Abstract
In mammalian cells, active sodium transport and its derived functions (e.g., plasma membrane potential) are dictated by the activity of the Na+,K+-ATPase (NK), whose regulation is essential for maintaining cell volume and composition, as well as other vital cell functions. Here we report the existence of a salt-inducible kinase-1 (SIK1) that associates constitutively with the NK regulatory complex and is responsible for increases in its catalytic activity following small elevations in intracellular sodium concentrations. Increases in intracellular sodium are paralleled by elevations in intracellular calcium through the reversible Na+/Ca2+ exchanger, leading to the activation of SIK1 (Thr-322 phosphorylation) by a calcium calmodulin-dependent kinase. Activation of SIK1 results in the dephosphorylation of the NK α-subunit and an increase in its catalytic activity. A protein phosphatase 2A/phosphatase methylesterase-1 (PME-1) complex, which constitutively associates with the NK α-subunit, is activated by SIK1 through phosphorylation of PME-1 and its dissociation from the complex. These observations illustrate the existence of a distinct intracellular signaling network, with SIK1 at its core, which is triggered by a monovalent cation (Na+) and links sodium permeability to its active transport.
Keywords: cell volume; Na+/Ca2+ exchanger; Na+,K+-ATPase; protein phosphatase 2A
Mammalian cells are endowed with the ability to maintain a tightly regulated cell volume. This function, among many others, entails maintaining the adequate ionic composition of the intracellular milieu in response to variations in the composition of the extracellular compartment (1). Because the plasma membrane is highly permeable to water, it is the concentration of ions across this membrane that is, in the short term, critical for maintaining an adequate cell volume (1). The plasma membrane Na+,K+-ATPase (NK) is important in this process because it provides the driving force for active sodium and potassium transport into and out of the cell, with water following isosmotically. Increases in sodium permeability require concomitant increments in NK-mediated outward sodium transport to prevent a disproportionate increase in the intracellular sodium concentration ([Na+]i)/osmotic pressure and, consequently, cell swelling. These adjustments in cell volume control occur in parallel with other interdependent processes within the cell, such as potassium and chloride transport (1). Studies performed in cell-free systems estimate that, in intact cells at basal [Na+]i, the NK operates at about one third of its maximal capacity (2). Because of this finding, it has been assumed that increases in [Na+]i would be paralleled by a simultaneous increase in NK activity and its immediate extrusion from the cell. However, this hypothesis does not take into account space distribution, time, and amplitude of this phenomenon in intact cells, nor the cellular mechanisms responsible for transforming the signal derived from small changes in [Na+]i to proportionally much larger increases in NK activity.
Specific signaling networks control NK activity in response to G protein-coupled receptors (3, 4) or mitochondria-generated reactive oxygen species (5). The cellular organization of such networks is characterized by their assembly with the target (NK) in time and space (either at the plasma membrane or intracellular endosomes) to ensure specificity and magnitude of the physiological response. Similarly, several intracellular mediators (i.e., kinases) specifically associate with NK molecules in response to digitalis and ouabain-like factors and are able to promote, independently and/or by small increases in intracellular sodium, changes in cell motility, proliferation, and gene expression (6).
In the present work, we searched for potential signaling targets that could sense accurately small increases in intracellular sodium and translate this signal to the NK molecule, which would lead to rapid changes in its catalytic activity.
Results
Identification of a Protein That Interacts with the NK in Renal Epithelial Cells.
To identify proteins that could associate with the NK and participate in its regulation in response to changes in [Na+]i, the NK bearing a GFP tag in its α-subunit (7) was immunoprecipitated (IP) from opossum kidney (OK) cells under basal conditions, and the material was analyzed by SDS/PAGE. A protein of ≈Mr 83 was excised from the gel and subjected to in-gel trypsin digestion [see supporting information (SI) Materials and Methods]. The peptides produced were analyzed by MALDI-TOF mass spectrometry, and the protein was identified as the salt-inducible kinase-1 [(SIK1) NP_067725].
Attention was focused on SIK1 for several reasons: (i) SIK1 has been found in several transporting epithelia (8), (ii) NK activity and intracellular trafficking are regulated by a phosphorylation–dephosphorylation process (9, 10), and (iii) SIK1 expression is regulated by an increase in salt intake (11). Thus, SIK1 might provide a functional link between changes in [Na+]i and responses in NK activity. Different SIK isoforms have been described [SIK1, SIK2 (QIK), and SIK3 (QSK)] (11). SIK isoforms' mRNAs are transcribed in the kidney (8) and OK cells (SI Fig. 5A Left). This association was further confirmed by using a SIK1 antibody (12) in the IP material with an NK antibody (SI Fig. 5A Right) and by immunofluorescence (SI Fig. 5B). The SIK2 and SIK3 isoforms do not associate with the NK (data not shown). In the present investigation, we chose not to focus on the structural aspects of such interaction (i.e., which domains in each molecule are involved, whether it requires the presence of an intermediate protein or an interacting module), but rather to concentrate on its network integration at the molecular level and its physiologic relevance.
SIK1 Participates in the Regulation of NK Activity in Response to Changes in Intracellular Sodium.
Studies were performed in OK cells bearing the native NK. The interaction of SIK1 with the NK was not increased further by rising [Na+]i (SI Fig. 5A Right) with monensin (Mon) as a sodium ionophore (13). The 5 μM concentration of Mon caused a rapid increase in [Na+]i without altering the OK cell architecture (13). Increases in NK activity in response to elevated [Na+]i (Fig. 1A) were time-dependent and, after 5 min, proportionally higher than the increments in [Na+]i [incubation with 6 μM Mon represents specific increments in [Na+]i over basal (≈9 mM): 5 min, ≈12 mM; 10 min, ≈15 mM; 15 min, ≈17 mM; 30 min, ≈22 mM] (13). This effect represents a true increase in its catalytic activity because cell-surface expression of the enzyme units did not significantly change during gradual increases in [Na+]i (Fig. 1A Inset), which is in agreement with previous observations (14). Considering the time-dependency of NK activation by sodium (Fig. 1A), we next examined whether it was associated with increases in SIK1 activity. Enzyme activity, determined as the degree of its autophosphorylation or by phosphorylation of another substrate (TORC2) (12), demonstrates that Mon induced a time-dependent increase in SIK1 activity (Fig. 1B). To examine whether SIK1 is of relevance during sodium-dependent regulation of NK activity, a SIK1-negative mutant (replaced Lys-56 → Met) lacking catalytic activity and the wild type (12) were expressed in OK cells (Fig. 1C Upper). In the presence of elevated [Na+]i, the NK activity was significantly increased in mock and wild-type SIK1, but not in K56M-transfected cells (Fig. 1C Lower), indicating that SIK1 is required during stimulation of NK activity by [Na+]i. Furthermore, HepG2 cells transfected with a plasmid for the corresponding SIK1-siRNA (15) demonstrated a significant reduction in the ability of Mon to increase NK activity, compared with cells transfected with plasmid having only the H1 promoter (Fig. 1D), further suggesting the wide biological significance of this phenomenon. In a cell-free system, neither sodium (Fig. 2A Lower and SI Fig. 6) nor Mon (Fig. 2A Upper) increased SIK1 activity, which suggests the need for an additional intracellular signaling partner that would translate the sodium signal into SIK1 activation.
Fig. 1.
SIK1 mediates the regulation of NK activity in response to high [Na+]i. In all conditions, cells were incubated with 5 μM Mon at 23°C. (A) Time-dependent increase in NK activity in the presence of Mon. Each point is the mean + SEM (n = 3–5). (Inset) NK α-subunit abundance at the plasma membrane after treatment with Mon for different periods of time. Representative WB (Upper) and quantitative analysis (Lower) of five to seven experiments. Bars represent the mean + SEM. (B) GST-SIK1 activity (autophosphorylation and TORC2 phosphorylation) in OK cells incubated with Mon for a different period. A representative autoradiogram is shown (n = 4). (C) (Upper) Expression of HA-SIK1 wild type (WT), the HA-SIK1 mutant (K56M) cDNA, or mock in lysates of OK cells. (Lower) NK activity was determined in the presence (filled bar) or absence (open bar) of Mon (10 min). Each bar represents the mean + SEM of five experiments. *, P < 0.05; **, P < 0.01; ns, not significant. (D) (Lower) Effect of Mon (10 min) on NK activity in HEPG2 cells transfected with 4.5 μg of either SIK1-siRNA or scRNA as control. NK activity was expressed as percentage stimulation. Each bar represents the mean + SEM (n = 6). ***, P < 0.001. (Upper) Representative WB from HEPG2 cells expressing the SIK1-siRNA (siRNA) or the H1 promoter (scRNA).
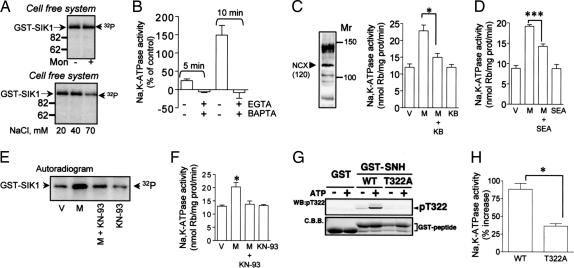
Fig. 2.
Sodium-induced calcium signals and SIK1 activity. (A) Purified GST-SIK1 was incubated with or without 5 μM Mon at 23°C for 15 min (Upper) or in the presence of 20, 40, or 70 mM NaCl for 15 min (Lower). A representative autoradiogram is shown. (B) OK cells previously exposed to 4 mM EGTA at 23°C for 60 min and 50 μM AM-BAPTA at 23°C for 30 min were incubated in the presence or absence of 5 μM Mon at 23°C for either 5 or 10 min. NK activity was expressed as percentage change. Each bar represents the mean + SEM (n = 3–4). (C) (Left) The NCX (Mr 120 subunit) was identified in 300 μg of OK cells postnuclear supernatant. (Right) NK activity was examined in OK cells pretreated with 7 μM NCX inhibitor KB-R7943 at 23°C for 30 min and incubated in the presence (M) or absence (V) of 5 μM Mon at 23°C for 10 min. Each bar represents the mean + SEM (n = 3). *, P < 0.05. (D) NK activity was examined in OK cells pretreated with 2 μM NCX1/NCX2 inhibitor SEA0400 at 23°C for 30 min and incubated in the presence (M) or absence (V) of 5 μM Mon at 23°C for 10 min. Each bar represents the mean + SEM (n = 3–4). ***, P < 0.001. (E and F) SIK1 (E) and NK (F) activity was determined in OK cells pretreated with 25 μM CaMK inhibitor KN-93 at 23°C for 30 min and incubated in the presence (M) or absence (V) of 5 μM Mon at 23°C for 10 min. (E) Representative autoradiogram is shown. (F) Each bar represents the mean + SEM (n = 4). *, P < 0.05. (G) In vitro CaMK1 phosphorylation of SIK1-SNH peptide wild type (WT) and SIK1-SNH peptide bearing a mutation in Thr-322 (T322A). Representative blot is shown. C.B.B., Coomassie brilliant blue. (H) NK activity was determined in OK cells transiently expressing the SIK1-WT or T322A mutant. Each bar represents the mean + SEM (n = 4). *, P < 0.05 T322 vs. WT.
Mon increases intracellular Ca2+ in OK cells (13). Hence, this cation might have triggered the signals required for stimulation of NK activity. Because elevation in intracellular Ca2+ was dependent on the extracellular Ca2+ concentration (13), the effect of Mon on NK activity was examined in the presence of EGTA and by quenching intracellular Ca2+ (13). Under these conditions, increases in [Na+]i failed to stimulate NK (Fig. 2B). In smooth muscle cells, changes in NK activity followed by changes in Ca2+ influx occurs through the Na+/Ca2+ exchanger (NCX) (16) localized in close proximity to NK molecules (17, 18). The NCX is present in OK cells (Fig. 2C Left), and the presence of an NCX inhibitor (19) prevented the increase in NK activity induced by Mon (Fig. 2C Right). SEA0400 (16) also significantly reduced the effect of Mon (Fig. 2D), further confirming the involvement of the NCX in this process and suggesting either the NCX1 and/or NCX2 isoform as possible mediators.
Because incubation of purified SIK1 with increasing Ca2+ concentrations did not result in higher SIK1 activity, we hypothesized that a Ca2+-dependent target could provide the link between the sodium-induced increase in intracellular Ca2+ and activation of SIK1. Calmodulin-dependent protein kinases (CaMK) are primary targets decoding Ca2+ signals. The presence of a CaMK inhibitor (KN-93) (20) prevented the increase in SIK1 (Fig. 2E) and NK activity (Fig. 2F) induced by high [Na+]i, suggesting that SIK1 might be under the control of CaMK by direct phosphorylation. We identified within the SIK1 protein sequence (IDRQRT322V) a consensus site (21) for CaMK-dependent phosphorylation. Indeed, CaMK1 phosphorylates in vitro the SIK1-SNH (sucrose nonfermenting homologue) wild-type peptide, but not the SIK1-SNH carrying the T322A mutation (Fig. 2G). The functional significance of CaMK1-dependent phosphorylation of SIK1 at T322 residue was highlighted in OK cells expressing a SIK1 mutant (T322A). Elevated [Na+]i increased NK activity in cells expressing wild-type SIK1, whereas this effect was significantly reduced in cells expressing the T322A mutant (Fig. 2H). Pharmacological increases in intracellular Ca2+ do not result in increased SIK1 activity (SI Fig. 7), further indicating the need for an integrated network triggered by elevated intracellular sodium.
SIK1-Dependent Activation of a Protein Phosphatase (PPase) Results in NK α-Subunit Dephosphorylation and Increases Its Catalytic Activity.
Raising [Na+]i and activation of SIK1 resulted in the dephosphorylation of the NK α-subunit (Fig. 3A). Dephosphorylation occurred in a time-dependent manner (Fig. 3B). Studies in the presence of a SIK1-negative mutant established that SIK1 was necessary for promoting dephosphorylation of the NK α-subunit (Fig. 3C). Additional evidence indicating that sodium-induced increases in NK activity requires activation of a PPase in an SIK1-dependent manner was further obtained: (i) in experiments using a PPase 2a (PP2A)-negative mutant (L199P) (22), it was possible to prevent the increase in NK activity in response to elevated [Na+]i (Fig. 3D); (ii) taking advantage of the fact that staurosporine inhibits SIK1 activity at ≤5 nM concentrations (23), which do not interfere with the activity of other kinases such as PKC, it was observed that increases in [Na+]i, which led to an ≈30% increase in PPase activity, were completely blocked by pretreatment with staurosporine (Fig. 3E); and (iii) overexpressing SIK1 resulted in elevated (2.5-fold) PPase activity in OK cells compared with nontransfected cells (arbitrary units per microgram of protein: 127 ± 16; n = 3) (Fig. 3F) (increasing the [Na+]i further did not result in additional increases in PPase activity), whereas overexpressing the SIK1 mutant was not associated with an increase in PPase activity over the basal.
Fig. 3.
SIK1 increases PPase activity and dephosphorylates the NK α-subunit in response to high [Na+]i. (A) NK α-subunit phosphorylation in OK cells incubated with (M) or without (V) 5 μM Mon at 23°C for 15 min. Representative WB (n = 5). (B) OK cells were incubated with 5 μM Mon at 23°C for different periods of time, and phosphorylation was analyzed as in A. The degree of α-subunit phosphorylation was established as a ratio between the phosphorylated signal and the amount of IP material and expressed as a percentage of control (without Mon). Each point represents the mean ± SE (n = 3–5). (C) NK α-subunit phosphorylation was examined in OK cells transiently expressing the SIK1 wild type (WT) or the SIK1 mutant (K56M) in the presence (M) or absence (V) of 5 μM Mon at 23°C for 15 min. A representative WB (Left) and the quantitative analysis (mean + SEM, n = 5) (Right) are shown. (D) Basal (open bars) and 5 μM Mon at 23°C for 10 min (filled bars) NK activity was determined in OK cells transiently expressing the PP2A wild type (WT), mutant (L199P), or mock transfected. Each bar represents the mean + SEM (n = 5). *, P < 0.001; ns, not significant. (E) PPase activity in OK cells treated with (M) or without (V) 5 μM Mon at 23°C for 10 min and with Mon in cells previously treated with 5 nM staurosporine (M + STO 5) at 23°C for 30 min. Each bar represents the mean + SEM (n = 4). *, P < 0.01. (F) Basal (nonstimulated) PPase activity was determined in OK cells transiently expressing SIK1 (WT) or mutant (K56M) cDNA. Each bar represents the mean + SEM (n = 5–6). **, P < 0.01.
SIK1-Dependent Activation of PP2A Is Mediated by PME-1 Phosphorylation and Its Dissociation from the PP2A/NK Complex.
Because direct phosphorylation of the PPase has generally been described as a process resulting in inhibition of its activity (24), attention was focused on methylation of PPase subunits as a potential regulatory mechanism. The presence of a leucine carboxy methyltransferase and a protein phosphatase methylesterase-1 (PME-1) that associates with the catalytic subunit could activate or inactivate the PPase. The presence of PME-1 catalyzes the demethylation and inactivation of PP2A (25, 26). A protein with an Mr of ≈45 present in the immune complex that coprecipitated with the NK α-subunit is recognized by a specific PME-1 antibody (Fig. 4A). Similarly, PP2A also was co-IP with the NK/PME-1 complex (Fig. 4A). PP2A has been reported to be associated with the NK α-subunit (27), which would imply that the PP2A will naturally be in an active state and, consequently, the NK will be permanently dephosphorylated unless an inhibitory mechanism is in place. PME-1 associates with the PP2AC α- or β-subunit and suppresses its activity, which raises the possibility that an increase in [Na+]i could favor a dissociation of PME-1 from the NK/PP2A complex and thereby promote NK α-subunit dephosphorylation, leading to its higher catalytic activity. Indeed, increases in [Na+]i with Mon promote the dissociation of PME-1 from the NK/PP2APR65 complex in the IP material with an NK antibody (Fig. 4 A and B), whereas the association of PP2APR65 subunit to the NK remained unchanged (Fig. 4A). The ≈20% dissociation measured is within the IP NK (and its associated PP2APR65). Therefore, the quantity represents a significant change within the fraction of total enzyme that is specifically undergoing regulation. Similarly, a reduced amount of PP2A was IP with a PME-1 antibody (Fig. 4C) from cells treated with Mon. In OK cells transiently transfected with the SIK1-negative mutant, Mon failed to promote the dissociation of PME-1 from the NK α-subunit complex, compared with cells transfected with the wild-type SIK1 (Fig. 4D). These observations suggest that PME-1 could be under the control of SIK1 by direct phosphorylation. To test this hypothesis, PME-1 phosphorylation was examined (Fig. 4E) by using the back-phosphorylation technique (10, 28), where the signal is inversely proportional to the degree of phosphorylation. Indeed, it was observed that PME-1 was phosphorylated in the presence of SIK1 and that Mon treatment decreased the amount of dephospho-PME-1(Fig. 4E Right).
Fig. 4.
SIK1 activates PPase activity in response to high [Na+]i by phosphorylating PME-1. (A) The association between PME-1 and PP2A65PR with the NK was examined in OK cells in the presence (M) or absence (V) of 5 μM Mon at 23°C for 5 min. A representative WB performed with antibodies against PP2A65PR (5 μg/ml), PME-1 (1:500), or α-subunit (1:1,000) is shown. (B) The association of PME-1 with the NK α-subunit was examined in OK cells incubated in the presence or absence of 5 μM Mon at 23°C for 5- or 15-min incubation. Each point represents the mean ± SEM (n = 5). (C) The association of PME-1 with PP2A was examined in the IP material with a PME-1 antibody from OK cells incubated with/without Mon (M) as indicated in A. Representative WB (n = 3). (D) The association of PME-1 with the NK α-subunit was examined in OK cells transiently expressing the SIK1 wild type (WT) or mutant (K56M). A representative WB (Left) and the quantitative analysis (mean + SEM, n = 4) (Right) are shown. (E) OK cells were treated with (M) or without (V) 5 μM Mon at 23°C for 2.5 min. A sample without addition of SIK1 was included (−). The IP PME-1 was subjected to in vitro phosphorylation in the presence of purified SIK1. Proteins were analyzed by autoradiography (Autoradiog.) and WB. (Left) A representative experiment is depicted. (Right) The amount of phospho-PME-1 was corrected for the amount of IP PME-1 and expressed in a percentage of control (n = 5). *, P < 0.05.
Discussion
Physiological increases in [Na+]i are associated with parallel increases in NK activity. Contrary to predictions, this study demonstrates that the mechanisms by which increased [Na+]i triggers the stimulation of NK in intact cells is not mediated by sodium per se, but by a Na+-dependent activation of a distinct intracellular signaling network (SI Fig. 8). At the core of this network is a sucrose nonfermenting-1-related serine/threonine kinase (SIK1) that associates with the NK and participates in the regulation of its catalytic activity. The current knowledge that acute increases in [Na+]i would only have direct regulatory properties on NK activity may have to be revisited when applied to intact cells. Although this effect may modulate the housekeeping role of NK, it is less likely to be involved during short-term adaptations of the NK activity to challenges in sodium gradients in intact cells. This view is supported by experiments in which suppressed SIK1 (by using siRNA) or SIK1 mutants (or backward/forward modulators of SIK1 activation) modulate the increases in NK activity and α-subunit dephosphorylation induced by elevated [Na+]i. Observations that SIK1 mutants do not affect the basal (nonstimulated) NK activity may indicate that SIK1 associates with a limited pool of NK molecules that can be subject to short-term regulation. Similarly, that this association was not increased by rising [Na+]i suggests a limited/selected pool of NK that can be regulated under this specific circumstance.
The presence of a proportion of NK molecules in a phosphorylated state (under basal nonstimulated conditions) that are regulated by SIK1-PP2A may suggest a rationale for having a (dormant) pool of NK molecules therein that can be activated in a rapid manner for quick adjustments to changes in sodium gradients. Assuming that dephosphorylation is a necessity, the Ca2+-SIK1-PP2A signaling could provide the basis for a mechanism that otherwise could not take place solely by increasing [Na+]i. The fact that an elevated [Na+]i is targeting only a proportion of enzyme molecules (i.e., those that are phosphorylated) may suggest that regulatory signals would only affect a different set of NK α1-isoforms (those not responsible for housekeeping functions) that may or may not have a particular distribution within the cells (close association with NCX).
A transient calcium gradient that is activated by sodium throughout the NCX within a time frame that precedes the activation of SIK1 and the increase in NK activity could link the sodium signals to SIK1. Moreover, studies in the absence of Ca2+ and the presence of CaMK inhibitors revealed that SIK1 activity is under the control of a Ca2+-dependent pathway. This mechanism bears strong similarities to that used by plant cells to adapt to a saline environment, where a vacuolar proton pump is activated by SOS2 (a sucrose nonfermenting kinase) (29, 30) via SOS3 (a Ca2+-dependent kinase) (31).
Sodium gradients regulate PP2A activity, and this effect is essential for controlling NK activity. PP2A has been previously shown to be associated constitutively with the NK α-subunit (27) possibly to prevent phosphorylation and thereby inactivation/endocytosis of NK molecules. Nevertheless, the results from this study appear to suggest that the presence of PP2A also serves as a key signal linking SIK1 to the target. Blocking SIK1 activation prevents the increase in PPase activity induced by sodium, and transient overexpression of SIK1 results in a 2.5-fold increase in PPase activity, compared with that of an inactive SIK1. Whereas phosphorylation/dissociation of PME-1 from PP2AC has been suggested, in this study we identified a physiological signal (elevated [Na+]i) mediating (by SIK1) PME-1 phosphorylation and its dissociation from the NK/PP2A complex. The absence of PME-1 may shift the equilibrium of PP2AC toward a methylated/activated state, as reported in a cell-free system.
In conclusion, SIK1 might represent a mechanism present in mammalian cells that senses changes in [Na+]i and transforms this event into a signaling cascade that links sodium transport out of the cells by increasing NK activity.
Materials and Methods
Antibodies and Reagents.
Detailed sources of antibodies and reagents can be found in SI Materials and Methods. Polyclonal antibodies against SIK1 (12), SIK2 (32), and SIK3 (23) have been reported.
Cell Culture and Transfection.
OK cells were grown in DMEM (Gibco/Invitrogen, Carlsbad, CA), and HepG2 cells (American Type Culture Collection, Manassas, CA) were grown in GlutaMAX-DMEM (Gibco/Invitrogen) with 10% FBS and penicillin/streptomycin. Transient transfections were performed by using LipofectAMINE 2000 (Invitrogen). cDNAs for wild-type PP2A and L199P were kindly provided by B. A. Hemmings (Friedrich Miescher Institute, Basel, Switzerland). Site-directed mutagenesis of SIK1 (K56M) was performed (12) by using a QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The SIK1 mutant lacking a CaMK regulatory residue was generated by exchanging nucleotides as follows: T322A (ACA-GCA). HA and GST tags were inserted at the N-terminal region of the SIK1 molecule (33). An expression plasmid of pCMVsport6-mouse PME-1 (IMAGE 5062326) was purchased from Invitrogen. All constructs were verified by DNA sequence analysis.
Immunoprecipiation.
Cells were washed in cold PBS. After the addition of immunoprecipitation buffer [50 mM Tris, 100 mM NaCl, 30 mM NaF, 2 mM EDTA, 2 mM EGTA, 1% Triton X-100, and protease inhibitors (1 mM PMSF, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 10 μg/ml aprotinin], the cells were homogenized and centrifuged, and the postnuclear supernatant was precleared. Equal amounts of protein were incubated with an appropriate antiserum for 16 h at 4°C. The proteins were analyzed by SDS/PAGE (34) and detected with Western blot (WB) analysis by chemiluminescence (ECL Plus; GE Healthcare, Chalfont St. Giles, U.K.).
NK Activity.
NK-mediated active transport was determined as described in ref. 13 by using ouabain-sensitive 86Rb+ transport. Each experiment was performed independently and in triplicate determinations.
Phosphorylation of NK α-Subunit.
After treatment, crude membranes were prepared (35). Cells were homogenized in 50 mM mannitol/5 mM Hepes-Tris buffer (pH 7.6) and centrifuged to remove the cell debris. Crude membranes were isolated by centrifugation of the supernatant at 25,000 × g for 30 min at 4°C and resuspended in immunoprecipitation buffer (including 30 mM Na4P2O7). The IP material (α-5 antibody against the NK) was analyzed by SDS/PAGE and WB analysis. The state of NK α-subunit phosphorylation was determined by using an antiphosphoserine antibody (1:500). Membranes were stripped and retested with an antibody against the NK (1:1,000). WB signals were quantitated by using the ImageJ software (http://rsb.info.nih.gov/ij). The state of NK phosphorylation was determined as the ratio between the phosphorylated signal and the amount of NK IP, and changes were expressed as a percentage of control.
Determination of SIK1 Activity.
SIK1 activity was determined in OK cells expressing the GST-SIK1 isoform as described in ref. 12. Equal aliquots of isolated GST-SIK1 were incubated with SIK phosphorylation buffer [50 mM Tris (pH 7.4), 10 mM MnCl2, 0.3–0.5 μCi/μl [γ-32P]ATP (PerkinElmer, Norwalk, CT)] and TORC2 substrate at 30°C for 30 min with constant shaking. The reaction was terminated by adding sample buffer. Proteins were separated on SDS/PAGE, and phosphorylated GST-SIK1 and TORC2 were examined by autoradiography.
Determination of Protein Phosphatase Activity.
Phosphatase activity was assayed by using the EnzoLyte MFP Protein Phosphatase Assay system (AnaSpec, San Jose, CA). After treatment, cells were homogenized and centrifuged at 100,000 × g at 4°C for 20 min, and enzymatic activity was assayed in the pellet. Dephosphorylation of 3-O-methylfluorescein phosphate was monitored by measuring the fluorescence of methylfluorescein product every 15 min for 1 h in a 96-well fluorescence plate reader (MicroLumat Plus LB; excitation, 485 nm; emission, 535 nm). Phosphatase activity was calculated as the slope of fluorescence recordings and expressed as arbitrary fluorescence units per microgram of protein.
Back Phosphorylation.
OK cells overexpressing the wild-type PME-1 were incubated with or without 5 μM Mon at 23°C for 2.5 min. The incubation was stopped by transferring the samples to ice and adding the immunoprecipitation buffer as described earlier with the addition of 30 mM Na4P2O7 and 2 mM activated Na3VO4. PME-1 was IP and phosphorylated by using ≈10 ng of purified GST-SIK1 in the presence of 50 mM Tris·HCl (pH 7.4), 10 mM MgCl2, 1 mM DTT, 250 μM Na2ATP, and 0.5 μCi/μl [γ-32P]ATP (PerkimElmer) for 30 min at 30°C. The reaction was stopped by the addition of sample buffer, and proteins were separated on SDS/PAGE and transferred to PVDF membranes. The radioactivity incorporated in the PME-1 was analyzed by autoradiography and the amount of PME-1 by WB.
In Vitro Phosphorylation of SIK1 by CaMK1.
To prepare an SIK1-SNH peptide, a cDNA fragment of the SIK1-SNH domain (amino acids 301–354) was amplified by PCR with primers linked with a BamHI site in the forward primer and a NotI site in the reverse primer. To express the SNH domain as a GST-fusion protein in Escherichia coli, the amplified product was ligated into the BamHI–NotI site of pGEX-6P3. An active CaMK was prepared by using an expression plasmid (pSport6-CaMK1, IMAGE 4483612; Invitrogen). To convert a native CaMK1 into a constitutive active form, a stop codon was inserted at the amino acid 296 position by site-directed mutagenesis. The resultant cDNA was ligated into the BamHI–NotI site of pEBG vector, and the active GST-CaMK1 was expressed in COS-7 cells. The purified GST-CaMK1 was mixed with GST, GST-SNH (wild type), or GST-SNH (T322A mutant) and incubated with or without 0.1 mM ATP at 30°C for 30 min in a CaMK1 reaction buffer [50 mM Tris (pH 7.4), 10 mM MgCl2]. The reaction was terminated by the addition of sample buffer and an aliquot subjected to SDS/PAGE, followed by WB analysis using an anti-pT322 antibody.
Cell-Surface Biotinylation.
After the experimental protocols, OK cells were placed on ice and washed with ice-cold PBS (1 mM Ca2+, 0.5 mM Mg2+), followed by the addition of 1.5 mg/ml biotin at 4°C for 30 min on a shaking platform protected from light. Thereafter, the cells were washed with ice-cold PBS (100 mM glycine, 1 mM Ca2+, 0.5 mM Mg2+) and ice-cold PBS without glycine. Subsequently, the PBS was replaced with RIPA buffer containing 50 mM Tris·HCl, 150 mM NaCl, 1% IGEPAL CA-630 (Nonidet P-40), 1% sodium deoxycholate, and protease inhibitors (1 mM PMSF, 5 μg/ml leupeptin, 5 μg/ml antipain, 5 μg/ml pepstatin A, and 10 μg/ml aprotinin), and cells were then scraped, vortexed, and centrifuged at 14,000 × g at 4°C for 1 min. Streptavidin was added to the supernatant (250–350 μg of protein) and incubated at 4°C with end-over-end rotation overnight. Proteins were analyzed by SDS/PAGE and WB analysis with a NK α-subunit (1:1,000).
Statistical Analysis.
All statistical tests were performed with the unpaired Student t test. P values <0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Marie-Odile Revel for technical assistance, Profs. Anders Hamsten and Mitsuhiro Okamoto for generous support, and Drs. Akemichi Baba and Toshio Matsuda (Osaka University, Osaka, Japan) for providing SEA0400. This work was supported by Swedish Research Council Grants 32X-10860 and 32P-14879, National Institutes of Health Grant HL 48129, the Swedish Heart and Lung Foundation, the Swedish Foundation for Kidney Research, and the Torsten och Ragnar Söderberg Foundation.
Abbreviations
- CaMK
calmodulin-dependent protein kinase
- IP
immunoprecipitated
- Mon
monensin
- NCX
Na+/Ca2+ exchanger
- NK
Na+,K+-ATPase
- OK
opossum kidney
- PME-1
phosphatase methylesterase-1
- PPase
protein phosphatase
- PP2A
PPase 2A
- SIK1
salt-inducible kinase 1
- WB
Western blot.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0706838104/DC1.
References
- 1.Lang F, Busch GL, Ritter M, Völkl H, Waldegger S, Gulbins E, Häussinger D. Physiol Rev. 1998;78:247–306. doi: 10.1152/physrev.1998.78.1.247. [DOI] [PubMed] [Google Scholar]
- 2.Skou JC. Methods Enzymol. 1988;156:1–25. doi: 10.1016/0076-6879(88)56004-4. [DOI] [PubMed] [Google Scholar]
- 3.Ogimoto G, Yudowski GA, Barker CJ, Köhler M, Katz AI, Féraille E, Pedemonte CH, Berggren P-O, Bertorello AM. Proc Natl Acad Sci USA. 2000;97:3242–3247. doi: 10.1073/pnas.060025597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yudowski GA, Efendiev R, Pedemonte CH, Katz AI, Berggren P-O, Bertorello AM. Proc Natl Acad Sci USA. 2000;97:6556–6561. doi: 10.1073/pnas.100128297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dada LA, Chandel NS, Ridge KM, Pedemonte CH, Bertorello AM, Sznajder JI. J Clin Invest. 2003;111:1057–1064. doi: 10.1172/JCI16826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Z, Askari A. Eur J Biochem. 2002;269:2434–2439. doi: 10.1046/j.1432-1033.2002.02910.x. [DOI] [PubMed] [Google Scholar]
- 7.Doné SC, Leibiger IB, Efendiev R, Katz AI, Leibiger B, Berggren P-O, Pedemonte CH, Bertorello AM. J Biol Chem. 2002;277:17108–17111. doi: 10.1074/jbc.M201326200. [DOI] [PubMed] [Google Scholar]
- 8.Feldman JD, Vician L, Crispino M, Hoe W, Baudry M, Herschman HR. J Neurochem. 2000;74:2227–2238. doi: 10.1046/j.1471-4159.2000.0742227.x. [DOI] [PubMed] [Google Scholar]
- 9.Carranza ML, Rousselot M, Chibalin AV, Bertorello AM, Favre H, Féraille E. J Physiol. 1998;511:235–243. doi: 10.1111/j.1469-7793.1998.235bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chibalin AV, Ogimoto G, Pedemonte CH, Pressley TA, Katz AI, Féraille E, Berggren P-O, Bertorello AM. J Biol Chem. 1999;274:1920–1927. doi: 10.1074/jbc.274.4.1920. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto M, Takemori H, Katoh Y. Trends Endocrinol Metab. 2004;15:21–26. doi: 10.1016/j.tem.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Lin X, Takemori H, Katoh Y, Doi J, Horike N, Makino A, Nonaka Y, Okamoto M. Mol Endocrinol. 2001;15:1264–1276. doi: 10.1210/mend.15.8.0675. [DOI] [PubMed] [Google Scholar]
- 13.Efendiev R, Bertorello AM, Zandomeni R, Cinelli AR, Pedemonte CH. J Biol Chem. 2002;277:11489–11496. doi: 10.1074/jbc.M108182200. [DOI] [PubMed] [Google Scholar]
- 14.Vinciguerra M, Deschênes G, Hasler U, Mordasini D, Rousselot M, Doucet A, Vandewalle A, Martin PY, Féraille E. Mol Biol Cell. 2003;14:2677–2688. doi: 10.1091/mbc.E02-11-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, et al. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 16.Iwamoto T, Kita S, Zhang J, Blaustein MP, Arai Y, Yoshida S, Wakimoto K, Komuro I, Katsuragi T. Nat Med. 2004;10:1193–1199. doi: 10.1038/nm1118. [DOI] [PubMed] [Google Scholar]
- 17.Moore ED, Etter EF, Philipson KD, Carrington WA, Fogarty KE, Lifshitz LM, Fay FS. Nature. 1993;365:657–660. doi: 10.1038/365657a0. [DOI] [PubMed] [Google Scholar]
- 18.Dostanic I, Schultz JJ, Lorenz JN, Lingrel JB. J Biol Chem. 2004;279:54053–54061. doi: 10.1074/jbc.M410737200. [DOI] [PubMed] [Google Scholar]
- 19.Iwamoto T, Watano T, Shigekawa M. J Biol Chem. 1996;271:22391–22397. doi: 10.1074/jbc.271.37.22391. [DOI] [PubMed] [Google Scholar]
- 20.Ishida A, Kameshita I, Okuno S, Kitani T, Fujisawa H. Biochem Biophys Res Commun. 1995;212:806–812. doi: 10.1006/bbrc.1995.2040. [DOI] [PubMed] [Google Scholar]
- 21.White RR, Kwon Y-G, Taing M, Lawrence DS, Edelman AM. J Biol Chem. 1998;273:3166–3172. doi: 10.1074/jbc.273.6.3166. [DOI] [PubMed] [Google Scholar]
- 22.Evan DRH, Myles T, Hofsteenge J, Hemmings BA. J Biol Chem. 1999;274:24038–24046. doi: 10.1074/jbc.274.34.24038. [DOI] [PubMed] [Google Scholar]
- 23.Katoh Y, Takemori H, Lin XZ, Tamura M, Muraoka M, Satoh T, Tsuchiya Y, Min L, Doi J, Miyauchi A, et al. FEBS J. 2006;273:2730–2748. doi: 10.1111/j.1742-4658.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 24.Janssens V, Goris J. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogris E, Du X, Nelson KC, Mak EK, Yu XX, Lane WS, Pallas DC. J Biol Chem. 1999;274:14382–14391. doi: 10.1074/jbc.274.20.14382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longin S, Jordens J, Martens E, Stevens I, Janssens V, Rondelez E, De Baere I, Derua R, Waelkens E, Goris J, Van Hoof C. Biochem J. 2004;380:111–119. doi: 10.1042/BJ20031643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lecuona E, Dada LA, Sun H, Butti ML, Zhou G, Chew TL, Sznajder JI. FASEB J. 2006;20:2618–2620. doi: 10.1096/fj.06-6503fje. [DOI] [PubMed] [Google Scholar]
- 28.Nestler E, Greengard P. Proc Natl Acad Sci USA. 2004;77:7479–7483. doi: 10.1073/pnas.77.12.7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Ishitani M, Halfter U, Kim C-S, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3730–3734. doi: 10.1073/pnas.060034197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Zhu J-K. Science. 1998;280:1943–1945. doi: 10.1126/science.280.5371.1943. [DOI] [PubMed] [Google Scholar]
- 31.Halfter U, Ishitani M, Zhu J-K. Proc Natl Acad Sci USA. 2000;97:3735–3740. doi: 10.1073/pnas.040577697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horike N, Takemori H, Katoh Y, Doi J, Min L, Asano T, Sun XJ, Yamamoto H, Kasayama S, Muraoka M, et al. J Biol Chem. 2003;278:18440–18447. doi: 10.1074/jbc.M211770200. [DOI] [PubMed] [Google Scholar]
- 33.Katoh Y, Takemori H, Min L, Muraoka M, Doi J, Horike N, Okamoto M. Eur J Biochem. 2004;271:4307–4319. doi: 10.1111/j.1432-1033.2004.04372.x. [DOI] [PubMed] [Google Scholar]
- 34.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 35.Khundmiri SJ, Bertorello AM, Delamere NA, Lederer ED. J Biol Chem. 2004;279:17418–17427. doi: 10.1074/jbc.M311715200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.