Abstract
The biogenesis of thylakoid membranes, an indispensable event for the photoautotrophic growth of plants, requires a significant increase in the level of the unique thylakoid membrane lipid monogalactosyldiacylglycerol (MGDG), which constitutes the bulk of membrane lipids in chloroplasts. The final step in MGDG biosynthesis occurs in the plastid envelope and is catalyzed by MGDG synthase. Here we report the identification and characterization of an Arabidopsis mutant showing a complete defect in MGDG synthase 1. The mutant seeds germinated as small albinos only in the presence of sucrose. The seedlings lacked galactolipids and had disrupted photosynthetic membranes, leading to the complete impairment of photosynthetic ability and photoautotrophic growth. Moreover, invagination of the inner envelope, which is not seen in mature WT chloroplasts, was observed in the mutant, supporting an old hypothesis that envelope invagination is a major event in early chloroplast biogenesis. In addition to the defective seedling phenotype, embryo development was arrested in the mutant, although seeds with impaired embryos could germinate heterotrophically. These results demonstrate the importance of galactolipids not only in photosynthetic growth but also in embryogenesis.
Keywords: glycosyltransferase, thylakoid membrane, monogalactosyldiacylglycerol, monogalactosyldiacylglycerol synthase
The photosynthetic reactions of higher plants rely on a well developed thylakoid membrane system inside chloroplasts. The biogenesis of thylakoid membranes, an indispensable event for photoautotrophic growth, is closely linked to the development of chloroplasts from other plastids such as proplastids. This drastic morphological change within chloroplasts requires a significant increase in the levels of the nonphosphorus glyco lipids monogalactosyldiacylglycerol (MGDG) and digalactosyl diacylglycerol (DGDG), which account for ≈50 and 25 mol% of total thylakoid lipids, respectively (1). These relative abundance levels are also found in cyanobacteria, which suggests that MGDG and DGDG are important for all oxygenic photosynthetic organisms (2). These galactolipids are also major lipid constituents of the inner and outer envelope membranes of plastids and are rarely detected in other cell membranes (1). MGDG has a small galactose head group and splayed polyunsaturated fatty acid tails, which together give this molecule a cone-like shape and the ability to induce curvature in lamellar membranes (3). In contrast to MGDG, which forms nonbilayer hexagonal phases, pure DGDG forms bilayer membranes (3). These unique galactolipid characteristics may be important for the organization of highly stacked thylakoid membranes (3). In addition, x-ray crystallographic analyses of photosystems I and II (PSI and PSII) revealed that galactolipids are tightly bound to these reaction centers (4–6), which suggests that these lipids are required not only as bulk constituents of photosynthetic membranes but also for the photosynthetic reaction itself.
The biosynthesis of both MGDG and DGDG is critical for thylakoid biogenesis. The final step in MGDG biosynthesis occurs in the plastid envelope and is catalyzed by MGDG synthase (UDP-galactose:1,2-diacylglycerol 3-β-d-galactosyltransferase) (7), and DGDG is subsequently synthesized by galactosylation of MGDG (8). Thus, MGDG synthase is the key enzyme for the biosynthesis of both of these galactolipids and, hence, for the biogenesis of chloroplast membranes (2). In Arabidopsis, three functional MGDG synthases have been identified, namely, MGD1, MGD2, and MGD3, which differ substantially with respect to their substrate specificity, subcellular localization, and gene expression profiles (9, 10). Whereas MGD1 is regarded as the major isoform localized to the inner envelope membranes of chloroplasts, MGD2 and MGD3 are targeted to outer envelopes and are considered to be less important for thylakoid membrane biogenesis (11). Although a knockdown mutant for MGD1 has been isolated in Arabidopsis in which total galactolipids were decreased by 20% compared with the WT plant (12), no other mutants have been reported that show a greater decrease in galactolipid content, presumably because of substantive developmental defects (11). To understand the function of galactolipids in plant development and growth, we isolated a knockout mutant of MGD1 in Arabidopsis. Characterization of this MGD1 knockout mutant demonstrates that MGD1 mediation of inner envelope MGDG synthesis is indispensable for thylakoid membrane biogenesis and early embryogenesis. The data also provide evidence supporting an old hypothesis that envelope invagination is a major event in early chloroplast biogenesis.
Results
Mutant Analyses During Embryogenesis.
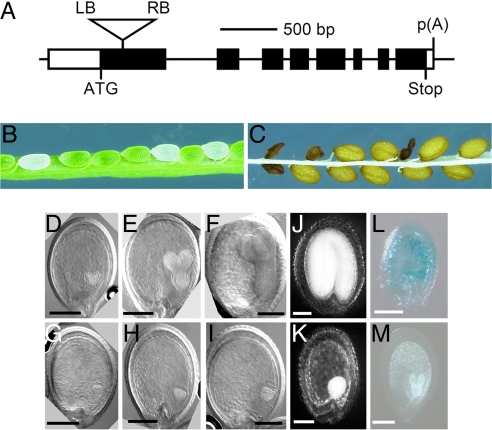
A plant carrying a portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells (T-DNA) insertion in atMGD1 (SALK_002620) was obtained from The Salk Institute Genomic Analysis Laboratory (La Jolla, CA). The T-DNA insertion locus was sequenced and, in combination with information available from The Salk Insertion Sequence Database (http://signal.salk.edu/tabout.html), the insertion site was determined to be located between +145 and +199 bp from the translation initiation site of atMGD1 (Fig. 1A). We further characterized this mutant, which was named mgd1-2. The first visible effect of mgd1-2 occurred during embryogenesis. Within the immature siliques of the self-fertilizing heterozygous mgd1-2 mutant, abnormal white seeds were detected at a frequency of ≈25% (white seed to green seed ratio of 83:242; Fig. 1B). Presumably, these white embryos developed into the wrinkled seeds that were observed at a similar frequency (wrinkled seed to normal seed ratio of 65:185; Fig. 1C). The segregation ratios suggest that these seed phenotypes are associated with one recessive mutation and that the aborted seeds represented mgd1-2 homozygous mutants. To characterize morphogenetic phenotypes in the aborted seeds, embryo development was examined by Nomarski microscopy (Fig. 1 D–I). Before the greening stage of the embryo, it was difficult to distinguish the mutant seeds in the heterozygous mgd1-2 siliques. At the greening stage, the homozygous mgd1-2 embryo, which was identifiable by its albino phenotype, was stunted at the globular stage, whereas green embryos corresponding to WT or heterozygous mgd1-2 formed heart-shaped embryos (compare Fig. 1 G with D). In older siliques, embryo development of the homozygous mutant was arrested without obvious differentiation at the heart stage, although the embryo size increased to some extent (Fig. 1 H and I). As a result, seed dehydration occurred without enough embryo maturation in the aborted seeds (Fig. 1K). Histochemical β-glucuronidase analysis of MGD1::GUS transformants (10) revealed that MGD1 promoter activity transiently increased in the growing embryos at the globular stage (Fig. 1 L and M); MGD2 and MGD3 promoters showed no detectable activity during embryo development (data not shown). These findings demonstrate that MGD1 and, thus, galactolipids play an indispensable role in early embryo development, particularly at the transition from the globular to the heart stage when thylakoid membranes start to develop.
Fig. 1.
Identification of the MGD1 knockout mutant mgd1-2. (A) Schematic diagram showing the T-DNA insertion site in mgd1-2. Exons and untranslated regions are represented by filled and open boxes, respectively. Lines between the boxes correspond to introns. The T-DNA insertion site is represented by the inverted triangle in the first exon. LB, left border of the T-DNA; RB, right border of the T-DNA; ATG, translation initiation codon; Stop, translation termination codon; p(A), polyadenylation site. (B and C) Light microscopic images of the dissected green silique (B) and mature seeds (C) of heterozygous mgd1-2 plants. (D–I) Nomarski microscopic images of green (D–F) and white (G–I) seeds removed from self-pollinated siliques of heterozygous mgd1-2 plants. The embryos are observed at heart (D and G), torpedo (E and H) and bent cotyledon (F and I) stages. (J and K) Light microscopic images of the normal (J) and aborted (K) seeds removed from dehydrated siliques of heterozygous mgd1-2 plants. (L and M) Histochemical analysis of the expression of MGD1::GUS (β-glucuronidase) constructs at the globular (L) and heart (M) stages. Developing immature seeds of MGD1::GUS transformants (10) were used for GUS analysis. (Scale bars: 100 μm.)
MGD1 Knockout Causes Dwarf and Albino Phenotypes.
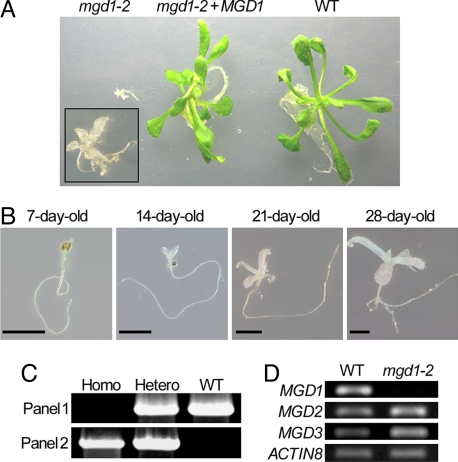
To test the germinating capacity of the aborted mgd1-2 seeds, the wrinkled seeds were collected and sown on sucrose-containing medium. Although the arrest of embryo growth generally causes embryonic lethality, the abnormal seeds in fact germinated, yielding seedlings with combined dwarf and albino phenotypes (Fig. 2A Left). Under sucrose-depleted conditions, the wrinkled seeds were unable to germinate, indicating an inability to grow photoautotrophically. Even on the sucrose-containing medium, mutant seed growth was very slow, particularly in the shoot, and the plants became stunted by ≈4 weeks (Fig. 2B). Genomic PCR analysis revealed that these albino seedlings were homozygous for mgd1-2 (Fig. 2C), confirming the association between this homozygous mutant genotype and the embryonic phenotypes shown in Fig. 1. To examine transcript levels of three atMGD genes in mgd1-2 homozygous seedlings, semiquantitative RT-PCR analyses were carried out by using ACTIN8 as a loading control (Fig. 2D). Transcript levels of both MGD2 and MGD3 were not significantly affected by the mutation in mgd1-2. By contrast, an amplified fragment corresponding to MGD1 was observed in the WT but was not detectable in the mgd1-2 mutant, demonstrating that MGD1 expression was disrupted in the mutant. To confirm that the dwarf and albino phenotypes of this mutant were actually attributable to loss of MGD1 function, mgd1-2 was complemented with an exogenously introduced MGD1 cDNA fused with the green fluorescent protein gene, GFP. The resulting plants, mgd1-2 + MGD1, exhibited a green and healthy phenotype, demonstrating that the mgd1-2 phenotypes are caused by MGD1 knockout (Fig. 2A). MGD1 cDNA without GFP also complemented mgd1-2 seedlings in the same manner as MGD1–GFP, eliminating the effect of GFP fusion (data not shown).
Fig. 2.
Characterization of mgd1-2 seedlings germinated from aborted seeds. (A) mgd1-2, mgd1-2 + MGD1, and WT seedlings (21 days old) on sucrose-containing medium. (Inset) Higher magnification of the mgd1-2 seedling. (B) Growth phenotypes of the mgd1-2 mutant. Homozygous mgd1-2 plants were grown in rotated liquid medium for the indicated times. (Scale bars: 2.5 mm.) (C) PCR analysis of genomic DNA from WT, heterozygous (Hetero), and homozygous (Homo) lines of mgd1-2. (Upper) Amplified bands representing the sequence between the translation initiation site and the second exon of MGD1. (Lower) Amplified bands representing the sequence between the right end of the T-DNA and the second exon of MGD1. (D) Semiquantitative RT-PCR analysis of three atMGD genes in WT and mgd1-2 homozygous plants. ACTIN8 transcripts were analyzed as a loading control.
Absence of Galactolipids in the mgd1-2 Mutant.
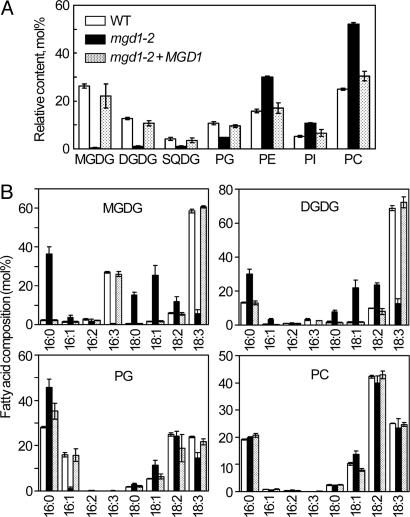
To examine the effect of MGD1 knockout on membrane lipid homeostasis, we analyzed the composition of total lipids extracted from mgd1-2, WT, and MGD1-complemented plants (Fig. 3A). In the mgd1-2 mutant, the amount of MGDG was reduced by ≈98% compared with the WT and the complemented plants, indicating that MGD1 function underlies the bulk of MGDG biosynthesis and that MGD2/MGD3 cannot compensate for loss of MGD1 function. In parallel with the reduction of MGDG content, a dramatic decrease in DGDG content was observed in the mutant, demonstrating that MGD1 is also predominantly responsible for DGDG synthesis, presumably by supplying MGDG as a substrate. In addition to galactolipids, the amount of sulfoquinovosyldiacylglycerol (SQDG) and phosphatidylglycerol (PG) was reduced in mgd1-2 (Fig. 3A). Because SQDG and PG are also components of thylakoid membrane lipids, this result suggests an overall decrease in thylakoid membrane content in the mutant. The mgd1-2 mutation also affected the fatty acid composition of the photosynthetic membrane lipids (Fig. 3B). Levels of hexadecatrienoic (16:3) and α-linolenic (18:3) fatty acids, which are typically found in chloroplast galactolipids, were dramatically decreased in the mutant. In addition, trans-Δ3-hexadecenoic acid (trans16:1), which is a PG-specific fatty acid found only in thylakoid membranes, was reduced to an undetectable level in the mutant although total PG content was only reduced by half. These data also suggest that thylakoid membrane lipid metabolism is defective in mgd1-2. By contrast, the fatty acid composition of phosphatidylcholine (PC), which is scarce in thylakoid membranes, was similar between the WT and mgd1-2, which suggests that lipid metabolism in the endoplasmic reticulum is not affected by the loss of MGD1 function.
Fig. 3.
Lipid analyses of the mgd1-2 mutant. (A) Composition of polar glycerolipids in WT, mgd1-2, and mgd1-2 + MGD1 plants. SQDG, sulfoquinovosyldiacylglycerol; PG, phosphatidylglycerol; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PC, phosphatidylcholine. Values are mean ± SE from three independent measurements. (B) Fatty acid composition of each lipid extracted from WT (open bars), mgd1-2 (filled bars), and mgd1-2 + MGD1 (stippled bars) plants. Values are mean ± SE from six independent measurements for mgd1-2 and three measurements for WT and mgd1-2 + MGD1 plants.
Impairment of the Photosynthetic System in mgd1-2.
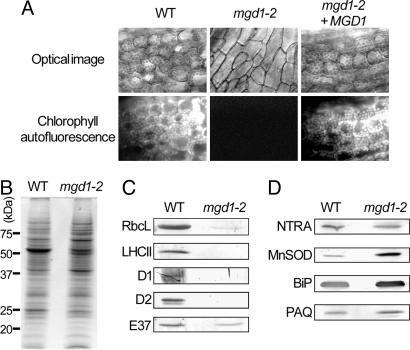
The sucrose requirement for seed germination of mgd1-2 demonstrated that the mutant cannot grow photoautotrophically. As expected, mgd1-2 seedlings showed no chlorophyll autofluorescence, demonstrating that the mutant did not accumulate chlorophyll (Fig. 4A). Measurement of the effective quantum yield of PSII, [maximum fluorescence (Fm′) − fluorescence at a given time (Ft)]/Fm′, revealed that photosynthetic activity was also completely disrupted in the mutant leaves (data not shown). To investigate the effect of the galactolipid deficiency on chloroplast proteins, protein analysis was performed on extracts of mgd1-2 seedlings. Coomassie blue staining of total plant protein was generally similar in mgd1-2 and the WT with some exceptions (Fig. 4B). However, immunoblot analysis revealed that the major photosynthetic proteins of thylakoid membranes, such as D1, D2, and the light-harvesting chlorophyll-protein complex II (LHCII) were undetectable in the mutant (Fig. 4C). The large subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), major stromal proteins, were also largely decreased in the mutant. On the other hand, a major protein of plastid inner envelope membranes, E37, was clearly detected in the mutant although the abundance was reduced compared with that in the WT. In contrast to the plastidic proteins, proteins in cytosol [NADPH-dependent thioredoxin reductase A (NTRA)], mitochondria [manganese-containing superoxide dismutase (MnSOD)], endoplasmic reticulum (BiP), and plasma membranes [plasma membrane aquaporin (PAQ)] were strongly detected even in the mutant (Fig. 4D). These data show that loss of galactolipids results in the disruption of the photosynthetic apparatus but does not affect extraplastidic proteins.
Fig. 4.
Impairment of the photosynthetic system in mgd1-2. (A) Fluorescence microscopic analysis of WT, mgd1-2, and mgd1-2 + MGD1 plants. Bright fluorescence represents chlorophyll accumulation in the chloroplasts. mgd1-2 plants showed no fluorescence. (B) Coomassie brilliant blue staining of an SDS/PAGE of total proteins extracted from WT and mgd1-2 plants. (C and D) Immunoblot analysis of plastidic (C) and extraplastidic (D) proteins from WT and mgd1-2 plants. RbcL, large subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase; NTRA, NADPH-dependent thioredoxin reductase A; MnSOD, manganese-containing superoxide dismutase; PAQ, plasma membrane aquaporin.
Thylakoid Membrane Disruption in mgd1-2.
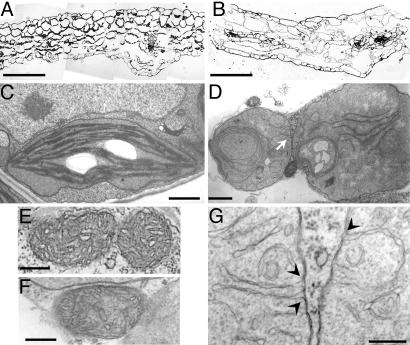
Fig. 2 A and B show that mgd1-2 can form leaves, albeit with severely retarded growth. To further characterize the effect of the MGD1 knockout on leaf morphology, we investigated the leaf structure of the mgd1-2 mutant by light microscopy. Unlike WT leaves, mutant leaves had large intercellular spaces and greatly reduced numbers of mesophyll cells (Fig. 5 A and B). Because photosynthetic cell development is thought to be associated with chloroplast biogenesis (13), it is likely that the chloroplast defect in the mutant caused the abnormal leaf morphology. To assess the effect of galactolipid loss on chloroplast biogenesis, plastid morphology in the mutant was analyzed by transmission electron microscopy (Fig. 5 C and D). The mutant leaves did not contain mature chloroplasts; rather, there were abnormal plastids with severely underdeveloped or no internal membrane structures. All abnormal plastids were round and lacked the lens-like structure typically observed in mature chloroplasts. Because the morphology of other organelles, such as the endoplasmic reticulum and the mitochondria, was similar in the mutant and WT (Fig. 5 E and F and data not shown), this suggests that the MGD1 disruption specifically affected chloroplast biogenesis in the leaves. Although galactolipids account for 80% and 50% of total lipids in the inner and outer envelope of chloroplasts, respectively (1), the plastid envelope appeared normal in mgd1-2, consistent with the immunoblot analysis of E37 (Fig. 4C). The detection of MGDG, DGDG, and SQDG in the mutant (Fig. 3A) provides further evidence that the plastid envelope, which is the only site for synthesis of these lipids, was functional (11, 14). These findings suggest either that galactolipids are not necessary for plastid envelope formation and function or that the remaining galactolipids in the mutant, which result from MGD2/MGD3 activity (9), are sufficient for envelope biogenesis. In contrast to plastid envelopes, proper alignment and stacking of internal chloroplast membranes were scarcely detected in mgd1-2, although unstacked long and short membrane fragments were observed inside the plastids. The long internal membrane fragments often produced scroll-like structures without stacking of the membrane (Fig. 5D). In the mutant plastids, invagination of inner envelope membranes was also observed (Fig. 5G). Such invagination was not seen in WT mature chloroplasts. These observations indicate that MGD1 is indispensable for proper thylakoid membrane biogenesis. Because it has been suggested that interactions between galactolipids and LHCII are required for stacking of thylakoid membranes (3), the deficiency of these components in the mgd1-2 mutant may affect proper assembly and development of thylakoid membranes.
Fig. 5.
Leaf morphology and ultrastructure of mgd1-2 plastids. (A and B) Light microscopy of WT (A) and mgd1-2 mutant (B) leaf sections. (Scale bars: 100 μm.) (C and D) Electron micrographs of plastids from WT (C) and mgd1-2 (D) leaves. (Scale bars: 1.0 μm.) (E and F) Electron micrographs of mitochondria from WT (E) and mgd1-2 (F). (Scale bars: 0.2 μm.) (G) A close-up of the membrane structures indicated by an arrow in D. Arrowheads indicate the sites of inner envelope invagination. (Scale bar: 0.2 μm.)
Discussion
The mgd1-2 seedlings showed a lack of galactolipids and disruption of photosynthetic membranes, leading to the complete impairment of photosynthetic ability and photoautotrophic growth. In Arabidopsis, although there are two galactolipid synthetic pathways in the chloroplast, the MGD1-mediated inner envelope pathway and the MGD2/MGD3-mediated outer envelope pathway (11), the data clearly demonstrate that the inner envelope pathway is predominantly responsible for galactolipid biosynthesis and thylakoid membrane biogenesis in chloroplasts. These findings are fully consistent with previous results from the expression analyses of MGD genes (9, 10). In the mgd1-2 seedlings, neither chlorophyll accumulation nor photosynthetic activity was detected, although a few membrane structures were observed in the plastids (Fig. 5 D and G). X-ray crystallographic analyses of photosynthetic proteins in cyanobacteria demonstrated that MGDG is associated with the core of the reaction center of both PSI and PSII, which may be important for electron transfer in the PS complexes (4–6). Another chloroplast galactolipid, DGDG, has also been found in the crystallized PSII complex and in LHCII (5, 15). Analyses of a dgd1 mutant in which DGDG content is significantly depleted indicated that DGDG plays a crucial role in PSII activity and the stabilization of trimeric LHCII (16–18). The dgd1 mutant also demonstrated the importance of DGDG for the stability and activity of the PSI complex (19, 20). Taken together, these data indicate the direct involvement of MGDG and DGDG in the photosynthetic reaction. The MGD1-null mutation in mgd1-2, which causes a severe depletion of galactolipids, may therefore directly affect the photosynthetic reaction in addition to thylakoid membrane organization.
The biogenesis of thylakoid membranes, an indispensable event for photoautotrophic growth, is closely linked to the development of chloroplasts from other plastids. However, it remains unclear how such a well developed membrane system is constructed inside chloroplasts. The vesicle-inducing protein in plastid 1 (VIPP1) is thought to be involved in thylakoid membrane formation through vesicle transport from the plastid envelope to the thylakoid membranes; indeed, Kroll et al. (21) reported that VIPP1 is a major factor in thylakoid membrane formation because a vipp1 mutant displayed an undeveloped thylakoid membrane phenotype. However, it is uncertain whether vesicle transport organized by VIPP1 is solely responsible for thylakoid membrane formation or whether another system is also required for the formation of the internal membrane core structure. Our results allowed us to revisit an old observation that young undifferentiated plastids displayed inner envelope invagination similar to that of mgd1-2 (22, 23). Transmission electron microscopy analysis of mgd1-2 leaves showed not only unstacked membrane fragments but also inner envelope invagination inside the aberrant chloroplasts (Fig. 5 D and G). Although we cannot eliminate the possibility that inner envelope invagination in mgd1-2 plastids is an artifact created by the inability of the chloroplasts to form proper thylakoids, our results suggest the possibility that invagination of the inner envelope initiates the formation of thylakoid membranes from undifferentiated proplastids.
In addition to seedling growth, embryo development was also severely impaired in the mgd1-2 mutant, indicating the importance of galactolipids during embryogenesis (Fig. 1). No photosynthetic activity was observed in the mgd1-2 white seeds, whereas developing green seeds of the WT possessed substantial photosynthetic capability (data not shown). However, prior studies that examined a chlorophyll-deficient mutant (24) and in vitro embryo development (25) have shown that photosynthesis is not required for embryogenesis; therefore, the defective embryo development observed in the mgd1-2 plants must be independent of the photosynthetic dysfunction caused by any impairment of thylakoid membranes. Because embryogenesis is not relevant to cyanobacteria, the suspected origin of chloroplasts, this suggests that, aside from photosynthetic reactions, higher plants have acquired a requirement for galactolipids during embryogenesis once chloroplasts have developed.
Materials and Methods
Plant Material and Growth Conditions.
All plants were the Columbia ecotype of Arabidopsis thaliana. Sterilized seeds were germinated on liquid or 0.8% (wt/vol) agar-solidified Murashige and Skoog (MS) medium (26) containing 1% (wt/vol) sucrose under continuous light. For comparative analyses, except for the experiment in Fig. 2A, we used 28-day-old seedlings of mgd1-2 and 12-day-old seedlings of WT and MGD1 cDNA-complemented plants (mgd1-2 + MGD1), all of which were at the same developmental stage (i.e., when the fourth leaf was just emerging).
Identification of the T-DNA Insertion Mutant of MGD1.
The T-DNA insertion mutant, mgd1-2 (SALK_002620), was obtained from The Salk Institute Genomic Analysis Laboratory. A T-DNA right border (forward) primer (5′-AATATGAGACTCTAATTGGATACCGAGGGG-3′) and an MGD1-specific (reverse) primer (5′-GAATTCCCACAGAGTTCCATGTTTCACC-3′) were used to screen for plants carrying a T-DNA insert in MGD1 and to determine the insertion site. These primers were also used for genotype analysis, which was performed by 40 cycles of PCR in combination with another MGD1-specific (forward) primer (5′-CATATGCAAAACCCTTCAACGGTAACC-3′).
RT-PCR Analysis.
Total RNA was extracted from 28-day-old mgd1-2 and 12-day-old WT seedlings by using the RNeasy Plant Mini kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. RT was performed by using a RT-PCR kit, RNA PCR kit version 3.0 (Takara Bio, Otsu, Japan) and an oligo(dT)12 primer. The PCR for three atMGD genes was performed as described (9). ACTIN8 transcripts were analyzed as a loading control by using the primers 5′-CTTAGGTATTGCAGACCGTATGAGC-3′ (forward) and 5′-GTTTTTATCCGAGTTTGAAGAGGCT-3′ (reverse). The PCR-amplified samples were electrophoresed by using 1.2% (wt/vol) agarose gels and detected with 0.1 mg/liter ethidium bromide.
Complementation of the mgd1-2-Null Mutant.
To complement the mgd1-2-null mutation, MGD1 cDNA fused with GFP (9) was introduced into pBI121, and the resulting plasmid was transformed into heterozygous mgd1-2 plants according to methods previously described (10). Homozygous mgd1-2 plants carrying MGD1–GFP were isolated by screening the seeds obtained from the self-fertilizing heterozygote on Murashige and Skoog (MS) medium containing 50 μg/ml kanamycin.
Analyses of Embryogenesis.
For embryonic development analyses, ovules from different developmental stages were treated with a clearing solution of chloral hydrate, water, and glycerol (8:2:1, vol/vol) for 1 h. Then the developing embryos were analyzed under a Nomarski microscope (Eclipse 80i; Nikon, Tokyo, Japan). For the light microscopy in Fig. 1 J and K, the cleared seeds were treated with 5% (vol/vol) sodium hypochlorite for an additional 1 h to bleach out the brown pigment of the seed coat, and then the samples were observed under a stereomicroscope (Multi-Viewer System VB-6010 Series; Keyence, Osaka, Japan). To detect β-glucuronidase activity of the MGD1::GUS transformants during embryogenesis, developing ovules were subject to histochemical glucuronidase analysis according to methods previously described (10). The GUS staining was observed under the stereomicroscope (Multi-Viewer System VB-6010 Series).
Lipid Analyses.
Total lipids were extracted from 28-day-old mgd1-2, 12-day-old WT, and 12-day-old MGD1-complemented plants as described (27). The lipids were separated by two-dimensional thin-layer chromatography by using the following solvent systems: chloroform/methanol/7 M ammonia, 15:10:1 (vol/vol), for the first dimension; and chloroform/methanol/acetic acid/water, 170:20:17:3, for the second dimension. Fatty acid detection and measurement of membrane lipid content was carried out as described (28).
Protein Analyses.
Total protein extracts were prepared from mgd1-2 and WT plants by suspension of homogenized seedling tissue in SDS/PAGE sample buffer followed by boiling. The total protein content was quantified as described (29) with BSA as a standard. Total protein (20 μg) was subjected to 12.5% (wt/vol) SDS/PAGE followed by staining with 0.25% (wt/vol) Coomassie brilliant blue. For immunoblot analysis, the separated proteins were electrotransferred onto PROTRAN nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany) and incubated with antibodies against the large subunits of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), LHCII (AgriSera, Vännäs, Sweden), D1, D2 (30), E37 (kindly provided by M. A. Block, Laboratoire de Physiologie Cellulaire Végétale, Grenoble, France), manganese-containing superoxide dismutase (MnSOD) (kindly provided by M. Shibasaka, Okayama University, Okayama, Japan), BiP (31), plasma membrane aquaporin (PAQ) (kindly provided by M. Maeshima, Nagoya University, Nagoya, Japan), and NADPH-dependent thioredoxin reductase A (NTRA) (kindly provided by T. Hisabori, Tokyo Institute of Technology, Yokohama, Japan). Immunoreactive bands were detected by using secondary antibodies coupled to alkaline phosphatase and an alkaline phosphatase substrate kit (Vector Laboratories, Burlingame, CA).
Microscopic Analyses of mgd1-2 Leaves.
To detect chlorophyll autofluorescence, leaf samples were examined by using a fluorescence microscope (Axioplan 2; Carl Zeiss, Oberkochen, Germany) with a long-pass 590-nm filter set (Filter Set 15; Carl Zeiss). For microscopic analyses of leaf sections, leaf segments from 4-week-old mgd1-2 plants were fixed with 4% (vol/vol) paraformaldehyde and 1% (vol/vol) glutaraldehyde in 50 mM cacodylate buffer (pH 7.4) by infiltration under vacuum. Samples were then cut into 1-mm-thick slices and treated for another 2 h with the same fixative. Postfixation treatment of the samples was performed with 0.25% (vol/vol) osmium tetroxide in the same cacodylate buffer for 2 h at room temperature. After washing with the same buffer, the specimens were stained in 0.5% (wt/vol) uranyl acetate for 2 h, with subsequent dehydration in a graded ethanol series at room temperature. The samples were treated with propylene oxide and infiltrated overnight with a 1:1 (vol/vol) solution of propylene oxide/Epon 812 resin (TAAB Laboratories, Reading, U.K.). The samples were then embedded in Epon resin that was allowed to polymerize at 60°C for 48 h. For light microscopic observation, samples were sectioned to 700 nm by using an ultramicrotome (Leica, Wetzlar, Germany). The sections were stained with 1% (wt/vol) toluidine blue and analyzed by using a light microscope (Axioplan 2; Carl Zeiss). For ultrastructural observation, the ultrathin sections (60–70 nm) were stained with 4% (wt/vol) uranyl acetate and 0.4% (wt/vol) lead citrate before analysis by using a transmission electron microscope (model H-7600; Hitachi High-Technologies, Krefeld, Germany) operated at 80 kV.
Acknowledgments
This work was supported by Grants-in-Aid 17051009 and 19039010 for Scientific Research on Priority Areas from the Ministry of Education, Sports, Science, and Culture in Japan. K.K. was supported by a research fellowship of the Japan Society for the Promotion of Science (2006).
Abbreviations
- MGDG
monogalactosyldiacylglycerol
- DGDG
digalactosyldiacylglycerol
- PSn
photosystem n
- LHCII
light-harvesting chlorophyll-protein complex II.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Block MA, Dorne AJ, Joyard J, Douce R. J Biol Chem. 1983;258:13281–13286. [PubMed] [Google Scholar]
- 2.Dörmann P, Benning C. Trends Plant Sci. 2002;7:112–118. doi: 10.1016/s1360-1385(01)02216-6. [DOI] [PubMed] [Google Scholar]
- 3.Lee AG. Curr Biol. 2000;10:R377–R380. doi: 10.1016/s0960-9822(00)00477-2. [DOI] [PubMed] [Google Scholar]
- 4.Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N. Nature. 2001;411:909–917. doi: 10.1038/35082000. [DOI] [PubMed] [Google Scholar]
- 5.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 6.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Biochim Biophys Acta. 2007;1767:509–519. doi: 10.1016/j.bbabio.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 7.Shimojima M, Ohta H, Iwamatsu A, Masuda T, Shioi Y, Takamiya K. Proc Natl Acad Sci USA. 1997;94:333–337. doi: 10.1073/pnas.94.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelly AA, Dörmann P. Curr Opin Plant Biol. 2004;7:262–269. doi: 10.1016/j.pbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 9.Awai K, Maréchal E, Block MA, Brun D, Masuda T, Shimada H, Takamiya K, Ohta H, Joyard J. Proc Natl Acad Sci USA. 2001;98:10960–10965. doi: 10.1073/pnas.181331498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kobayashi K, Awai K, Takamiya K, Ohta H. Plant Physiol. 2004;134:640–648. doi: 10.1104/pp.103.032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benning C, Ohta H. J Biol Chem. 2005;280:2397–2400. doi: 10.1074/jbc.R400032200. [DOI] [PubMed] [Google Scholar]
- 12.Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J. Proc Natl Acad Sci USA. 2000;97:8175–8179. doi: 10.1073/pnas.100132197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sulmon C, Gouesbet G, Couée I, Cabello-Hurtado F, Cavalier A, Penno C, Zaka R, Bechtold N, Thomas D, El Amrani A. Gene. 2006;382:88–99. doi: 10.1016/j.gene.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 14.Tietje C, Heinz E. Planta. 1998;206:72–78. [Google Scholar]
- 15.Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W. Nature. 2004;428:287–292. doi: 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- 16.Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. Plant Cell. 1995;7:1801–1810. doi: 10.1105/tpc.7.11.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hölzl G, Witt S, Kelly AA, Zähringer U, Warnecke D, Dörmann P, Heinz E. Proc Natl Acad Sci USA. 2006;103:7512–7517. doi: 10.1073/pnas.0600525103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steffen R, Kelly AA, Huyer J, Dörmann P, Renger G. Biochemistry. 2005;44:3134–3142. doi: 10.1021/bi048465f. [DOI] [PubMed] [Google Scholar]
- 19.Guo J, Zhang Z, Bi Y, Yang W, Xu Y, Zhang L. FEBS Lett. 2005;579:3619–3624. doi: 10.1016/j.febslet.2005.05.049. [DOI] [PubMed] [Google Scholar]
- 20.Ivanov AG, Hendrickson L, Krol M, Selstam E, Oquist G, Hurry V, Huner NP. Plant Cell Physiol. 2006;47:1146–1157. doi: 10.1093/pcp/pcj089. [DOI] [PubMed] [Google Scholar]
- 21.Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mühlethaler K, Frey-Wyssing A. J Biophys Biochem Cytol. 1959;6:507–512. [PMC free article] [PubMed] [Google Scholar]
- 23.Vothknecht UC, Westhoff P. Biochim Biophys Acta. 2001;1541:91–101. doi: 10.1016/s0167-4889(01)00153-7. [DOI] [PubMed] [Google Scholar]
- 24.Frick G, Su Q, Apel K, Armstrong GA. Plant J. 2003;35:141–153. doi: 10.1046/j.1365-313x.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- 25.Sauer M, Friml J. Plant J. 2004;40:835–843. doi: 10.1111/j.1365-313X.2004.02248.x. [DOI] [PubMed] [Google Scholar]
- 26.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 27.Bligh EG, Dyer WJ. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi K, Masuda T, Takamiya K, Ohta H. Plant J. 2006;47:238–248. doi: 10.1111/j.1365-313X.2006.02778.x. [DOI] [PubMed] [Google Scholar]
- 29.Bensadoun A, Weinstein D. Anal Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- 30.Shimada H, Ohno R, Shibata M, Ikegami I, Onai K, Ohto MA, Takamiya K. Plant J. 2005;41:627–637. doi: 10.1111/j.1365-313X.2004.02326.x. [DOI] [PubMed] [Google Scholar]
- 31.Hatano K, Shimada T, Hiraiwa N, Nishimura M, Hara-Nishimura I. Plant Cell Physiol. 1997;38:344–351. doi: 10.1093/oxfordjournals.pcp.a029172. [DOI] [PubMed] [Google Scholar]