 0.3 μg/μL poly(U) mRNA, and 1 μM [14C] Phe-tRNAPhe or [3H] Tyr-tRNAPhe synthesized with native E. coli tRNAPhe. Reaction was performed at 37°C and aliquots were spotted on 3 MM discs presoaked with 10% TCA, washed twice in 5% TCA at 90°C for 10 min each, dried, and scintillation counted.
ACKNOWLEDGMENTS
We thank Dr. B. Kraal (Leiden University, Leiden, The Netherlands) and Dr. D. Tirrell (California Institute of Technology, Pasadena, CA) for strains and plasmids, and Dr. K. Fredrick for helpful discussions and materials. We also thank H. Roy and C. Hausmann for critical reading of the manuscript, and D. Qin and S. Shoji for materials. This work was supported by National Science Foundation Grant MCB-0344002.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.684107.
REFERENCES
- Ahel, I., Korencic, D., Ibba, M., Söll, D. Trans-editing of mischarged tRNAs. Proc. Natl. Acad. Sci. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrogelly, A., Palioura, S., Söll, D. Natural expansion of the genetic code. Nat. Chem. Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- An, S., Musier-Forsyth, K. Cys-tRNAPro editing by Haemophilus influenzae YbaK via a novel synthetase/YbaK/tRNA ternary complex. J. Biol. Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- Asahara, H., Uhlenbeck, O.C. The tRNA specificity of Thermus thermophilus EF-Tu. Proc. Natl. Acad. Sci. 2002;99:3499–3504. doi: 10.1073/pnas.052028599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahara, H., Uhlenbeck, O.C. Predicting the binding affinities of misacylated tRNAs for Thermus thermophilus EF-Tu.GTP. Biochemistry. 2005;44:11254–11261. doi: 10.1021/bi050204y. [DOI] [PubMed] [Google Scholar]
- Bacher, J.M., De Crécy-Lagard, V., Schimmel, P.R. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc. Natl. Acad. Sci. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, H.D., Kern, D. Thermus thermophilus: A link in evolution of the tRNA-dependent amino acid amidation pathways. Proc. Natl. Acad. Sci. 1998;95:12832–12837. doi: 10.1073/pnas.95.22.12832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe, K., Ribas De Pouplana, L., Schimmel, P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe, K., Merriman, E., Ribas De Pouplana, L., Schimmel, P. A domain for editing by an archaebacterial tRNA synthetase. Proc. Natl. Acad. Sci. 2004;101:5958–5963. doi: 10.1073/pnas.0401530101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuning, P.J., Musier-Forsyth, K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc. Natl. Acad. Sci. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuta, A., Quiggle, K., Ott, T., Ringer, D., Chladek, S. Stereochemical control of ribosomal peptidyltransferase reaction. Role of amino acid side-chain orientation of acceptor substrate. Biochemistry. 1981;20:8–15. doi: 10.1021/bi00504a002. [DOI] [PubMed] [Google Scholar]
- Blanchard, S.C., Gonzalez, R.L., Kim, H.D., Chu, S., Puglisi, J.D. tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 2004;11:1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- Chapeville, F., Lipmann, F., von Ehrenstein, G., Weisblum, B., Ray W.J., Jr, Benzer, S. On the role of soluble ribonucleic acid in coding for amino acids. Proc. Natl. Acad. Sci. 1962;48:1086–1092. doi: 10.1073/pnas.48.6.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella, L., Green, R. An active role for tRNA in decoding beyond codon:anticodon pairing. Science. 2005;308:1178–1180. doi: 10.1126/science.1111408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, T., Uhlenbeck, O.C. Amino acid specificity in translation. Trends Biochem. Sci. 2005a;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Dale, T., Uhlenbeck, O.C. Binding of misacylated tRNAs to the ribosomal A site. RNA. 2005b;11:1610–1615. doi: 10.1261/rna.2130505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale, T., Sanderson, L.E., Uhlenbeck, O.C. The affinity of elongation factor Tu for an aminoacyl-tRNA is modulated by the esterified amino acid. Biochemistry. 2004;43:6159–6166. doi: 10.1021/bi036290o. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon, A., Sankaranarayanan, R., Romby, P., Caillet, J., Springer, M., Rees, B., Francklyn, C.S., Ehresmann, C., Moras, D. Transfer RNA-mediated editing in threonyl-tRNA synthetase. The class II solution to the double discrimination problem. Cell. 2000;103:877–884. doi: 10.1016/s0092-8674(00)00191-4. [DOI] [PubMed] [Google Scholar]
- Dock-Bregeon, A.C., Rees, B., Torres-Larios, A., Bey, G., Caillet, J., Moras, D. Achieving error-free translation; the mechanism of proofreading of threonyl-tRNA synthetase at atomic resolution. Mol. Cell. 2004;16:375–386. doi: 10.1016/j.molcel.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Döring, V., Mootz, H.D., Nangle, L.A., Hendrickson, T.L., De Crécy-Lagard, V., Schimmel, P., Marlière, P. Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science. 2001;292:501–504. doi: 10.1126/science.1057718. [DOI] [PubMed] [Google Scholar]
- Fahlman, R.P., Dale, T., Uhlenbeck, O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- Fredrick, K., Noller, H.F. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Fukai, S., Nureki, O., Sekine, S., Shimada, A., Tao, J., Vassylyev, D.G., Yokoyama, S. Structural basis for double-sieve discrimination of L-valine from L-isoleucine and L-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell. 2000;103:793–803. doi: 10.1016/s0092-8674(00)00182-3. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Rodnina, M.V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004a;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Rodnina, M.V. Streptomycin interferes with conformational coupling between codon recognition and GTPase activation on the ribosome. Nat. Struct. Mol. Biol. 2004b;11:316–322. doi: 10.1038/nsmb742. [DOI] [PubMed] [Google Scholar]
- Ibba, M., Söll, D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- Korencic, D., Ahel, I., Schelert, J., Sacher, M., Ruan, B., Stathopoulos, C., Blum, P., Ibba, M., Söll, D. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc. Natl. Acad. Sci. 2004;101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere, F.J., Wolfson, A.D., Uhlenbeck, O.C. Uniform binding of aminoacyl-tRNAs to elongation factor Tu by thermodynamic compensation. Science. 2001;294:165–168. doi: 10.1126/science.1064242. [DOI] [PubMed] [Google Scholar]
- Lee, J.W., Beebe, K., Nangle, L.A., Jang, J., Longo-Guess, C.M., Cook, S.A., Davisson, M.T., Sundberg, J.P., Schimmel, P., Ackerman, S.L. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Lin, S.X., Baltzinger, M., Remy, P. Fast kinetic study of yeast phenylalanyl-tRNA synthetase: Role of tRNAPhe in the discrimination between tyrosine and phenylalanine. Biochemistry. 1984;23:4109–4116. doi: 10.1021/bi00313a015. [DOI] [PubMed] [Google Scholar]
- Lincecum, T.L., Tukalo, M., Yaremchuk, A., Mursinna, R.S., Williams, A.M., Sproat, B.S., Van Den Eynde, W., Link, A., Van Calenbergh, S., Grøtli, M., et al. Structural and mechanistic basis of pre- and post-transfer editing by leucyl-tRNA synthetase. Mol. Cell. 2003;11:951–963. doi: 10.1016/s1097-2765(03)00098-4. [DOI] [PubMed] [Google Scholar]
- Ling, J., Roy, H., Ibba, M. Mechanism of tRNA-dependent editing in translational quality control. Proc. Natl. Acad. Sci. 2007;104:72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle, L.A., De Crécy-Lagard, V., Döring, V., Schimmel, P. Genetic code ambiguity. Cell viability related to the severity of editing defects in mutant tRNA synthetases. J. Biol. Chem. 2002;277:45729–45733. doi: 10.1074/jbc.M208093200. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Kjeldgaard, M., Thirup, S., Polekhina, G., Reshetnikova, L., Clark, B.F., Nyborg, J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- Nureki, O., Vassylyev, D.G., Tateno, M., Shimada, A., Nakama, T., Fukai, S., Konno, M., Hendrickson, T.L., Schimmel, P., Yokoyama, S. Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science. 1998;280:578–582. doi: 10.1126/science.280.5363.578. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–732. doi: 10.1016/s0092-8674(02)01086-3. [DOI] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodnina, M.V. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome. EMBO J. 1998;17:7490–7497. doi: 10.1093/emboj/17.24.7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodnina, M. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J. 1999;18:3800–3807. doi: 10.1093/emboj/18.13.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape, T., Wintermeyer, W., Rodnina, M.V. Conformational switch in the decoding region of 16S rRNA during aminoacyl-tRNA selection on the ribosome. Nat. Struct. Biol. 2000;7:104–107. doi: 10.1038/72364. [DOI] [PubMed] [Google Scholar]
- Roy, H., Ling, J., Irnov, M., Ibba, M. Post-transfer editing in vitro and in vivo by the β subunit of phenylalanyl-tRNA synthetase. EMBO J. 2004;23:4639–4648. doi: 10.1038/sj.emboj.7600474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, H., Ling, J., Alfonzo, J., Ibba, M. Loss of editing activity during the evolution of mitochondrial phenylalanyl-tRNA synthetase. J. Biol. Chem. 2005;280:38186–38192. doi: 10.1074/jbc.M508281200. [DOI] [PubMed] [Google Scholar]
- Roy, H., Becker, H.D., Mazauric, M.-H., Kern, D. Structural elements defining elongation factor Tu mediated suppression of codon ambiguity. Nucleic Acids Res. 2007;35:3420–3430. doi: 10.1093/nar/gkm211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing, T.M., Huang, K.S., Strobel, S.A., Steitz, T.A. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Silvian, L.F., Wang, J., Steitz, T.A. Insights into editing from an Ile-tRNA synthetase structure with tRNAIle and mupirocin. Science. 1999;285:1074–1077. [PubMed] [Google Scholar]
- Stanzel, M., Schon, A., Sprinzl, M. Discrimination against misacylated tRNA by chloroplast elongation factor Tu. Eur. J. Biochem. 1994;219:435–439. doi: 10.1111/j.1432-1033.1994.tb19956.x. [DOI] [PubMed] [Google Scholar]
- Starck, S.R., Qi, X., Olsen, B.N., Roberts, R.W. The puromycin route to assess stereo- and regiochemical constraints on peptide bond formation in eukaryotic ribosomes. J. Am. Chem. Soc. 2003;125:8090–8091. doi: 10.1021/ja034817e. [DOI] [PubMed] [Google Scholar]
- Vorstenbosch, E.L., Potapov, A.P., de Graaf, J.M., Kraal, B. The effect of mutations in EF-Tu on its affinity for tRNA as measured by two novel and independent methods of general applicability. J. Biochem. Biophys. Methods. 2000;42:1–14. doi: 10.1016/s0165-022x(99)00032-9. [DOI] [PubMed] [Google Scholar]
- Wang, L., Xie, J., Schultz, P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- Wolfson, A.D., Uhlenbeck, O.C. Modulation of tRNAAla identity by inorganic pyrophosphatase. Proc. Natl. Acad. Sci. 2002;99:5965–5970. doi: 10.1073/pnas.092152799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, F.C., Beuning, P.J., Silvers, C., Musier-Forsyth, K. An isolated class II aminoacyl-tRNA synthetase insertion domain is functional in amino acid editing. J. Biol. Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- Xie, J., Schultz, P.G. A chemical toolkit for proteins—An expanded genetic code. Nat. Rev. Mol. Cell Biol. 2006;7:775–782. doi: 10.1038/nrm2005. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Brunelle, J.L., Kochaniak, A.B., Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]