Abstract
Nuclear receptor 2E1 (NR2E1) is expressed in human fetal and adult brains; however, its role in human brain–behavior development is unknown. Previously, we have corrected the cortical hypoplasia and behavioral abnormalities in Nr2e1−/− mice using a genomic clone spanning human NR2E1, which bolsters the hypothesis that NR2E1 may similarly play a role in human cortical and behavioral development. To test the hypothesis that humans with abnormal brain–behavior development may have null or hypomorphic NR2E1 mutations, we undertook the first candidate mutation screen of NR2E1 by sequencing its entire coding region, untranslated, splice site, proximal promoter and evolutionarily conserved non-coding regions in 56 unrelated patients with cortical disorders, namely microcephaly. We then genotyped the candidate mutations in 325 unrelated control subjects and 15 relatives. We did not detect any coding region changes in NR2E1; however, we identified seven novel candidate regulatory mutations that were absent from control subjects. We used in silico tools to predict the effects of these candidate mutations on neural transcription factor binding sites (TFBS). Four candidate mutations were predicted to alter TFBS. To facilitate the present and future studies of NR2E1, we also elucidated its molecular evolution, genetic diversity, haplotype structure and linkage disequilibrium by sequencing an additional 94 unaffected humans representing Africa, the Americas, Asia, Europe, the Middle East and Oceania, as well as great apes and monkeys. We detected strong purifying selection, low genetic diversity, 21 novel polymorphisms and five common haplotypes at NR2E1. We conclude that protein-coding changes in NR2E1 do not contribute to cortical and behavioral abnormalities in the patients examined here, but that regulatory mutations may play a role.
Keywords: Cortex, ‘fierce’ mice, mental retardation, microcephaly, nuclear receptor, Tlx
Genes with expression patterns and developmental functions consistent with a role in regulating neurogenesis and cortical size are suitable for studying the genetic basis of human brain development and evolution (Gilbert et al. 2005; Kornack & Rakic 1998; Rakic 1995). To date, only a limited number of genes have been identified that are expressed at sites of cortical neurogenesis that are known to regulate neural stem cells, forebrain size and behavior. One such gene is the nuclear receptor 2E1 (Nr2e1; previously Mtll, Tailless, Tll and Tlx), for which a clear role in mouse brain–behavior development makes it an excellent candidate for genetic studies of human abnormal brain–behavior development and evolution.
NR2E1 is expressed in human fetal brain (Strausberg et al. 2002) and in mouse embryonic forebrain (Monaghan et al. 1995) and is also detected in the adult forebrains of humans and mice (Jackson et al. 1998; Shi et al. 2004). Nr2e1 is required for normal temporal regulation of cortical neurogenesis during embryonic development and regulates proliferation and differentiation of neural progenitor cells in the embryonic and adult mouse cortex (Roy et al. 2002, Roy, 2004; Shi et al. 2004). Mice deleted for both copies of Nr2e1 (Nr2e1−/−) show cortical hypoplasia, limbic system abnormalities, cognitive impairment, short stature, vision problems and abnormal social behaviors that include pathological violence (Christie et al. 2006; Kumar et al. 2004b; Land & Monaghan 2003; Miyawaki et al. 2004; Roy et al. 2002; Young et al. 2002).
Multiple additional lines of evidence support a role for NR2E1 in human brain development. First, we have recently corrected the cortical and behavioral abnormalities of Nr2e1−/− mice using a genomic clone spanning the human NR2E1 locus (Abrahams et al. 2005), providing robust evidence that human and mouse NR2E1 are functionally equivalent in mice. Second, members of the nuclear receptor superfamily have been implicated in disorders of human brain and behavior, including NR4A2 (Buervenich et al. 2000; Chen et al. 2001; Hering et al. 2004; Iwayama-Shigeno et al. 2003; Le et al. 2003; Smith et al. 2005) and the estrogen receptor (Westberg et al. 2003). Importantly, mutations in human and mouse NR2E3, a gene closely related to NR2E1, produce similar eye developmental abnormalities (Akhmedov et al. 2000; Haider et al. 2000), suggesting that human and mouse NR2E1 mutations might also cause the same phenotype. Third, some individuals with cortical abnormalities have de novo interstitial deletions encompassing the NR2E1 locus at 6q21. Chery et al. (1989) report a de novo interstitial deletion of 6q21 in a male with moderate microcephaly, facial dysmorphism and psychomotor retardation (Chery et al. 1989). In addition, patient 2 reported by Hopkin et al. (1997) has an interstitial deletion that includes 6q21 and presents with severe intrauterine growth retardation and severe congenital microcephaly (Hopkin et al. 1997).
NR2E1 hypomorphic mutations could underlie human cortical malformations. Mice deleted for a single copy of Nr2e1 (Nr2e1+/−) show premature neurogenesis during early corticogenesis that results in reduced neuron numbers that are intermediate to that produced in Nr2e1+/+ and Nr2e1−/− mice (Roy et al. 2004), providing strong support for dosage sensitivity for Nr2e1 during cortical development. Support for a hypomorphic mechanism is also provided by studies in mice that are double heterozygotes for Nr2e1 and Pax6, which result in altered regionalization of the cerebral cortex (Stenman et al. 2003). Mice heterozygous for either Nr2e1 or Pax6 alone do not show alterations in cortical gene expression at the pallial–subpallial boundary, indicating that normal cortical regionalization at this boundary involve a genetic interaction between Pax6 and Nr2e1 (Stenman et al. 2003). Importantly, human cortical malformations are known to result from PAX6 haploinsufficiency (Sisodiya et al. 2001). In addition, Glaser et al. (1994) describe a newborn boy with homozygous mutations of PAX6 that results in severe congenital microcephaly and polymicrogyria (Glaser et al. 1994). Taken together, mouse and human genetic studies support the proposal that some human cortical disorders may involve a single- or a multigene mechanism involving NR2E1 null or hypomorphic mutations.
In this study, we report the first genetic analyses of NR2E1 in patients. To test the hypothesis that humans with abnormal cortical development and mental retardation may have null or hypomorphic mutations in NR2E1, we searched for candidate mutations by sequencing the complete coding region, 5′- and 3′-untranslated (UTR), splice site, proximal promoter and evolutionarily conserved non-coding regions in 56 unrelated patients with unexplained congenital microcephaly, a neurodevelopmental disorder characterized by marked reduction in cortical size that may result from failure of neurogenesis (Dobyns 2002; Mochida & Walsh 2001). We genotyped candidate mutations in ethnically matched control subjects that included 137 Africans and 188 Europeans. To guide the present and future studies of NR2E1, we also elucidated its molecular evolution, genetic diversity, haplotype structure and linkage disequilibrium by sequencing an additional 94 unaffected humans representing Africa, the Americas, Asia, Europe, the Middle East and Oceania, as well as chimpanzee, gorilla, orangutan and macaque.
Materials and methods
Human and non-human primate samples
Approval for this study was obtained from The University of British Columbia (Certificate of Approval # C99-0524), Child & Family Research Institute of British Columbia (Certificate of Approval # W00-0005) and the Department of Medical Genetics (Certificate of Approval #6-3-20). The research followed the Canada’s Tri-Council Statement on ‘Ethical Conduct for Research Involving Humans’ (sections 2.5–2.7). We studied 56 unrelated patients with congenital microcephaly (with or without simplified gyral patterns) and additional features resembling Nr2e1−/− mice, including short stature, vision problems, cognitive impairment and abnormal social behaviors. Patient demographic and clinical data are reported in Table 1. For a subset of patients, unaffected and affected family members that included 14 parents and four siblings were also studied. The following control subjects without severe cortical malformations or known behavioral problems were studied: (1) 110 individuals of African descent obtained from the Coriell Cell Repository (http://coriell.umdnj.edu/); (2) 27 individuals of African descent obtained from Dr M. R. Hayden (University of British Columbia, Vancouver, Canada); (3) 94 Caucasians obtained from the Coriell Cell Repository (http://coriell.umdnj.edu/); and (4) 94 Caucasian patients diagnosed with Gilbert syndrome. For genetic diversity and molecular evolutionary studies, we examined an additional 94 ethnically diverse unaffected humans, who included African (African-American, Mbuti, Biaka), American (Cheyenne, Mayan, Quechua, Karitiana), Asian (Indo-Pakistani, Chinese, Japanese), European (Russian, Italian, Northern European, Icelandic), Middle Eastern (Ashkenazi Jewish, Druze Arab) and Oceanic people (Pacific and Melanesian). Ethnically diverse DNA samples were obtained from the Coriell Cell Repository (http://coriell.umdnj.edu/) and do not overlap with any of the Coriell ethnically matched control subjects described above. Great ape tissues were obtained from Dr E. Eichler (University of Washington, Seattle, USA). DNAs (three chimpanzees, three gorillas, three orangutans) were isolated from either lymphoblasts or fibroblasts using the Gentra Puregene kit (Minneapolis, MN, USA). Macaque DNAs (two rhesus macaques, two Japanese macaques) were obtained from Oregon Regional Primate Research Center (Beaverton, OR, USA).
Table 1:
Demographic and clinical information on patients with cortical malformations
| Patient ID | Ethnicity | Sex | Brain abnormality | MR | Seizures | Psychosis | Stature | Vision problems | Other |
|---|---|---|---|---|---|---|---|---|---|
| CMS 3226 | b | m | mic | Yes | Yes | No | Short | u | − |
| CMS 5041 | b | m | mic | Yes | No | Yes | Normal | u | − |
| CMS 5811 | b | m | mic | Yes | Yes | Yes | Short | u | − |
| CMS 5162 | w | m | mic | Yes | Yes | Yes | Short | u | − |
| CMS 4775 | w | m | mic | Yes | Yes | No | Short | u | − |
| CMS 5207 | b | m | mic | Yes | Yes | No | Normal | Yes | − |
| CMS 5315 | b | m | mic | Yes | Yes | No | Short | u | − |
| CMS 7456 | u | m | mic | Yes | No | Yes | u | u | − |
| CMS 5538 | b | m | mic | Yes | No | Yes | Normal | u | − |
| CMS 5838 | b | m | mic | Yes | No | Yes | Normal | u | − |
| CMS 5151 | w | m | mic | Yes | No | Yes | Normal | u | − |
| 12856 | u | m | mic | Yes | No | Yes | Normal | u | − |
| 17763 | w | m | mic | Yes | Yes | Yes | Normal | u | − |
| 8348 | b | m | mic | Yes | Yes | Yes | u | u | − |
| 11362 | w | m | mic | Yes | No | Yes | Normal | u | − |
| 29494 | w | m | mic | Yes | Yes | No | Short | u | − |
| LP95-042a2 | w | m | mic msg | Severe | Yes | u | u | No | Early death |
| LP97-105 | u | f | mic msg xax | u | u | u | u | u | − |
| LP98-038a1 | w | f | mic msg | Moderate | No | No | u | No | − |
| LP98-052 | w | m | mic msg pmg | Severe | Yes | u | u | No | Early death |
| LP98-095 | w | f | mic msg | Mild | No | No | u | No | − |
| LP99-035 | w | m | mic msg | Severe | u | u | u | u | Jejunal |
| LP99-0100a1 | w-me | f | mic msg | Severe | Yes | u | u | u | − |
| LP99-156 | w | m | mic msg bch | Severe | u | u | u | u | Early death |
| LR00-025 | u | m | mic msg | u | Yes | u | u | u | − |
| LR00-144 | w | m | mic msg | Severe | Yes | u | u | u | Early death |
| LR00-182a1 | w-ash j | f | mic msg | Severe | Yes | u | Normal | No | − |
| LR00-188 | w-me | m | mic msg | u | u | u | u | u | − |
| LR00-196 | u | m | mic msg acc | Severe | u | u | u | u | Jejunal |
| LR00-204 | u | f | mic msg | Severe | u | u | u | u | Jejunal |
| LR01-068 | w | f | mic msg | u | u | u | u | u | − |
| LR01-099 | u | f | mic msg bch xax acc | Severe | Yes | u | u | Optic atrophy | − |
| LR01-148 | u | f | mic msg bch xax | Severe | Yes | u | u | No | − |
| LR01-171 | w-me | m | mic msg | Mild | No | No | Normal | No | − |
| LR01-194 | w | m | mic msg bch acc | Severe | Yes | u | u | u | − |
| LR01-224 | w | m | mic msg xax | Moderate | No | No | Normal | Yes | − |
| LR01-265 | w | f | mic msg | Severe | Yes | u | u | No | − |
| LR01-271 | w | f | mic msg acc | u | Yes | u | u | u | − |
| LR01-314 | u | m | mic msg | u | Yes | u | Normal | No | − |
| LR01-338 | w | f | mic msg | u | No | u | u | No | − |
| LR01-356 | w-me | m | mic msg bch | u | u | u | u | u | − |
| LR02-005 | w | f | mic msg xax | u | u | u | u | u | − |
| LR02-016a3 | w | u | mic msg bch | u | u | u | u | u | − |
| LR02-046 | w | f | mic msg acc | u | No | u | u | u | − |
| LR02-080 | u | m | mic msg | u | u | u | u | u | − |
| LR02-085 | w | f | mic msg | Mod-severe | Yes | u | u | Amblyopia | − |
| LR02-112 | u | f | mic msg xax | u | No | u | u | u | − |
| LR02-153 | w-me | f | mic msg bch | u | u | u | u | u | − |
| LR02-154a1 | w | f | mic msg xax | u | Yes | No | u | Sclerocornea | − |
| LR02-171 | u | m | mic msg acc | u | u | u | u | micr scl | − |
| LR02-304 | u | m | mic msg | u | No | u | u | u | − |
| LR02-421 | w | m | mic msg | dd | u | u | u | u | − |
| LR03-059 | u | f | mic msg xax | dd | Yes | No | u | u | − |
| LR03-184a1 | u | m | mic msg bch | Severe | Yes | No | u | u | − |
| LR03-277 | u | m | mic msg xax | Severe | u | u | u | u | − |
| gEMS594 | u | m | mic | Severe | u | u | Short | Micropthalmia | − |
Ethnicity: b, black; w, white; w-me, white-Middle Eastern; w-ash j, white Ashkenazi jewish; u, unknown.
Sex: f, female; m, male.
Brain abnormality: acc, agenesis of the corpus callosum; bch, brainstem-cerebellar hypoplasia; mic, microcephaly; msg, microcephaly with simplified gyral pattern; pmg, polymicrogyria, xax, enlarged extra-axial space.
MR: MR, mental retardation; note that for some patients, MR was scored as being present (i.e. ’yes‘) whereas for other patients the severity of MR was noted (i.e. mild, moderate, moderate-severe (Mod-severe), or severe); dd, developmental delay.
Vision problems: micr scl, micropthalmia and sclerocornea.
Other: jejunal, jejunal atresia;−, no other phenotypes noted.
u, unknown.
DNA amplification and sequencing
We sequenced NR2E1 using DNA amplicons generated from 20 polymerase chain reaction (PCR) assays that covered the coding region (1146 bp), complete 5′- and 3′-UTRs (1973 bp) and exon-flanking regions including consensus splice sites (1719 bp). In addition, we sequenced six evolutionarily conserved non-coding regions including proximal promoter (1528 bp) as previously described (Abrahams et al. 2002). Human genomic NR2E1 sequence AL078596 (http://www.ncbi.nlm.nih.gov/) was used as the reference sequence. Polymerase chain reactions were performed in a 96-well microtitre plate thermal cycler. Polymerase chain reactions were prepared in a total volume of 20 μl using 10 ng of genomic template and the following reagents from Invitrogen (Burlington, Ontario, Canada): 1× buffer, 1 mm MgSO4, 0.2 mm dNTPs, 0.5 mm primer [each of forward and reverse (Table 2)] and 0.0125 units Pfx polymerase. Thermal cycling was performed as follows: 30 cycles, 94°C for 2 min, annealing T (58–63°C) for 30 seconds, 68°C for 1 min. Polymerase chain reaction products were purified using magnetic beads from Agencourt Bioscience Corporation (Beverly, MA, USA) as per manufacturer’s instructions. Non-human primate sequencing reactions used 10–20 ng of DNA under similar conditions. Sequencing reactions performed in 384-well plates were as follows: BD Ready Rxn Mix V3 (0.54 μl), 5× Reaction Buffer (0.43 μl), 5 μm Primer (0.26 μl; Table 2), 0.2 μm 18 MΩ ddH20 (0.77 μl) and DNA (5–100 ng). Sequences were visually inspected and scored by at least two individuals using either Consed (Gordon et al. 1998) or Sequencher (Gene Codes, Ann Arbor, MI, USA). Every human variant that was identified only once (i.e. singletons) was confirmed by repeating the PCR and sequencing process. The CA-repeat assay (D6S1594; GenBank Accession Z52880) was prepared in a total volume of 15 μl using 10–50 ng of genomic template and the following reagents from Invitrogen: 1× buffer, 2.5 mm MgSO4, 0.25 mm dNTPs and 0.04 units Pfx polymerase. Primers (0.5 mm) were fluorescently labeled with FAM (ABI, Foster City, CA, USA). Post-PCR products were diluted 1:30 with ddH2O and 1 μl was combined with a 9.5 μl mix of formamide and Gene Scan™ 400HD ROX as per manufacturer’s instructions (ABI). Samples were denatured at 95°C for 5 min and placed on ice until loaded onto the ABI 3100 Genetic Analyzer (ABI). Polymerase chain reaction fragments were analyzed using Gene Mapper 3.0 (ABI).
Table 2:
Polymerase chain reaction primers used to amplify NR2E1 sequences
| Forward* | Reverse† | |||
|---|---|---|---|---|
| Assay | Name | Sequence | Name | Sequence |
| CE11A | oEMS1988 | 5′-TACGCCTTAAATCCGAGGTC-3′ | oEMS1989 | 5′-CGATCAAGCATGGTGTCAAG-3′ |
| CE12A | oEMS1990 | 5′-TGACACCGAGTCTGGAGAAA-3′ | oEMS2031 | 5′-GTCGCCTCCATTATCTGCAC-3′ |
| CE13A | oEMS1994 | 5′-CAGCTCTGCTTGGGGGAAG-3′ | oEMS1995 | 5′-AAAACGCTTTTCCCCCTCT-3′ |
| CE14A | oEMS1998 | 5′-TCCTTCTTGCCGTGAAATATAC-3′ | oEMS2032 | 5′-GGAAAACTAGATTGCTGGGAAAT-3′ |
| 5′-UTRa | oEMS2033 | 5′-CCAGGGACGCCCTATTCC-3′ | oEMS2034 | 5′-GAGGAAGAAGGAAGAACAGCA-3′ |
| 5′-UTRb | oEMS2035 | 5′-CCCACACTCTGCATGCCTAT-3′ | oEMS2036 | 5′-GACAGGTGGGTGTCAGTCG-3′ |
| Exon1 | oEMS2037 | 5′-TGTGTCCATATCAAGCAGCA-3′ | oEMS2038 | 5′-CTCCACGAAATGCTCCAACT-3′ |
| CE17B | oEMS2011 | 5′-GGAGAGCAGAGCGATGTCAC-3′ | oEMS2012 | 5′-TCACGAGACAAGCTGGTTGA-3′ |
| CE19B | oEMS2013 | 5′-CCTCCCACAGCACAATCTC-3′ | oEMS2016 | 5′-GTCCCAGACTCGTCTCAGGT-3′ |
| Exon2 | oEMS1966 | 5′-TTCGGTGCTAATCCCTTCAG-3′ | oEMS1967 | 5′-AGAGGAAGGGAGAGGTCAGG-3′ |
| Exon3 | oEMS1968 | 5′-GGACTGGCCCTCTTGAAGTA-3′ | oEMS1969 | 5′-TCCCAGCATCTGGAAAGAAG-3′ |
| Exon4 | oEMS1970 | 5′-CTCCCTCAGATTCCCTCTCC-3′ | oEMS2039 | 5′-AACTGGGTGCGTCCCTCT-3′ |
| Exon5 | oEMS1972 | 5′-TACCCACCAATGTCAACTGC-3′ | oEMS1973 | 5′-AACCCACAGGAAGAAGCAAG-3′ |
| Exon6 | oEMS1974 | 5′-TGGGAAAATAAGGGAAAGCTAGA-3′ | oEMS1975 | 5′-ATTTAAATAACAATGCAAGCAGTCA-3′ |
| Exon7 | oEMS1976 | 5′-CTTTCATACAATATAGCCGGTTTACA-3′ | oEMS1977 | 5′-AACATGCAGGTTCCCATAGC-3′ |
| Exon8 | oEMS1978 | 5′-GATTACAGACACATGCCACCAT-3′ | oEMS1979 | 5′-CACCCACCCTGAGAGATAGG-3′ |
| Exon9 | oEMS2040 | 5′-TTCAAGTGTAAGACGTTAGTTTCCA-3′ | oEMS2041 | 5′-CTGTGGCAACCCCCAGTT-3′ |
| 3′-UTRa | oEMS2042 | 5′-AAAGCATTCCAGTAGCTATGACC-3′ | oEMS2043 | 5′-GTTGCCTGGCCTATGGTATT-3′ |
| 3′-UTRb | oEMS2044 | 5′-CATTATTAAGTGGCCTTCAGAACT-3′ | oEMS2045 | 5′-CAGTTTTCGGAAAGGCATTG-3′ |
| 3′-UTRc | oEMS2046 | 5′-CCAGACAGGAAACGAATATGG-3′ | oEMS2047 | 5′-CCTTGTTTCTGGTGGGTGAG-3′ |
5′-TGTAAAACGACGGCCAGT-3′ sequence (-21M13F) was added to the 5′ end of each forward primer to facilitate sequencing.
5′-CAGGAAACAGCTATGAC-3′ sequence (M13R) was added to the 5′ end of each reverse primer to facilitate sequencing.
Transcription factor binding site (TFBS) analyses
To predict whether genetic variants at NR2E1 (i.e. candidate mutations, polymorphisms and human-specific nucleotides) alter experimentally validated consensus-binding sequences for neural transcription factors, we performed TFBS analyses using MatInspector (Quandt et al. 1995). We analyzed the minor and major alleles at each variant site together with 50 bp of surrounding sequence using the Optimized Matrix Similarity thresholds. We focused specifically on transcription factors with brain-relevant roles that include cortical patterning, neural cell proliferation and differentiation, neuronal apoptosis, neuronal survival and synaptic plasticity.
Evolutionary, nucleotide diversity and genetic differentiation analyses
The following standard measures of genetic diversity were calculated using DnaSP version 3.0 (Rozas & Rozas 1999): S (the number of segregating sites); and θW and π (nucleotide diversity). The following statistical tests of selection were performed using DnaSP version 3.0 (Rozas & Rozas 1999): Tajima’s D-test (which compares the number of nucleotide polymorphisms (θW) with the mean pairwise difference between sequences (π); Fu and Li’s D* (which compares the number of derived nucleotide variants observed only once in a sample with the total number of derived nucleotide variants); Fu and Li’s F* (which compares the number of derived nucleotide variants observed only once in a sample with the mean pairwise differences between sequences) and Fay and Wu’s H (which compares the number of derived nucleotide variants observed only once in a sample with the mean pairwise differences between sequences). Non-human primate outgroups were used to infer the ancestral and derived states of human variants. The P values for Tajima’s D and Fay and Wu’s H were estimated from 10 000 coalescent simulations of an infinite site locus that conditioned on the sample size. Human and non-human primate sequence data were aligned using MEGA version 3.0 (Kumar et al. 2004c) and human-specific variants were identified visually and confirmed by at least two individuals.
Haplotype and linkage disequilibrium reconstruction
We reconstructed haplotypes and estimated their frequencies by implementing PHASE (V. 2.0). We calculated haplotype diversity for each population as 2n(1−Σxi2)/(2n−1), where xi is the frequency of haplotype i and n is the sample number. Pairwise linkage disequilibrium (LD) between each common SNP was computed as |D’| and r2 using DnaSP version 3.0 (Rozas & Rozas 1999). We did not analyze the indels because gaps are excluded from the LD analyses (Rozas & Rozas 1999). Significance of LD was tested using Fisher’s exact test after Bonferroni adjustment for multiple tests.
Results
Candidate NR2E1 mutations identified in patients with cortical abnormalities
In total, we generated approximately 368 220 bp of NR2E1 sequence data. We did not detect any synonymous or non-synonymous coding variants. Nine out of the 56 patients (16%) were homozygous across all sites sequenced, which spanned 25.5 kb. We identified 11 patients harboring 15 novel non-coding variants (i.e. variants that have not been previously reported (http://www.ncbi.nlm.nih.gov/projects/SNP/; Build 124) (Table 3). Each of these variants (herein referred to as ‘patient variants’) was present in the heterozygote state. Thirty-three percent of the patient variants resided within the proximal promoter, 33% within a UTR and 33% within intronic sequence. Transitions and transversions accounted for 47% and 53% of all variants, respectively.
Table 3:
Characterization of 15 NR2E1 patient variants in families and control subjects
| Genotype§ | |||||||
|---|---|---|---|---|---|---|---|
| Frequency of patient variant in control chromosomes¶ | |||||||
| Patient ID* | Location† | Nucleotide variant‡ | Patient | Unaffected father | Unaffected mother | Sibling | |
| LR00-144 | CE11A | g.-2945A>G | A/G | A/A | A/G | n/a | 0/330 (0%) |
| LR00-144 | PPR | g.-1767G>T | G/T | G/G | G/T | n/a | 0/518 (0%) |
| LR00-144 | 3′-UTR | g.21502TG>C | T/C | T/C | T/T | n/a | 0/344 (%) |
| LR03-184a1 | PPR | g.-1431C>A | C/A | C/C | C/A | C/C | 6/528 (1.1%) |
| LR03-184a1 | Intron 1 | g.151T>A | T/A | T/T | T/A | T/T | 6/350 (1.7%) |
| LR00-204 | PPR | g.-1453C>G | C/G | C/C | C/G | n/a | 1/528 (0.2%) |
| LR00-204 | Intron 5 | g.11559C>T | C/T | C/T | C/C | n/a | 2/550 (0.4%) |
| LR03-277 | 3′-UTR | g.21762C>A | C/A | C/A | C/C | n/a | 1/352 (0.3%) |
| LR03-277 | 3′-UTR | g.21796G>A | G/A | G/G | G/A | n/a | 1/352 (0.3%) |
| LR02-304 | CE12A | g.-1726C>A | C/A | C/A | C/C | n/a | 0/528 (0%) |
| LP98-052 | PPR | g.-1453C>G | C/G | C/G | C/C | n/a | 1/528 (0.2%) |
| CMS5151 | 5′-UTR | g.-555C>T | C/T | n/a | n/a | n/a | 2/540 (0.4%) |
| 8348 | Intron 3 | g.8213T>C | T/C | n/a | n/a | n/a | 0/146 (0%) |
| 12856 XS | Intron 7 | g.14617A>C | A/C | n/a | n/a | n/a | 1/558 (0.2%) |
| LR01-194 | Intron 7 | g.14718C>T | C/T | C/C | C/T | n/a | 1/558(0%) |
| LR01-148 | 3′-UTR | g.20765C>A | C/A | n/a | n/a | n/a | 0/362 (0%) |
Note that patients LP98-052 and LR00-204 both harboured identical variants (i.e. g.-1453C>G). Thus, a total of 15 novel variants were identified.
PPR, proximal promoter region (defined as a 2.0-kb region upstream of the initiator Met codon); CE, evolutionary conserved element within PPR (as described in Abrahams et al. 2002); UTR, untranslated region.
g, genomic; numbering based on Antonarakis and the Nomenclature Working Group [1998], where A of the initiator Met codon in exon 1 is denoted nucleotide +1. Human genomic NR2E1 sequence: NCBI AL078596.
Sibling of LR03-184a1 is affected with microcephaly with simplified gyral pattern.
numbers represent the total number of successfully sequenced chromosomes and not the total number of chromosomes screened.
n/a, not available.
Four patients harbored multiple patient variants, including patients LR00-44, LR03-184a1, LR00-204 and LR03-277, in whom we identified three, two, two and two patient variants, respectively. Patients LR00-204 and LP98-052 both harbored the g.-1453C>G substitution.
We amplified and sequenced the regions corresponding to the 15 novel patient variants in 15 additional family members (Table 3). Fourteen parents were available for typing for 12 of the 15 novel patient variants, including both parents for the two unrelated patients having the identical g.-1453C>G patient variant. In all 12 cases, at least one unaffected parent harbored the patient variant, indicating that none of these variants were de novo. For four patient variants, parents were unavailable for typing; therefore, we cannot exclude the possibility that these variants are de novo. An affected sibling was studied for both patient variants identified in patient LR03-184a1; neither of the two variants was identified in the sibling, suggesting that the two patient variants do not predict the cortical phenotypes in these siblings.
We amplified and sequenced the regions corresponding to the 15 novel patient variants in ethnically matched controls of African (274 chromosomes) and European (376 chromosomes) descent. If the ethnicity of the patient was unknown, the patient variants were genotyped in chromosomes of African and European descent (650 chromosomes). None of the control subjects were reported to have cortical malformations. Of the 15 novel variants identified in the patients, seven (g.-2945A>G, g.-1767G>T, g.-1726C>A, g.8213T>C, g.14718C>T, g.20765C>A and g.21502T>C) were not detected in any control subject (Table 3). These seven patient variants will now be referred to as ‘candidate mutations’. Three of the seven candidate mutations (g.-2945A>G, g.-1767G>T and g.21502T>C) were identified in patient LR00-144: two of these (g.-2945A>G and -1767G>T) were maternal and one (g.21502T>C) was paternal. Consequently, patient LR00-144 is a compound heterozygote for NR2E1 mutations. Importantly, both g.-2945A>G and g.-1767G>T reside within the proximal promoter (PPR) and g.21502T>C resides within the 3′-UTR, which makes each of these variants reasonable candidates for putatively regulatory hypomorphic mutations. The four remaining candidate mutations were identified individually in unrelated patients. Two of these reside within putative regulatory regions (g.-1726C>A in a 100-bp element in the PPR that is conserved between mouse and human; and g. 20765C>A that resides in the 3′-UTR); the remaining two candidate mutations were identified in intronic regions outside the consensus splice site.
Two additional patients were compound heterozygotes for patient variants of NR2E1. Patient LR00-204 harbored g.-1453C>G (maternal) and g.11559C>T (paternal), each present in the general population at 0.2% and 0.4%, respectively. Patient LR03-277 harbored g.21762C>A (paternal) and g.21796G>A (maternal), both present in the general population at 0.3%. We did not identify a single control subject bearing either g.-1453C>G / g.11559C>T or g.21762C>A / g.21796G>A allelic pairs. We therefore consider these variants as candidates for rare functional polymorphisms.
Predicted alterations of consensus transcription factor binding sites by NR2E1 candidate mutations
To predict the impact of the seven candidate mutations on transcription factor binding, we performed in silico analyses on experimentally-validated consensus sequences for TFBS. We restricted our analyses to transcription factors expressed in the brain. Of the seven candidate mutations, four (g.-1767G>T, g.-1726C>A, g.8213T>C, g.14718C>T) were predicted to create or abolish binding of transcription factors known to have roles in neuronal proliferation and survival, cortical patterning, neuronal differentiation and synaptic plasticity (Table 4). Of the four functional polymorphisms, one (g.-1453C>G) was predicted to create binding of two neural transcription factors (Table 4).
Table 4:
NR2E1 patient variants predicted to alter neural transcription factor consensus-binding sites
| Variant type | Nucleotide variant | Location | Transcription factor binding site | Transcription factor (s) | Role in brain | Orthologous major allele in other species* | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Human | Apes | Macaque | Mouse | Fugu | ||||||
| Candidate mutation | g.-1767G>T | PPR | Created | lA-1 | Regulator of neuronal development | G | G | G | na | na |
| Created | NRSE | Repressor of multiple neuronal genes | ||||||||
| Candidate mutation | g.-1726C>A | CE12A | Abolished | SP1 | Regulator of neuronal survival | C | C | C | C | C |
| Candidate mutation | g.8213T>C | Intron 3 | Abolished | OCT-1 | Regulator of neuronal differentiation | T | T | T | T | T |
| Created | BRN-5 | Regulator of neuronal differentiation | ||||||||
| Created | PAX-6 | Regulator of neuronal Proliferation and fate | ||||||||
| Candidate mutation | g.14718C>T | Intron 7 | Created | TBX5 | Regulator of eye morphogenesis | C | N | na | na | na |
| Candidate mutation | g.2945A>G | CE11A | No effect | n/a | n/a | A | A | A | A | na |
| Candidate mutation | g.20765C>A | 3′-UTR | No effect | n/a | n/a | C | C | C | C | na |
| Candidate mutation | g.21502T>C | 3′-UTR | No effect | n/a | n/a | T | T | T | na | na |
| Functional polymorphism | g.-1453C>G | PPR | Created | EGR3 | Regulator of synaptic plasticity | C | C | C | na | na |
| Created | NUDR | Regulator of 5-HT1A receptor in neurons | ||||||||
| Functional polymorphism | g.11559C>T | Intron 5 | No effect | n/a | n/a | C | C | C | C | na |
| Functional polymorphism | g.21762C>A | 3′-UTR | No effect | n/a | n/a | C | C | C | na | na |
| Functional polymorphism | g.21796G>A | 3′-UTR | No effect | n/a | n/a | G | − | A | na | na |
See Table 3 for definitions.
EGR3, Early growth response gene 3 product.
n/a, not applicable.
apes include chimpanzee, gorilla, and orangutan. na, ortholgous region does not align with human sequence (in the case of ‘Macaque’, this region was sequenced but does not align; N, refers to nucleotide variability among apes; −, sequence data not available.
To determine whether functional constraint may exist at the sites corresponding to the seven candidate mutations, we determined the orthologous major allele at each of the sites in chimpanzee, gorilla, orangutan, macaque, mouse and Fugu. Notably, in two instances (g.-1726C>A and g.8213T>C), the major human nucleotide was conserved to Fugu (Table 4). The absence of nucleotide variability at these two non-coding sites between human and Fugu, which are separated by 900 million years (Kumar & Hedges 1998), suggests strong functional constraint and supports the proposal that these sites may represent putative regulatory regions.
To determine whether the NR2E1 candidate mutations may reside within cis-acting UTR motifs that are known to be critical for many aspects of gene expression and regulation (Mignone et al. 2002), we searched for the presence of experimentally validated functional motifs in the 5′- and 3′-UTR of NR2E1 using UTRscan (Mignone et al. 2005). We identified three motifs in the 5′-UTR (15-LOX-DICE, IRES, Brd-Box) and two in the 3′-UTR (IRES, Brd-Box); however, none of these motifs included a candidate mutation. To determine whether any of the candidate mutations may alter 3′-UTR binding for microRNAs (miRNA), which are known to regulate genes (Bartel 2004), we aligned the 3′-UTR of NR2E1 against known miRNA motifs (Xie et al. 2005). We detected two motifs; however, neither included a candidate mutation.
Strong purifying selection and low nucleotide diversity at NR2E1 in ethnically diverse humans
Genetic diversity and molecular evolutionary studies of neural genes in humans and non-human primates represent powerful tools for understanding cortical development (Enard & Paabo 2004; Gilbert et al. 2005). We therefore sought to gain additional insight into the extent and patterns of genetic variation at NR2E1 by systematically resequencing the same coding and non-coding regions as described above in 94 unaffected, ethnically diverse humans representing Africa, the Americas, Asia, Europe, the Middle East and Oceania; none of these humans was studied as part of the data set used as controls in our previous analyses. In addition, we studied NR2E1 in chimpanzee, gorilla, orangutan, Japanese macaque and rhesus macaque. The human sample size chosen was sufficient to detect alleles with minor allele frequencies of 10% or greater with 90% power.
We did not detect a single non-synonymous or synonymous change in the coding region of any human sample. We also did not detect a single non-synonymous change in any non-human primate sample (2–3 individuals from five species). The complete lack of synonymous variation among humans and the complete absence of non-synonymous variation between humans and non-human primates suggests that NR2E1 has experienced strong functional constraint (i.e. purifying selection).
In this ethnically diverse sample, we observed a total of 25 non-coding variants (Fig. 1a; variants 1–25). Twenty-three of the 95 subjects (24%) were homozygous across all sequenced sites. Twenty of the 25 variants were novel (http://www.ncbi.nlm.nih.gov/projects/SNP/; dbSNP Build 124) (Fig. 1b). None of the seven candidate mutations identified in patients were detected in this unaffected ethnically diverse human panel. Consequently, if we include control subjects of mixed ethnic origin into our analyses of candidate mutations, the total number of unaffected chromosomes not harboring candidate mutations is as follows: g.-2945A>G (518 chromosomes), g.-1767G>T (706 chromosomes), g.-1726C>A (716 chromosomes), g.8213T>C (334 chromosomes), g.14718C>T (746 chromosomes), g.20765C>A (550 chromosomes) and g.21502T>C (532 chromosomes).
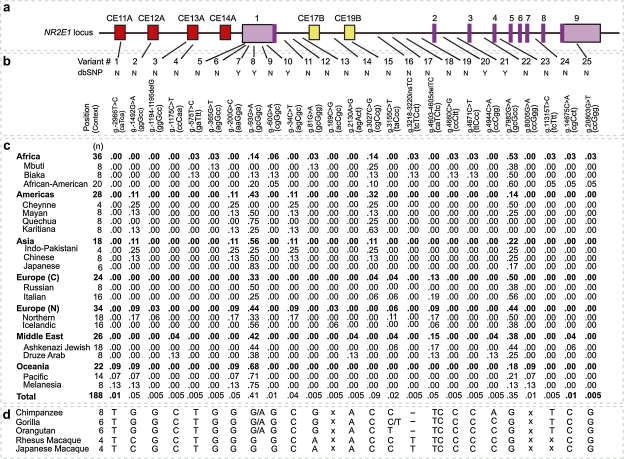
Figure 1:
A few common and many rare NR2E1 variants detected in human populations representative for global diversity. (a) Functional and putatively functional regions of NR2E1 were resequenced, including coding (dark purple boxes), 5′- and 3′-untranslated (light purple boxes), and human non-coding regions that are conserved (Abrahams et al. 2002) in mouse (CE-A; red boxes) and mouse and Fugu (CE-B; yellow boxes). (b) A total of 26 variants was identified (variants 1–25 and CA-repeat, see Fig. 2). The nucleotide in the first position represents the human major (i.e. consensus) allele. Numbering based on Antonarakis and the Nomenclature Working Group (Antonarakis 1998), where A of the initiator Met codon in exon 1 is denoted nucleotide +1. Human genomic NR2E1 sequence: AL078596 (http://www.ncbi.nlm.nih.gov/). Variants catalogued in dbSNP (‘Y’) are distinguished from those that are newly discovered here (‘N’). The DNA context of each variant is shown. (c) The number of chromosomes surveyed (n) and minor allele frequencies for each variable site are indicated for 18 world populations. (d) The corresponding chimpanzee, gorilla, orangutan, rhesus macaque, and Japanese macaque alleles are indicated. ‘x’ indicates that no sequence was obtained due to failed PCR or sequencing reaction. ‘−’ indicates that no corresponding nucleotide was present at that position in the non-human primate.
We determined the frequencies of all 25 variants in each ethnic group. Only six of the 25 variants (numbers 2, 7, 8, 14, 17 and 21) were common (i.e. minor allele frequency (MAF) ≥5%) (Fig. 1c). For each human variant, we also inferred the ancestral and derived states by comparing it to the orthologous non-human primate sequence (Fig. 1d). Interestingly, chimpanzee, gorilla and orangutan were all polymorphic for the same G > A transition (variant 8) observed in humans. This is the first report of a human polymorphic site that is also polymorphic for the same alleles across these three great apes.
We estimated the levels of human nucleotide diversity by computing θW, which is based on the proportion of segregating sites (S) in a population and π, which is based on the average number of nucleotide differences per site between two sequences randomly drawn from the population (Hartl 1997) (Table 5). The total human estimates for θW (5.7 × 10−4± 0.17 × 10−4) and π (2.6 × 10−4± 0. 20 × 10−4) for NR2E1 fall at the lower 20% and 30% of previous studies, respectively (Przeworski et al. 2000).
Table 5:
Human nucleotide diversity and Tajima’s D at NR2E1
| Population | n | S | θW (±SD) | π (±SD) | ηS | Tajima’s D |
|---|---|---|---|---|---|---|
| Africa | 36 | 12 | 0.00045 | 0.00024 | 8 | −1.45 |
| (0.00018) | (0.00004) | |||||
| Americas | 28 | 6 | 0.00024 | 0.00029 | 0 | 0.61 |
| (0.00012) | (0.00005) | |||||
| Asia | 18 | 6 | 0.00027 | 0.00027 | 0 | −0.36 |
| (0.00014) | (0.00006) | |||||
| Europe (C) | 24 | 3 | 0.00013 | 0.00017 | 1 | 0.83 |
| (0.00008) | (0.00002) | |||||
| Europe (N) | 34 | 7 | 0.00027 | 0.00027 | 1 | −0.04 |
| (0.00013) | (0.00004) | |||||
| Middle-East | 26 | 7 | 0.00029 | 0.00022 | 5 | −0.75 |
| (0.00014) | (0.00003) | |||||
| Oceania | 22 | 5 | 0.00022 | 0.00020 | 0 | −0.18 |
| (0.00004) | (0.00004) | |||||
| Total human | 188 | 21 | 0.00057 | 0.00026 | 11 | −1.50 |
| (0.00017) | (0.00002) | |||||
n, number of alleles; ηS, number of singleton mutations; S, number of segregating sites.
Evidence of non-neutral evolution at NR2E1
Genes that have been implicated in severe cortical malformations, including ASPM (Evans et al. 2004b) and Microcephalin(Evans et al. 2004a), show robust molecular signatures of positive Darwinian selection; consequently, the identification of signatures of selection in candidate neural genes such as NR2E1 may strengthen their proposed role in human cortical disorders. To elucidate the human molecular evolution of NR2E1, we first used the nucleotide diversity measures θW and π to calculate Tajima’s D (Tajima 1989) (Table 5). Positive and negative values of this test correspond to departures from the neutral expectations of molecular evolution. We obtained a negative value for Tajima’s D in the ethnically diverse population, which is consistent with another report that obtained a negative Tajima’s D at NR2E1 (Stephens et al. 2001).
To further evaluate the role of natural selection at NR2E1, we used the ancestral and derived states of each variant from the ethnically diverse population to perform three additional tests of molecular neutrality: Fu and Li’s D* (Fu & Li 1993), Fu and Li’s F* (Fu & Li 1993) and Fay and Wu’s H (Fay & Wu 2000). We obtained statistically significant negative values for Fu and Li’s D* and F* (Table 6), which may indicate genetic hitchhiking or background selection. However, based on these tests alone, we cannot exclude the possibility that demographic factors such as population bottlenecks may also explain deviations from neutrality observed at NR2E1 (Fu 1997; Kreitman 2000).
Table 6:
Neutrality tests using chimpanzee as outgroup
| Population | Fu and Li’s D* | Fu and Li’s F* | Fay and Wu’s H |
|---|---|---|---|
| Africa | −2.77† | −2.79‡ | 0.89 |
| Americas | 1.27 | 1.26 | 1.14 |
| Asia | 1.33 | 1.11 | 0.86 |
| Europe (C) | −0.24 | 0.07 | 0.31 |
| Europe (N) | 1.24 | 1.13 | 0.77 |
| Middle East | −1.97 | −1.81 | 0.69 |
| Oceania | 1.22 | 0.96 | 0.19 |
| Total human | −3.08† | −2.63‡ | 0.98 |
P < 0.02.
P < 0.05.
Human-specific NR2E1 sites identified
Insight into the evolution of human-specific traits, such as enlarged cerebral cortex, may be gained by the identification of human-specific sites (i.e. nucleotides that are fixed among all humans but absent from non-human species) (Enard & Paabo 2004). To identify such sites, we aligned 6137 bp of human and non-human primate coding and non-coding sequences. We identified 26 human-specific (divergent) sites (data not shown). Of these 26, five resided within functional (i.e. exons) or putatively functional (i.e. evolutionarily conserved non-coding) regions of NR2E1: one synonymous coding variant and four putative regulatory variants (Table 7). We extended our analysis to mouse and determined that four divergent sites still remained (the 3′-UTRs between human and mouse NR2E1 could not be aligned) (Table 7). To determine whether these four variants may disrupt or create TFBS, we performed in silico TFBS analyses. We did not detect any alterations of TFBS for transcription factors expressed in the brain.
Table 7:
NR2E1 sites that are fixed among all humans but differ in non-human species
| Region | Location* (bp) | Humans† | Great apes‡ | Old world monkeys§ | Mouse¶ |
|---|---|---|---|---|---|
| CE11A | −2994 | G | A | A | A |
| 5′-UTR | −542 | T | C | C | C |
| 5′-UTR | −498 | A | T | T | T |
| Exon 4†† | 9843 | A | T | T | T |
| 3′-UTR | 21090 | C | T | T | na |
Numbering adopted from Antonarakis et al. (1998), where A of the initiator Met codon in exon 1 is denoted nucleotide +1. Human genomic NR2E1 sequence: NCBI AL078596.
includes all humans examined (African, Asia, Americas, Europe, Middle East, Oceania).
includes all chimpanzees, gorillas, orangutans examined.
includes all rhesus and Japanese macaques examined.
na, orthologous region does not align with human sequence.
CCA_(Pro) to CCT_(Pro).
NR2E1 haplotype and LD structure provide effective tools for disease-mapping studies
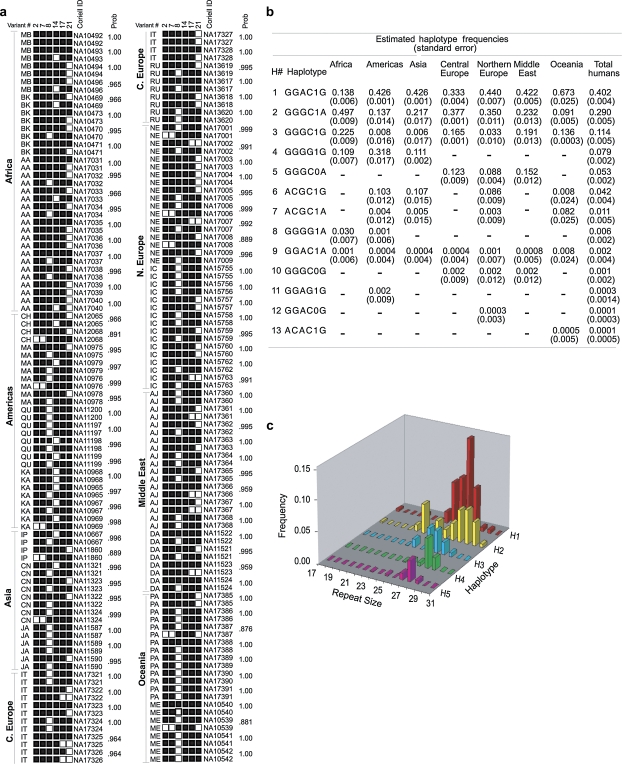
To inform future association and linkage-based studies of NR2E1 in disorders of brain and behavior, we elucidated haplotype structure and LD using a subset of the 21 novel variants identified in our analyses of ethnically diverse and unaffected humans. To characterize the haplotype structure of human NR2E1, we inferred haplotypes using bi-allelic variants whose MAFs were 5% or greater. Genotypes of all markers were in Hardy–Weinberg equilibrium (data not shown). For each individual, we inferred haplotypes (Fig. 2a) and estimated the population haplotype frequencies for all seven human populations (Fig. 2b). We also typed a 12-allele CA-repeat in the 3′-UTR (dinucleotide repeat range 17–31; data not shown). We then re-constructed haplotypes using the CA-repeat data and the five most common haplotypes (Fig. 2c). Our characterization of haplotype structure in NR2E1 identified five haplotypes and CA-repeat alleles that would be useful for disease-mapping studies.
Figure 2:
Five common SNP-based NR2E1 haplotypes account for the majority of chromosomes examined for global diversity. (a) The SNP-based haplotypes for both chromosomes of every individual are illustrated. Each row represents one chromosome. Each column represents one variable site, the number of which is indicated above each column (see Fig. 1b). Black boxes indicate the major allele; white boxes represent the minor allele. Coriell Cell Repositories ID codes are indicated. ‘SNP’ refers to single nucleotide polymorphism (i.e. single nucleotide substitutions with minor allele frequencies ≥1%). ‘Prob’ is the probability of haplotype assignment, where 1.00 = 100% probable (i.e. individual is either homozygous at all sites or a heterozyote for only one site). MB, Mbuti; BK, Biaka; AA, African-American; CH, Cheyenne; MA, Mayan; QU, Quechua; KA, Karitiana; IP, Indo-Pakistani; CN, Chinese; JA, Japanese; IT, Italian; RU, Russian; NE, Northern European; IC, Icelandic; AJ, Ashkenazi Jewish; DA, Druze Arab; PA, Pacific Islanders; ME, Melanesian. (b) Estimated population haplotype frequencies of the 13 most frequent SNP-based NR2E1 haplotypes. ‘−’ indicates that the haplotype is absent from the population. ‘1’ and ‘0’ represent present and absence of TC indel, respectively. (c) The frequency (y-axis) of the CA-repeat allele (x-axis) with the five most common NR2E1 haplotypes (z-axis) is plotted for the global diversity population.
To empirically estimate the degree of non-random association between NR2E1 variants, we calculated LD using two statistics: Lewontin’s coefficient |D’| and Pearson’s correlation r2 (Ardlie et al. 2002). We used all the variants with MAFs equal to or greater than 5% except indel variant 17 for technical reasons (see Materials and methods). Despite our markers being only a few kilobases apart, in most cases we observed weak LD in this region (data not shown); however, we note that small sample sizes may underestimate the extent of significant LD. The only substantial LD in NR2E1 was between variants 2 and 7 (|D’|= 0.894; r2= 0.880; Fisher P < 0.001, significant using the conservative Bonferroni correction). We also examined the relationship between LD and distance using both |D’| and r2, which indicated a general decrease in the level of LD with increasing distance (data not shown).
Discussion
The present study represents the first genetic report of NR2E1 in clinical samples. In addition, it provides the most comprehensive evolutionary study of NR2E1 reported to date. Our studies of NR2E1 are noteworthy in several respects. First, we used a direct resequencing approach, which is the most reliable, complete and impartial means of mutation and polymorphism discovery; however, one limitation of using this approach alone is its inability to distinguish between homozygosity across loci vs. large deletions. Second, our experiments were designed to identify candidate mutations and polymorphisms in both coding and key non-coding regions, such as evolutionarily conserved sequences that may harbor functionally important and disease-causing variants (Drake et al. 2006). Third, we studied a diverse collection of human genomic DNAs representing the world’s major continental populations as a means to thoroughly assess the natural genetic variation at this locus.
Our candidate mutation screen demonstrated that protein-coding mutations in NR2E1 do not contribute to cortical and behavioral abnormalities in the patients examined here. In addition, we detected individuals homozygous across the NR2E1 locus, but given the overall lack of variability at the locus and the fact that these cases were not enriched in the patient sample, this data does not argue for the presence of large deletions. We did identify and characterize seven candidate non-coding mutations and four candidate functional polymorphisms. Of particular interest is patient LR00-144, who is a compound heterozygote for candidate NR2E1 mutations, which is consistent with the recessive inheritance of the cortical phenotype in Nr2e1−/− mice. Strikingly, patient LR00-144 harbored three of the seven candidate mutations. The chances of observing three candidate mutations in a single patient are extremely rare [(7 candidate mutations/56 patients)3= 1.9 × 10−3]. The g.-1767G>T candidate mutation identified in this patient is predicted to create two transcription factor binding sites (TFBS). One of these is for the zinc finger protein insulinoma-associated 1 (IA-1), which is present in fetal brain tissue and functions as a transcriptional repressor during neuronal development (Breslin et al. 2002). IA-1 binding sites have been identified in the 5′-flanking regions of several genes including Pax6 and NeuroD/β2 (Breslin et al. 2002). The g.-1767G>T substitution is also predicted to create a neural-restrictive-silencer element (NRSE). NRSE motifs are known to bind the neural-restrictive silencer factor (NRSF) that functions as a transcriptional repressor of multiple neuronal genes such as NR2B, which contains five NRSE motifs in its 5′-flanking region (Qiang et al. 2005). The highest expression of NRSF is observed in the mouse embryonic cortex at E14, but is also detected in the adult mouse brain as well as in cultured cortical neurons (Qiang et al. 2005). Taken together, the creation of at least two transcriptional repressor binding sites in the proximal promoter of NR2E1 in patient LR00-144 supports the proposal that the g.-1767G>T candidate mutation could contribute to the cortical phenotype and severe mental retardation observed in this patient.
Patients LR00-204 and LR03-277 also represent compound heterozygotes for patient variants of NR2E1. Interestingly, LR00-204, who had microcephaly, was also diagnosed with optic nerve hypoplasia, a phenotype observed in Nr2e1−/− mice (Young et al. 2002; Yu et al. 2000). The specific pair of candidate functional polymorphisms observed in each patient was absent in all controls examined; thus, these particular combinations of alleles were specific to cortical disorders. Each of these variants may act through a hypomorphic mechanism that involves reduced levels of NR2E1 transcription. Such a mechanism is supported by the demonstration that Nr2e1+/− mice show altered neurogenesis early during cortical development (Roy et al. 2004), which indicates dosage sensitivity for Nr2e1.
Patients 8348 and LR01-148 harbored candidate mutations g.8213T>C and g.20765C>A, respectively. Parents of both these patients were unavailable to study; consequently, we cannot exclude the possibility that both these variants may represent de novo mutations. The candidate mutation g.8213T>C identified in intron 3 in patient 8348 is predicted to abolish a binding site for OCT1, a regulator of neuronal differentiation and is also predicted to create binding sites for BRN5, another regulator of neuronal differentiation and PAX6, a regulator of neuronal proliferation and fate. The major allele T at this site is conserved between human and Fugu, which strengthens the proposal that a nucleotide substitution at this site may be pathogenic.
Finally, the two candidate mutations g.-1726C>A and g.14718C>T identified in patients LR02-304 and LR01-194, respectively, may also underlie cortical disorders. However, as each was also present in a parent, we propose a multigenic mechanism underlying the cortical phenotypes in each case. Such a proposal receives support from mice that are double heterozygotes for mutations at Nr2e1 and Pax6, which interact genetically to alter normal forebrain development (Stenman et al. 2003). Both candidate mutations are predicted to alter neural TFBS and one of these (g.-1726C>A) resides in the promoter region, which together strengthens their candidacy for disease.
Our genetic diversity and evolutionary analyses of NR2E1 in ethnically diverse humans and non-human primates will inform future NR2E1 studies of human brain–behavior disorders. Our data indicate strong evolutionary constraint (i.e. purifying selection) in the coding region of NR2E1 that is higher in comparison to many other genes examined for genetic diversity (Cargill et al. 1999; Dorus et al. 2004; Freudenberg-Hua et al. 2003) (http://genebank.nibio.go.jp/gbank/qfbase/index.html). Studying additional non-human primates would serve to further strengthen this conclusion. The implication of strong functional constraint is that any future identification of an NR2E1 coding variant in a patient with a brain–behaviour phenotype is likely to be related to the disorder. Importantly, the striking absence of synonymous changes, which are typically considered to be selectively neutral (Enard & Paabo 2004), may suggest a functional constraint that operates at the RNA level to maintain its secondary structure or stability, as described for other genes (Capon et al. 2004; Chamary & Hurst 2005; Duan et al. 2003).
We provide evidence of adaptive evolution at NR2E1 that may act on regulatory sites, which constitute an important class of non-coding sequences that are potential targets of Darwinian selection (Wray et al. 2003). We observed an excess of rare, derived NR2E1 variants, as indicated by the significantly negative Fu and Li’s D* and F* values, which could be evidence of a ‘selective sweep’ (i.e. the rare variants have ‘hitch-hiked’ along with a variant on which positive selection has occurred) (Fay & Wu 2000). In this regard, it is conceivable that one or more of the human-specific NR2E1 sites identified in the present study may have been fixed by positive selection in a manner similar to that proposed for ASPM, which is mutated in some patients with microcephaly (Bond et al. 2003; Kumar et al. 2004; Woods et al. 2005).
The knowledge of the genetic architecture of NR2E1 generated in this study in ethnically diverse humans and non-human primates provides additional tools for future disease-mapping studies of brain–behavior disorders. Our results expand those of one other study that examined normal genetic architecture at NR2E1 (Stephens et al. 2001); however, our analyses employed over twice as much sequence data (including evolutionarily conserved regions not previously examined) from a more diverse set of humans and non-human primate species. The identification and characterization of common SNPs, microsatellites, and haplotypes in multiple ethnic groups will benefit future association analyses of brain–behavior disorders by helping to reduce or eliminate false-positive and negative associations that can arise as a result of population stratification, which is a well established confound in human disease-mapping efforts (Freedman et al. 2004; Kang et al. 1999). We also provide the first example of a human polymorphic site that is also polymorphic for the same alleles in chimpanzee, gorilla and orangutan. Therefore, we strongly recommend that multiple non-human primate species be used to robustly infer ancestral states of human polymorphisms.
In conclusion, our analysis of human NR2E1 has identified candidate regulatory mutations and rare putative functional polymorphisms. Future work will involve testing these alleles for abnormal function using whole-animal assessment as proposed by Abrahams et al. (2005). In this study, we selected patients enriched for features resembling Nr2e1−/− mice in addition to microcephaly (e.g. four patients with agenesis of the corpus callosum, one patient with optic atrophy). Future research may benefit by focusing on other region-specific malformations present in the Nr2e1−/− mice, such as preferential reduction of superficial cortical layers II and III, in an effort to enhance detection of NR2E1 mutations. Our genetic diversity and evolutionary analyses provide the foundation to facilitate future examination of the role of NR2E1 in additional human disorders of brain and behavior.
Acknowledgments
We thank Evan Eichler and Sean McGrath (University of Washington, USA) for providing us with non-human primate DNA samples. The authors are grateful to Dr Sylvie Langlois (University of British Columbia, Canada) for critical discussions about patient recruitment criteria and to Kathleen G. Banks (University of British Columbia, Canada) for assistance in typing the microsatellite repeat. We also thank Tracey D. Weir and Nichole Sturwold (Centre for Molecular Medicine and Therapeutics, Canada) and Sarah Otto (University of British Columbia, Canada) for helpful comments on the manuscript. This work was supported by grants from the Jack and Doris Brown Foundation and British Columbia Institute for Children‘s & Women’s Health (to R. A. K.); Harry Frank Guggenheim Foundation (to B. S. A.); South Carolina Department of Disabilities and Special Needs (SCDDSN) (to C. E. S.) and Canadian Institutes for Health Research (CIHR) and Canada Research Chair in Genetics and Behaviour (to E. M. S.).
References
- Abrahams BS, Mak GM, Berry ML, Palmquist DL, Saionz JR, Tay A, Tan YH, Brenner S, Simpson EM, Venkatesh B. Novel vertebrate genes and putative regulatory elements identified at kidney disease and NR2E1/fierce loci. Genomics. 2002;80:45–53. doi: 10.1006/geno.2002.6795. [DOI] [PubMed] [Google Scholar]
- Abrahams BS, Kwok MC, Trinh E, Budaghzadeh S, Hossain SM, Simpson EM. Pathological aggression in ‘fierce’ mice corrected by human nuclear receptor 2E1. J Neurosci. 2005;25:6263–6270. doi: 10.1523/JNEUROSCI.4757-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, Nusinowitz S, Heckenlively JR, Roderick TH, Kozak CA, Danciger M, Davisson MT, Farber DB. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci U S A. 2000;97:5551–5556. doi: 10.1073/pnas.97.10.5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE. Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bond J, Scott S, Hampshire DJ, Springell K, Corry P, Abramowicz MJ, Mochida GH, Hennekam RC, Maher ER, Fryns JP, Alswaid A, Jafri H, Rashid Y, Mubaidin A, Walsh CA, Roberts E, Woods CG. Protein-truncating mutations in ASPM cause variable reduction in brain size. Am J Hum Genet. 2003;73:1170–1177. doi: 10.1086/379085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin MB, Zhu M, Notkins AL, Lan MS. Neuroendocrine differentiation factor, IA-1, is a transcriptional repressor and contains a specific DNA-binding domain: identification of consensus IA-1 binding sequence. Nucleic Acids Res. 2002;30:1038–1045. doi: 10.1093/nar/30.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buervenich S, Carmine A, Arvidsson M, Xiang F, Zhang Z, Sydow O, Jonsson EG, Sedvall GC, Leonard S, Ross RG, Freedman R, Chowdari KV, Nimgaonkar VL, Perlmann T, Anvret M, Olson L. NURR1 mutations in cases of schizophrenia and manic-depressive disorder. Am J Med Genet. 2000;96:808–813. doi: 10.1002/1096-8628(20001204)96:6<808::aid-ajmg23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Capon F, Allen MH, Ameen M, Burden AD, Tillman D, Barker JN, Trembath RC. A synonymous SNP of the corneodesmosin gene leads to increased mRNA stability and demonstrates association with psoriasis across diverse ethnic groups. Hum Mol Genet. 2004;13:2361–2368. doi: 10.1093/hmg/ddh273. [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75.1–R75.12. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Tsai MT, Shaw CK, Chen CH. Mutation analysis of the human NR4A2 gene, an essential gene for midbrain dopaminergic neurogenesis, in schizophrenic patients. Am J Med Genet. 2001;105:753–757. doi: 10.1002/ajmg.10036. [DOI] [PubMed] [Google Scholar]
- Chery M, Formiga LF, Mujica P, Andre M, Stehelin D, Dozier C, Gilgenkrantz S. Interstitial deletion of the long arm of chromosome 6. Ann Genet. 1989;32:82–86. [PubMed] [Google Scholar]
- Christie BR, Li AM, Redila VA, Booth H, Wong BK, Eadie BD, Ernst C, Simpson EM. Deletion of the nuclear receptor Nr2e1 impairs synaptic plasticity and dendritic structure in the mouse dentate gyrus. Neuroscience. 2006;137:1031–1037. doi: 10.1016/j.neuroscience.2005.08.091. [DOI] [PubMed] [Google Scholar]
- Dobyns WB. Primary microcephaly: new approaches for an old disorder. Am J Med Genet. 2002;112:315–317. doi: 10.1002/ajmg.10580. [DOI] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, Hirschhorn JN. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nat Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Enard W, Paabo S. Comparative primate genomics. Annu Rev Genomics Hum Genet. 2004;5:351–378. doi: 10.1146/annurev.genom.5.061903.180040. [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum Mol Genet. 2004a;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Gilbert SL, Malcom CM, Dorus S, Lahn BT. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum Mol Genet. 2004b;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, Gabriel SB, Topol EJ, Smoller JW, Pato CN, Pato MT, Petryshen TL, Kolonel LN, Lander ES, Sklar P, Henderson B, Hirschhorn JN, Altshuler D. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Freudenberg-Hua Y, Freudenberg J, Kluck N, Cichon S, Propping P, Nothen MM. Single nucleotide variation analysis in 65 candidate genes for CNS disorders in a representative sample of the European population. Genome Res. 2003;13:2271–2276. doi: 10.1101/gr.1299703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SL, Dobyns WB, Lahn BT. Genetic links between brain development and brain evolution. Nat Rev Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Haider NB, Jacobson SG, Cideciyan AV, Swiderski R, Streb LM, Searby C, Beck G, Hockey R, Hanna DB, Gorman S, Duhl D, Carmi R, Bennett J, Weleber RG, Fishman GA, Wright AF, Stone EM, Sheffield VC. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–131. doi: 10.1038/72777. [DOI] [PubMed] [Google Scholar]
- Hartl C. Principles of Population Genetics. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- Hering R, Petrovic S, Mietz EM, Holzmann C, Berg D, Bauer P, Woitalla D, Muller T, Berger K, Kruger R, Riess O. Extended mutation analysis and association studies of Nurr1 (NR4A2) in Parkinson disease. Neurology. 2004;62:1231–1232. doi: 10.1212/01.wnl.0000118285.18383.90. [DOI] [PubMed] [Google Scholar]
- Hopkin RJ, Schorry E, Bofinger M, Milatovich A, Stern HJ, Jayne C, Saal HM. New insights into the phenotypes of 6q deletions. Am J Med Genet. 1997;70:377–386. [PubMed] [Google Scholar]
- Iwayama-Shigeno Y, Yamada K, Toyota T, Shimizu H, Hattori E, Yoshitsugu K, Fujisawa T, Yoshida Y, Kobayashi T, Toru M, et al. Distribution of haplotypes derived from three common variants of the NR4A2 gene in Japanese patients with schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2003;118:20–24. doi: 10.1002/ajmg.b.10053. [DOI] [PubMed] [Google Scholar]
- Jackson A, Panayiotidis P, Foroni L. The human homologue of the Drosophila tailless gene (TLX): characterization and mapping to a region of common deletion in human lymphoid leukemia on chromosome 6q21. Genomics. 1998;50:34–43. doi: 10.1006/geno.1998.5270. [DOI] [PubMed] [Google Scholar]
- Kang AM, Palmatier MA, Kidd KK. Global variation of a 40-bp VNTR in the 3’-untranslated region of the dopamine transporter gene (SLC6A3) Biol Psychiatry. 1999;46:151–160. doi: 10.1016/s0006-3223(99)00101-8. [DOI] [PubMed] [Google Scholar]
- Kornack DR, Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc Natl Acad Sci U S A. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreitman M. Methods to detect selection in populations with applications to the human. Annu Rev Genomics Hum Genet. 2000;1:539–559. doi: 10.1146/annurev.genom.1.1.539. [DOI] [PubMed] [Google Scholar]
- Kumar A, Blanton SH, Babu M, Markandaya M, Girimaji SC. Genetic analysis of primary microcephaly in Indian families: novel ASPM mutations. Clin Genet. 2004a;66:341–348. doi: 10.1111/j.1399-0004.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- Kumar RA, Chan KL, Wong AH, Little KQ, Rajcan-Separovic E, Abrahams BS, Simpson EM. Unexpected embryonic stem (ES) cell mutations represent a concern in gene targeting: lessons from “fierce” mice. Genesis. 2004b;38:51–57. doi: 10.1002/gene.20001. [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004c;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Land PW, Monaghan AP. Expression of the transcription factor, tailless, is required for formation of superficial cortical layers. Cereb Cortex. 2003;13:921–931. doi: 10.1093/cercor/13.9.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le WD, Xu P, Jankovic J, Jiang H, Appel SH, Smith RG, Vassilatis DK. Mutations in NR4A2 associated with familial Parkinson disease. Nat Genet. 2003;33:85–89. doi: 10.1038/ng1066. [DOI] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:1–0004. doi: 10.1186/gb-2002-3-3-reviews0004. REVIEWS0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignone F, Grillo G, Licciulli F, Iacono M, Liuni S, Kersey PJ, Duarte J, Saccone C, Pesole G. UTRdb and UTRsite: a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 2005;33:D141–D146. doi: 10.1093/nar/gki021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki T, Uemura A, Dezawa M, Yu RT, Ide C, Nishikawa S, Honda Y, Tanabe Y, Tanabe T. Tlx, an orphan nuclear receptor, regulates cell numbers and astrocyte development in the developing retina. J Neurosci. 2004;24:8124–8134. doi: 10.1523/JNEUROSCI.2235-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida GH, Walsh CA. Molecular genetics of human microcephaly. Curr Opin Neurol. 2001;14:151–156. doi: 10.1097/00019052-200104000-00003. [DOI] [PubMed] [Google Scholar]
- Monaghan AP, Grau E, Bock D, Schutz G. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development. 1995;121:839–853. doi: 10.1242/dev.121.3.839. [DOI] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A. Adjusting the focus on human variation. Trends Genet. 2000;16:296–302. doi: 10.1016/s0168-9525(00)02030-8. [DOI] [PubMed] [Google Scholar]
- Qiang M, Rani CS, Ticku MK. Neuron-restrictive silencer factor regulates the N-methyl-D-aspartate receptor 2B subunit gene in basal and ethanol-induced gene expression in fetal cortical neurons. Mol Pharmacol. 2005;67:2115–2125. doi: 10.1124/mol.104.010751. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- Roy K, Kuznicki K, Wu Q, Sun Z, Bock D, Schutz G, Vranich N, Monaghan AP. The Tlx gene regulates the timing of neurogenesis in the cortex. J Neurosci. 2004;24:8333–8345. doi: 10.1523/JNEUROSCI.1148-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy K, Thiels E, Monaghan AP. Loss of the tailless gene affects forebrain development and emotional behavior. Physiol Behav. 2002;77:595–600. doi: 10.1016/s0031-9384(02)00902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Rozas R. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics. 1999;15:174–175. doi: 10.1093/bioinformatics/15.2.174. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chichung Lie D, Taupin P, Nakashima K, Ray J, Yu RT, Gage FH, Evans RM. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature. 2004;427:78–83. doi: 10.1038/nature02211. [DOI] [PubMed] [Google Scholar]
- Sisodiya SM, Free SL, Williamson KA, Mitchell TN, Willis C, Stevens JM, Kendall BE, Shorvon SD, Hanson IM, Moore AT, van Heyningen V. PAX6 haploinsufficiency causes cerebral malformation and olfactory dysfunction in humans. Nat Genet. 2001;28:214–216. doi: 10.1038/90042. [DOI] [PubMed] [Google Scholar]
- Smith KM, Bauer L, Fischer M, Barkley R, Navia BA. Identification and characterization of human NR4A2 polymorphisms in attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;133:57–63. doi: 10.1002/ajmg.b.30127. [DOI] [PubMed] [Google Scholar]
- Stenman J, Yu RT, Evans RM, Campbell K. Tlx and Pax6 co-operate genetically to establish the pallio-subpallial boundary in the embryonic mouse telencephalon. Development. 2003;130:1113–1122. doi: 10.1242/dev.00328. [DOI] [PubMed] [Google Scholar]
- Stephens JC, Schneider JA, Tanguay DA, Choi J, Acharya T, Stanley SE, Jiang R, Messer CJ, Chew A, Han JH, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293:489–493. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westberg L, Melke J, Landen M, Nilsson S, Baghaei F, Rosmond R, Jansson M, Holm G, Bjorntorp P, Eriksson E. Association between a dinucleotide repeat polymorphism of the estrogen receptor alpha gene and personality traits in women. Mol Psychiatry. 2003;8:118–122. doi: 10.1038/sj.mp.4001192. [DOI] [PubMed] [Google Scholar]
- Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KA, Berry ML, Mahaffey CL, Saionz JR, Hawes NL, Chang B, Zheng QY, Smith RS, Bronson RT, Nelson RJ, Simpson EM. Fierce: a new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav Brain Res. 2002;132:145–158. doi: 10.1016/s0166-4328(01)00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu RT, Chiang MY, Tanabe T, Kobayashi M, Yasuda K, Evans RM, Umesono K. The orphan nuclear receptor Tlx regulates Pax2 and is essential for vision. Proc Natl Acad Sci U S A. 2000;97:2621–2625. doi: 10.1073/pnas.050566897. [DOI] [PMC free article] [PubMed] [Google Scholar]