Abstract
Atom-resolved real-time studies of kinetic processes in proteins have been hampered in the past by the lack of experimental techniques that yield sufficient temporal and atomic resolution. Here we present band-selective optimized flip-angle short transient (SOFAST) real-time 2D NMR spectroscopy, a method that allows simultaneous observation of reaction kinetics for a large number of nuclear sites along the polypeptide chain of a protein with an unprecedented time resolution of a few seconds. SOFAST real-time 2D NMR spectroscopy combines fast NMR data acquisition techniques with rapid sample mixing inside the NMR magnet to initiate the kinetic event. We demonstrate the use of SOFAST real-time 2D NMR to monitor the conformational transition of α-lactalbumin from a molten globular to the native state for a large number of amide sites along the polypeptide chain. The kinetic behavior observed for the disappearance of the molten globule and the appearance of the native state is monoexponential and uniform along the polypeptide chain. This observation confirms previous findings that a single transition state ensemble controls folding of α-lactalbumin from the molten globule to the native state. In a second application, the spontaneous unfolding of native ubiquitin under nondenaturing conditions is characterized by amide hydrogen exchange rate constants measured at high pH by using SOFAST real-time 2D NMR. Our data reveal that ubiquitin unfolds in a gradual manner with distinct unfolding regimes.
Keywords: hydrogen/deuterium exchange, molecular kinetics, α-lactalbumin, ubiquitin
A detailed description of the structural changes occurring during the folding and unfolding of proteins still remains a challenging objective in biophysics. The understanding of the fundamental mechanisms of the folding process will also shed light on the factors leading to protein misfolding responsible for neurodegenerative diseases such as Alzheimer's and Parkinson's diseases (1). The ideal method to study protein folding/unfolding provides structural information at atomic resolution and is sensitive to changes occurring on time scales ranging from microseconds to minutes, a task that no single technique is able to fulfill. NMR spectroscopy is especially well adapted to obtain detailed atomistic information about the mechanisms, kinetics, and energetics of the folding/unfolding process for virtually every nuclear site in the protein. NMR methods are sensitive to molecular dynamics occurring over a wide range of time scales (Fig. 1a). Although steady-state NMR methods are well suited to characterize equilibrium molecular dynamics occurring on a subsecond time scale (2, 3), unidirectional processes are best studied by real-time NMR (4, 5), where a series of NMR spectra is recorded during the reaction (Fig. 1b). Two difficulties have so far hampered the widespread application of real-time NMR to the study of protein folding. The first problem is the low intrinsic sensitivity of NMR at ambient temperature, a consequence of the small transition energies involved, compared with other spectroscopic techniques. Recent progress in NMR technology (magnetic field strength, cryogenic probes, and electronics, for example), however, has greatly enhanced the sensitivity of NMR experiments, allowing acquisition of 1D spectra of proteins at submillimolar concentration at ≈1–10 s−1 rates. The second limitation concerns the time requirements to record higher-dimensional (≥2D) NMR spectra indispensable to resolve the large number of nuclear resonances in a protein. Whereas a 1D NMR spectrum can be obtained in <1 s within a single scan by physical detection of the NMR time domain signal induced in a receiver coil, the recording of a 2D NMR data set requires numerous repetitions (typically ≈100) of the basic pulse sequence to sample the evolution of the nuclear spins in the additional time dimension (6). In the past, this requirement has limited the application of real-time 2D NMR methods to relatively slow kinetic processes with characteristic time constants of minutes to hours. A major challenge for real-time NMR is therefore to achieve reduced acquisition times that will allow extension of real-time NMR methods to the missing NMR time window of seconds to minutes (Fig. 1a). Doing so will make the technique applicable to the study of a larger variety of molecular kinetics. Here we introduce band-selective optimized flip-angle short transient (SOFAST) real-time 2D NMR, a method that allows site-resolved studies of kinetic processes in proteins with a time resolution down to a few seconds. We demonstrate the potential of this method for the study of protein folding and unfolding reactions.
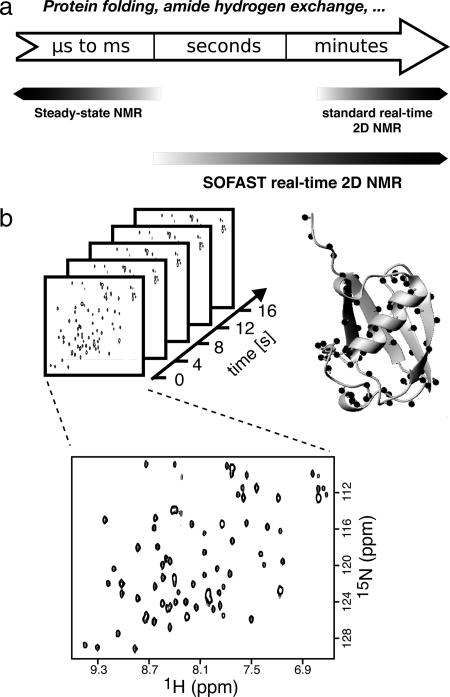
Fig. 1.
SOFAST real-time 2D NMR closes the seconds time gap for NMR studies of protein dynamics. (a) Time scales of biophysical processes such as protein folding or amide hydrogen exchange, and NMR methods available to study the kinetics of these processes at atomic resolution. (b) Principle of SOFAST real-time 2D NMR. A series of 2D FTA-SOFAST-HMQC spectra is recorded after initiating a kinetic change in the protein state. Each cross-peak reports on the local structure and dynamics at the site of a single amide group. The bottom spectrum has been recorded in an experimental time of 4 s on a 0.2 mM 15N-labeled sample of ubiquitin at a magnetic field strength of 18.8 T by using a cryogenically cooled probe.
Results and Discussion
SOFAST Real-Time 2D NMR Spectroscopy.
To overcome the inherent time limitation of 2D NMR spectroscopy, two conceptually different approaches can be envisaged. First, the number of scans required to sample the multidimensional time space may be reduced by using spectral aliasing, nonlinear data sampling techniques, spatial frequency encoding, or Hadamard-type frequency-space spectroscopy (7, 8). Second, accelerating the recovery of spin polarization between scans by means of optimized pulse sequences (9–11) or the addition of relaxation agents to the solution (12, 13) allows higher repetition rates of the pulse sequence. Here, we propose a fluid turbulence-adapted (FTA) version of 1H-15N 2D SOFAST-heteronuclear multiple quantum coherence (HMQC) (10, 14) to follow a kinetic process for individual amide sites in a protein [see supporting information (SI) Fig. 4]. Longitudinal relaxation optimization in FTA-SOFAST-HMQC allows the use of scan times of <100 ms without a significant reduction in sensitivity (signal to noise per unit time), resulting in experimental times on the order of 5 s per 2D 1H-15N correlation spectrum, which shows one correlation peak for each nonproline residue of the protein. The advantage of the SOFAST approach with respect to other fast 2D NMR techniques, such as Hadamard or spatial frequency encoding, is the higher sensitivity provided by this technique for the short acquisition times of a few seconds. High-quality 2D spectra are obtained by the FTA-SOFAST-HMQC experiment on a modern high-field NMR spectrometer for protein samples in the submillimolar concentration range, as illustrated in Fig. 1b for a 0.2 mM sample of ubiquitin recorded in a 4-s acquisition time. If faster sampling of the kinetic time domain is desired, this minimal experimental time required for FTA-SOFAST-HMQC can be reduced further to ≈1 s by using computer-optimized spectral aliasing in the 15N dimension (15).
To benefit fully from the increased time resolution provided by the FTA-SOFAST-HMQC experiment for real-time kinetic studies, a device is required that allows initiation of the kinetic event inside the magnet. The triggering of kinetic reactions can be achieved either by a sudden change of the protein state itself, e.g., by a photo-induced excitation or cleavage of chemical bonds (16, 17), or by a change of environment, such as the solvent composition, temperature, or pH (4, 18). Here, we opted for an initiation of the reaction by an abrupt change in the solvent conditions achieved by rapid mixing of two solutions inside the NMR magnet (5, 19). The fast mixing device used for this work (SI Fig. 5a) allows complete mixing in <300 ms as demonstrated by dye injection experiments into water solutions (SI Fig. 5b). A drawback of the fast injection is the presence of turbulences of the liquid over times significantly longer than the mixing time. Many modern NMR experiments make use of pairs of pulsed-field gradients for spectral artifact suppression. In the presence of bulk liquid motion, incomplete refocusing of spin coherence caused by translational diffusion during the time delay between the two pulsed-field gradients yields uniformly reduced NMR signal intensities. The FTA-SOFAST-HMQC experiment has been optimized for minimal signal loss in the presence of bulk liquid motion, yielding dead times after the injection of <2 s (SI Fig. 6). We have chosen two conceptually different applications to demonstrate the potential of SOFAST real-time 2D NMR to provide accurate measures of kinetic rate constants for a large number of amide sites. The first example concerns the folding of α-lactalbumin from a molten globular (MG) to the native state. In the second application, the unfolding kinetics experienced by individual residues of ubiquitin under native equilibrium conditions are investigated.
Conformational Transition Kinetics of α-Lactalbumin from an MG to the Native State Studied in Real Time.
The protein α-lactalbumin serves as a model system to investigate the structural transition from a partially folded to the native state by using SOFAST real-time 2D NMR. This protein has been used extensively as a model system for protein-folding studies. The structure of native α-lactalbumin (14 kDa) comprises two domains, one containing four α-helices and a short 310 helix, and the other containing a three-stranded β-sheet and another 310 helix. Here, we focused on the Ca-free (apo) form of α-lactalbumin. Under destabilizing conditions such as low pH, addition of cosolvents, high temperature, or combinations thereof, apo-α-lactalbumin exists in an MG state. The MG state is characterized by the absence of long-lived tertiary structure, but it still contains a high degree of secondary structure (20, 21), with a radius of gyration only ≈10% larger than the native state (22, 23). The MG state of α-lactalbumin has recently attracted considerable interest when it was realized that it may act as an important component causing apoptosis of tumor cells (24).
Pioneering work on the use of time-resolved NMR methods for monitoring protein refolding was performed on α-lactalbumin (5, 23, 25–27). One-dimensional 1H NMR spectra recorded during the refolding reaction of apo-α-lactalbumin provided evidence that the MG state, accumulated during the early stages of the folding reaction, shows similar spectral characteristics (poorly dispersed and broad resonances) and therefore similar dynamic properties of the conformational ensemble to those observed for the MG state stabilized at acidic pH (25). Additional diffusion-edited and NOE transfer NMR measurements allowed monitoring the compactness of the protein and the establishing of native tertiary contacts, respectively (23, 27). The folding kinetics of apo-α-lactalbumin have also been studied previously at a residue level by line shape analysis of individual cross-peaks detected in a single 2D NMR spectrum recorded during the refolding event (26). This technique, however, is limited to relatively slow kinetics (several minutes) and is prone to large experimental uncertainties. In the study of Balbach et al. (26), folding rates for a total of 25 backbone amides could be quantified with an estimated experimental error of ≈25%. Because of the small number and the high uncertainties of the measured rate constants, these data did not allow definite exclusion of the presence of differential folding kinetics along the polypeptide chain. To illustrate the improvements provided by SOFAST real-time 2D NMR for the measurement of residue-specific folding rates, we monitored the same refolding reaction as Balbach et al. from the MG state, formed at pH 2, to the native state. Refolding was initiated by a sudden pH jump (from 2 to 8), and the reaction was monitored by a series of FTA-SOFAST-HMQC spectra recorded at a 0.1 s−1 rate. Spectra acquired before injection, immediately after, and 2 min after injection are shown in Fig. 2a. The center spectrum in Fig. 2a provides a snapshot of the transient MG state under native conditions. The broad and weak signals in this spectrum are indicative of large-scale dynamics occurring on a micro- to millisecond time scale. These data are in agreement with this transient MG state being a compact, highly dynamic ensemble of conformational states. All NMR signals observed during the folding process can be assigned either to the MG or the native state. No additional peaks indicative of a significantly populated folding intermediate were detected, in agreement with previous findings (23, 26, 27). The refolding kinetics could be quantified for 92 of 121 backbone amide sites in the protein from intensity measurements of well resolved cross-peaks in the 1H-15N correlation spectra (Fig. 2b). In addition, and in contrast to the studies by Balbach et al. (23, 26, 27), the intensity decay of five cross-peaks characteristic for the MG state could be quantified. Under the experimental conditions chosen (15°C, pH 8, Ca2+-free), the NMR intensity decay and buildup curves can be fitted to monoexponential functions. Fitting the data to more complex, e.g., biexponential or stretched exponential functions, does not yield statistically significant improvements. The folding time constants measured for individual amide sites of the native protein and the MG state are identical (τf = 109 ± 5 s) within the experimental uncertainty. The obtained folding kinetics are consistent with results from fluorescence measurements (SI Fig. 7), yielding a characteristic time constant of 110 ± 10 s for the establishment of native tertiary structure. The good agreement with the fluorescence measurements demonstrates the accuracy obtained by SOFAST real-time 2D NMR experiments.
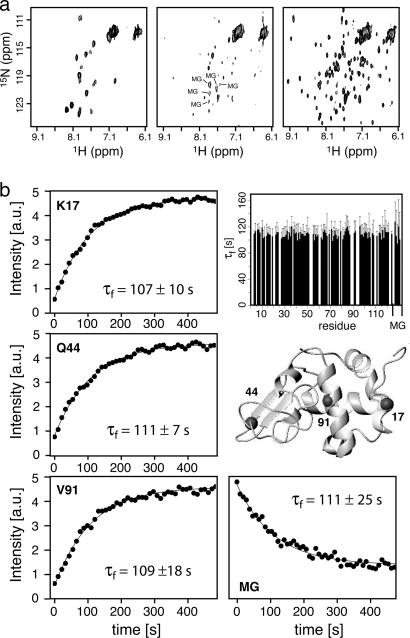
Fig. 2.
Folding of α-lactalbumin studied by SOFAST real-time 2D NMR. (a) FTA-SOFAST-HMQC spectra of bovine α-lactalbumin at pH 2.0 (Left), immediately after a sudden pH jump to pH 8.0 that triggers folding (Center), and 120 s after injection (Right). Each spectrum shows the sum of two acquisitions of 10.9-s duration. Peaks corresponding to the MG state that disappear during folding are annotated. (b) Refolding kinetics of bovine α-lactalbumin from the MG state to the native state. The measured peak intensities are plotted as a function of the folding time. Shown are three residues situated in loop (K17), β-sheet (Q44), and α-helical regions (V91) that are indicated on the structure (Protein Data Bank ID code 1F6R). In addition, the signal decay observed for a peak assigned to the MG state is shown. Solid lines represent best fits to a three-parameter exponential function. A histogram shows the measured folding time constants for 92 residues in the native state as well as the 5 rates measured for the disappearance of the MG state.
The MG state of apo-α-lactalbumin explores a large conformational space, possibly including the presence of native and nonnative secondary structures (20). Therefore, the transition from this MG state to the native state requires a large decrease in conformational entropy, known as entropy bottleneck effect (28, 29), which is the main reason for the significantly slower folding kinetics observed for the apo form compared with the holo form of α-lactalbumin (30). The high precision of rate constants (error of ≈5–10%) obtained in our work for a large number of individual amide sites allows us to draw some conclusions on the energy landscape and folding pathways. The finding of equal kinetic rates for the buildup of the native state and the disappearance of the MG state indicates a transition between only two states, the MG ensemble representing a large number of conformational substates interconverting on the micro- to millisecond time scale, and the native state. In other words, the establishment of the native tertiary structure is not accompanied by a change in the structural and dynamic properties of the MG ensemble as folding proceeds. These observations are in agreement with the assumption of a smooth energy landscape where the folding rate is controlled by a single transition state ensemble originating from the conformational entropy bottleneck effect. The existence of a unique transition state is in agreement with conclusions drawn from other biophysical folding experiments (30).
Ubiquitin Unfolding Kinetics Under Equilibrium Conditions Revealed by EX1 Amide Hydrogen Exchange Measurements.
To illustrate further the potential of SOFAST real-time 2D NMR to detect differential kinetic behavior along the polypeptide chain, we have investigated the unfolding kinetics of human ubiquitin under nondenaturing equilibrium conditions by amide hydrogen/deuterium (H/D) exchange methods. H/D exchange is a powerful tool for the study of protein structure, folding, unfolding, and binding (31, 32). The exchange between amide and solvent hydrogens requires that the amide group is in an exchange-competent, solvent-accessible conformation, which can be achieved by local, subglobal, or global protein unfolding fluctuations (“opening reactions”) that transiently break the exchange-protecting structures, e.g., hydrogen bonds. The measurement of amide H/D exchange rates thus provides residue-specific information on the otherwise invisible manifold of partially or globally unfolded excited conformational states populated at a very low level. Despite their low population under native conditions, such high-energy states may be crucial for protein function (33, 34), or they may represent intermediate states on the protein folding pathway (35). Especially interesting are H/D exchange measurements in the so-called EX1 regime, where the measured exchange rate constants directly reflect the kinetics of the unfolding reaction at individual amide sites (32). EX1 conditions are generally reached at high pH. However, for most proteins, H/D exchange at high pH is too fast to measure by conventional 2D real-time NMR. Therefore, previous EX1 studies had to focus on a small number of well protected amide sites. It has been shown that the EX1 exchange rates measured for a few slowly exchanging amide sites in ubiquitin correspond to the rate of global unfolding observed with other biophysical methods (36). The SOFAST real-time 2D NMR method, presented here, allows extension of the H/D exchange measurements under EX1 conditions to a significantly larger number of amide sites, yielding a more comprehensive picture of the potentially heterogeneous unfolding processes along the polypeptide chain. Representative exchange curves measured for the 76-residue protein ubiquitin at pH 11.95 and 25°C are shown in Fig. 3. For all amides, even in the most protected parts of the protein, exchange time constants τex <200 s were observed. The high quality of the data allowed accurate quantification of exchange kinetics down to τex ≈5 s, with a detection limit of τex ≈1 s. This time resolution proved to be sufficient to measure exchange rates for amides in secondary structural elements, whereas H/D exchange in loop regions was generally completed during the dead time of the experiment. In the top drawing of Fig. 3, the measured exchange rates are color-coded on the ribbon structure of ubiquitin. The slowest H/D exchange is observed for residues in strands I and II of the β-sheet (residues 3–5 and 15) and the central part of the α-helix (residue 27), whereas amide hydrogens located in other parts of the molecule, comprising strands III–V, exchange much faster. Interestingly, a gradual increase in the kinetics of opening fluctuations across the β-sheet is observed, indicating noncooperative unfolding events in this β-sheet, reminiscent of a hydrophobic zipper folding mechanism (37).
Fig. 3.
H/D exchange data obtained for human ubiquitin at pH 11.95 by using SOFAST real-time 2D NMR. (Upper) The measured exchange rates are color-coded on the ubiquitin structure. (Lower) Examples of exchange curves corresponding to different exchange regimes together with the fitted exchange time constants. For residues color-coded in white, no signal decay was observed because the cross-peak intensity has decreased to its plateau value during the dead time of the experiment.
Over the past 20 years, ubiquitin has been used as a model system for the study of protein stability and folding with a variety of biophysical techniques, denaturation methods, protein engineering studies, and computational approaches (38). Therefore, it is interesting to compare the results presented here with previous findings on the thermodynamic stability, folding, and unfolding kinetics of ubiquitin under different experimental conditions. H/D exchange rates measured under EX2 conditions (39, 40) provide a measure of the thermodynamic equilibrium constant K = kopen/kclose, rather than a kinetic rate constant as obtained under EX1 conditions. Interestingly, the residue-specific equilibrium constants also show differential stability among the secondary structural elements of ubiquitin, with notably the first two β-strands and the α-helix being the most stable parts. The kinetic information obtained from H/D exchange measurements under EX1 conditions complements these former results, indicating that the differential thermodynamic stability observed for ubiquitin is mainly determined by the unfolding kinetics (opening rates). Differential noncooperative unfolding has also been observed by NMR studies of ubiquitin under denaturing conditions. The populations of nonnative conformational states can be enhanced by changing the temperature, pressure, or pH, or by adding alcohol cosolvents. A cold-denaturation study by Wand et al. (41, 42) shows for reverse micelle-encapsulated ubiquitin that the mixed β-sheet is progressively destabilized from the C-terminal side in the temperature range from −20° to −30°C. In an NMR study by Cordier and Grzesiek (43), the change in hydrogen bond strength with increasing temperature was measured. The N terminus of β-strand V was found to be the least thermally stable, whereas strands I and II remain stable even at elevated temperatures. Spectral changes observed upon high-pressure denaturation at ambient temperature indicate the presence of a partially unfolded intermediate in which the C-terminal part (residues 70–76) as well as residues 33–42 and residue 8 are denatured (44). The higher stability of the secondary structural elements in the N-terminal part of ubiquitin (βI-βII strand and α-helix) is also evidenced by the observation of a partially structured state (A state) at low pH in a 60%/40% methanol/water mixture (45). In the A state, the N-terminal part of ubiquitin retains a native-like secondary structure, whereas the C-terminal part undergoes a conformational transition form the native state to an ensemble of conformational states with high helical propensity (46). Finally, a recent time-resolved infrared (IR) study (47) revealed a complex thermal unfolding behavior of ubiquitin spanning a wide range of time scales, indicating a gradual, rather than a cooperative, unfolding process. All experimental data from both equilibrium and transient unfolding studies support the picture of noncooperative unfolding events taking place in the native environment. The C-terminal part, comprising β-strands III–V, is significantly less stable, with unfolding rates that are up to 2 orders of magnitude higher than those observed for β-strands I and II, and the α-helix in the N-terminal part. The observed conformational dynamics under native conditions and the presence of partially unfolded conformational states may be of importance for the biological function of ubiquitin because it has been shown that incorporation of a disulfide bridge between residues 4 and 66, stabilizing the connection between strands I and V of the β-sheet, leads to a 70–80% decrease of activity in signaling proteolysis (48).
SOFAST real-time 2D NMR closes the gap (seconds to minutes) on the kinetic time scale (see Fig. 1), which makes possible H/D exchange measurements for a large number of amide sites at any desired pH by 2D NMR methods. The combined interpretation of H/D exchange rates measured under EX1 as well as the more common EX2 conditions presents an attractive tool to access residue-specific kinetic folding and unfolding rates of proteins under native (physiological) conditions. In concert with other NMR data, providing the same atomic resolution, e.g., spin relaxation measurements or NMR spectra recorded under denaturing conditions, H/D exchange measurements will help us to gain a deeper insight into the nature of partially unfolded conformational states present under native conditions and the rates of interconversion between those excited states and the native state.
In summary, we have demonstrated that SOFAST real-time 2D NMR allows the accurate measurement of kinetic rate constants up to ≈1 s−1 simultaneously for a large number of amide sites in a protein. The time frame ranging from a few seconds to roughly a minute used to be inaccessible to conventional 2D NMR methods. We have illustrated the potential of SOFAST real-time 2D NMR to provide accurate residue-specific kinetic information on protein folding and unfolding events either by direct real-time studies of folding/unfolding or indirectly by real-time measurements of H/D exchange kinetics. The kinetic rates measured for the refolding of α-lactalbumin are uniform throughout the protein, indicating a smooth energy landscape for the transition from the MG to the native state. On the contrary, the kinetic data collected for the protein ubiquitin indicate a highly noncooperative unfolding behavior under native conditions, in agreement with previous results from NMR and IR spectroscopy. The methodology presented here can be easily applied to protein systems with molecular masses ranging between ≈5 and ≈20 kDa where deuteration is not a prerequisite for obtaining well resolved 1H-15N correlation spectra. Here, we have focused on the application of SOFAST real-time 2D NMR to the study of protein folding and unfolding, but the method also offers opportunities for residue-specific kinetic measurements of other unidirectional events occurring on a time scale of seconds, such as binding, enzymatic reactions, and chemical exchange. Combined with recent advances toward NMR studies of proteins inside intact living cells (49, 50), SOFAST real-time 2D NMR may also become a powerful experimental tool for in situ time- and site-resolved observation of kinetic events, such as protein folding and posttranslational modifications in a cellular environment.
Materials and Methods
SOFAST Real-Time 2D NMR.
All NMR experiments were performed on an INOVA spectrometer (Varian, Palo Alto, CA) operating at 800-MHz 1H frequency and equipped with a cryogenic probe. To keep the dead time as short as possible, the acquisition of the first spectrum was started after the dead time of the mixing required to stabilize the fluid turbulence, which was ≈2 s (see SI Fig. 6). To achieve high time resolution while retaining good spectral quality without the need of advanced spectral processing, we used alternate phase cycling in subsequent spectra as follows. Each experiment was performed with only one scan per increment in t1, ensuring highest repetition rates of experiments, and the sign of the 15N excitation pulse phase and receiver phase in FTA-SOFAST-HMQC was alternated between subsequent experiments. Adding two succeeding spectra (1 and 2, 2 and 3, 3 and 4,…) along the kinetic time dimension removes spectral artifacts such as axial peaks or t1 noise at the water frequency and improves the base line, allowing a more accurate and precise measurement of spectral parameters, such as peak positions and intensities. This procedure does not change the kinetic information contained in the data and the time resolution as illustrated in SI Fig. 8. The only price to pay is an effective dead time that is longer by half the duration of one basic 2D experiment. All data were extracted as peak intensities and fit to a three-parameter exponential function. Uncertainties in the obtained fit parameters were estimated from Monte Carlo simulations by using 100 data sets generated by varying the intensity value of each data point within three times the spectral noise.
Real-Time Folding of Bovine α-Lactalbumin.
Nitrogen-15 labeled bovine α-lactalbumin (M90V) was expressed and purified as described previously (51). The MG state was prepared by dissolving 8.8 mg of protein in 350 μl of H2O/D2O (10:1) and adjusting the pH to 2.0 with 1 M HCl solution. Fifty microliters of a refolding buffer solution [H2O/D2O (10:1)] containing 800 mM Tris and 80 mM EDTA (pH 9.1) was loaded in the injection device, and refolding was initiated inside the spectrometer by rapid injection into the protein solution. The folding reaction was followed by a series of FTA-SOFAST-HMQC experiments of 10.9-s duration with a scan time of 130 ms and 40 complex data points in t1 (80 scans per 2D spectrum). The final pH measured after the folding reaction was 8.0, and the final protein concentration was 1.55 mM. NMR assignments were taken from ref. 27. The reproducibility of the real-time folding data has been evaluated by repeating the experiment twice. The difference in the rate constants obtained from the two measurement series is smaller than the experimental error estimated from Monte Carlo simulations of a single data set.
Amide Hydrogen Exchange Measurements in Ubiquitin.
For the SOFAST real-time 2D NMR amide hydrogen exchange measurements, 2.5 mg of human ubiquitin was dissolved in 50 μl of H2O buffer containing 50 mM glycine and loaded into the injection device. H/D exchange was initiated by injection of the protein solution into 350 μl of D2O buffer, resulting in a final protein concentration of 0.7 mM. The resulting pH, corrected for the electrode isotope effect, was 11.95. The decay of peak intensities caused by exchange was followed by recording a series of FTA-SOFAST-HMQC spectra of 6.7-s duration with a scan time of 112 ms, and recording 30 complex data points (60 scans). NMR assignments at pH 11.95 were obtained from published data at pH 6.6 (52), and a series of 1H-15N SOFAST-HMQC spectra was recorded in the pH range 7–12.
SOFAST NMR makes use of longitudinal amide 1H relaxation enhancement to increase the experimental sensitivity for high repetition rates of the experiment (10, 14). Shorter spin-lattice relaxation times are achieved by using amide-proton selective radiofrequency pulses that leave the polarization of other 1H spins, e.g., aliphatic and water 1H, unperturbed. The energy put into the system is then efficiently dissipated within the dipolar-coupled 1H spin network by spin diffusion (NOE) effects. In the course of H/D exchange monitored by real-time 2D NMR, amide hydrogens are progressively exchanged for deuterons. This exchange alters the 1H spin coupling network in the protein from one 2D experiment to the other, which influences the relaxation properties of the amide 1H spins and raises the question about the accuracy of the measured exchange rates by using SOFAST real-time 2D NMR. We have addressed this issue by both experiment and computer simulations. The results, shown in SI Fig. 9, indicate that the systematic error introduced by the changing 1H spin network remains small compared with the statistical uncertainty and can be safely neglected.
Supplementary Material
Acknowledgments
We thank Dr. Keith Brew (University of Miami) and Dr. Stephan Grzesiek (Biozentrum Basel) for providing the expression vectors of bovine α-lactalbumin and ubiquitin, and Isabel Ayala and Cécile Giustini (Institut de Biologie Structurale Grenoble) for help with the preparation of the isotope-labeled protein samples. This work was supported by the Commissariat à l'Energie Atomique, the Centre National de la Recherche Scientifique, the French Research Agency Agence National de la Recherche (ANR) Grant ANR-05-JCJC-0077, and European Commission (EU-NMR) Grant RII3-026145. P.S. acknowledges support from the French ministry of Education, Research, and Technology.
Abbreviations
- FTA
fluid turbulence-adapted
- HMQC
heteronuclear multiple quantum coherence
- MG
molten globule
- SOFAST
band-selective optimized flip-angle short transient.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702069104/DC1.
References
- 1.Dobson CM. Semin Cell Dev Biol. 2004;15:3–16. doi: 10.1016/j.semcdb.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Palmer AG. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 3.Mittermaier A, Kay LE. Science. 2006;312:224–228. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 4.Zeeb M, Balbach J. Methods. 2004;34:65–74. doi: 10.1016/j.ymeth.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 5.Van Nuland NAJ, Forge V, Balbach J, Dobson CM. Acc Chem Res. 1998;31:773–780. [Google Scholar]
- 6.Ernst R, Bodenhausen G, Wokaun G. Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Oxford: Oxford Univ Press; 1987. pp. 124–131. [Google Scholar]
- 7.Freeman R, Kupce E. J Biomol NMR. 2003;27:101–113. doi: 10.1023/a:1024960302926. [DOI] [PubMed] [Google Scholar]
- 8.Malmodin D, Billeter M. Prog Nucl Magn Reson Spectrosc. 2005;46:109–129. [Google Scholar]
- 9.Pervushin K, Vögeli B, Eletsky A. J Am Chem Soc. 2002;124:12898–12902. doi: 10.1021/ja027149q. [DOI] [PubMed] [Google Scholar]
- 10.Schanda P, Brutscher B. J Am Chem Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 11.Schanda P, Van Melckebeke H, Brutscher B. J Am Chem Soc. 2006;128:9042–9043. doi: 10.1021/ja062025p. [DOI] [PubMed] [Google Scholar]
- 12.Hiller S, Wider G, Etezady-Esfarjani T, Horst R, Wüthrich K. J Biomol NMR. 2005;32:61–70. doi: 10.1007/s10858-005-3070-8. [DOI] [PubMed] [Google Scholar]
- 13.Cai S, Seu C, Kovacs Z, Sherry AD, Chen Y. J Am Chem Soc. 2006;128:13474–13478. doi: 10.1021/ja0634526. [DOI] [PubMed] [Google Scholar]
- 14.Schanda P, Kupce E, Brutscher B. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 15.Lescop E, Schanda P, Rasia R, Brutscher B. J Am Chem Soc. 2007;129:2756–2757. doi: 10.1021/ja068949u. [DOI] [PubMed] [Google Scholar]
- 16.Rubinstenn G, Vuister GW, Mulder FAA, Dux PE, Boelens R, Hellingwerf KJ, Kaptein R. Nat Struct Biol. 1998;5:568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 17.Wenter P, Furtig B, Hainard A, Schwalbe H, Pitsch S. Angew Chem Int Ed Engl. 2005;44:2600–2603. doi: 10.1002/anie.200462724. [DOI] [PubMed] [Google Scholar]
- 18.Buevich AV, Dai QH, Liu XY, Brodsky B, Baum J. Biochemistry. 2000;39:4299–4308. doi: 10.1021/bi992584r. [DOI] [PubMed] [Google Scholar]
- 19.Mok KH, Nagashima T, Day IJ, Jones JA, Jones CJV, Dobson CM, Hore PJ. J Am Chem Soc. 2003;125:12484–12492. doi: 10.1021/ja036357v. [DOI] [PubMed] [Google Scholar]
- 20.Troullier A, Reinstadler D, Dupont Y, Naumann D, Forge V. Nat Struct Biol. 2000;7:78–86. doi: 10.1038/71286. [DOI] [PubMed] [Google Scholar]
- 21.Mok KH, Nagashima T, Day LJ, Hore PJ, Dobson CM. Proc Natl Acad Sci USA. 2005;102:8899–8904. doi: 10.1073/pnas.0500661102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kataoka M, Kuwajima K, Tokunaga F, Goto Y. Protein Sci. 1997;6:422–430. doi: 10.1002/pro.5560060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balbach J. J Am Chem Soc. 2000;122:5887–5888. [Google Scholar]
- 24.Gustafsson L, Leijonhufvud I, Aronsson A, Mossberg A, Svanborg C. N Engl J Med. 2004;350:2663–2672. doi: 10.1056/NEJMoa032454. [DOI] [PubMed] [Google Scholar]
- 25.Balbach J, Forge V, Van Nuland NAJ, Winder SL, Hore PJ, Dobson CM. Nat Struct Biol. 1995;2:865–870. doi: 10.1038/nsb1095-865. [DOI] [PubMed] [Google Scholar]
- 26.Balbach J, Forge V, Lau WS, Van Nuland NAJ, Brew K, Dobson CM. Science. 1996;274:1161–1163. doi: 10.1126/science.274.5290.1161. [DOI] [PubMed] [Google Scholar]
- 27.Forge V, Wijesinha RT, Balbach J, Brew K, Robinson CV, Redfield C, Dobson CM. J Mol Biol. 1999;288:673–688. doi: 10.1006/jmbi.1999.2687. [DOI] [PubMed] [Google Scholar]
- 28.Dill KA, Chan HS. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 29.Wolynes PG, Onuchic JN, Thirumalai D. Science. 1995;267:1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- 30.Bushmarina NA, Blanchet CE, Vernier G, Forge V. Protein Sci. 2006;15:659–671. doi: 10.1110/ps.051904206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishna MMG, Hoang L, Lin Y, Englander SW. Methods. 2004;34:51–64. doi: 10.1016/j.ymeth.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Ferraro DM, Robertson AD. Biochemistry. 2004;43:587–594. doi: 10.1021/bi035943y. [DOI] [PubMed] [Google Scholar]
- 33.Frauenfelder H, Sligar SG, Wolynes PG. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 34.Eisenmesser EZ, Bosco DA, Akke M, Kern D. Science. 2002;295:1520–1523. doi: 10.1126/science.1066176. [DOI] [PubMed] [Google Scholar]
- 35.Neudecker P, Zarrine-Afsar A, Choy WY, Muhandiram DR, Davidson AR, Kay LE. J Mol Biol. 2006;363:958–976. doi: 10.1016/j.jmb.2006.08.047. [DOI] [PubMed] [Google Scholar]
- 36.Sivaraman T, Arrington CB, Robertson AD. Nat Struct Biol. 2001;8:331–333. doi: 10.1038/86208. [DOI] [PubMed] [Google Scholar]
- 37.Dill KA, Fiebig KM, Chan HS. Proc Natl Acad Sci USA. 1993;90:1942–1946. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson SE. Org Biomol Chem. 2006;4:1845–1853. doi: 10.1039/b600829c. [DOI] [PubMed] [Google Scholar]
- 39.Pan YQ, Briggs MS. Biochemistry. 1992;31:11405–11412. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]
- 40.Bougault C, Feng LM, Glushka J, Kupce E, Prestegard JH. J Biomol NMR. 2004;28:385–390. doi: 10.1023/B:JNMR.0000015406.66725.30. [DOI] [PubMed] [Google Scholar]
- 41.Babu CR, Hilser VJ, Wand AJ. Nat Struct Mol Biol. 2004;11:352–357. doi: 10.1038/nsmb739. [DOI] [PubMed] [Google Scholar]
- 42.Pometun MS, Peterson RW, Babu CR, Wand AJ. J Am Chem Soc. 2006;128:10652–10653. doi: 10.1021/ja0628654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cordier F, Grzesiek S. J Mol Biol. 2002;317:739–752. doi: 10.1006/jmbi.2002.5446. [DOI] [PubMed] [Google Scholar]
- 44.Kitahara R, Akasaka K. Proc Natl Acad Sci USA. 2003;100:3167–3172. doi: 10.1073/pnas.0630309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson KD, Mayer AN. Arch Biochem Biophys. 1986;250:390–399. doi: 10.1016/0003-9861(86)90741-1. [DOI] [PubMed] [Google Scholar]
- 46.Brutscher B, Brüschweiler R, Ernst RR. Biochemistry. 1997;36:13043–13053. doi: 10.1021/bi971538t. [DOI] [PubMed] [Google Scholar]
- 47.Chung HS, Khalil M, Smith AW, Ganim Z, Tokmakoff A. Proc Natl Acad Sci USA. 2005;102:612–617. doi: 10.1073/pnas.0408646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ecker DJ, Butt TR, Marsh J, Sternberg E, Shatzman A, Dixon JS, Weber PL, Crooke ST. J Biol Chem. 1989;264:1887–1893. [PubMed] [Google Scholar]
- 49.Reckel S, Löhr F, Dötsch V. ChemBioChem. 2005;6:1601–1606. doi: 10.1002/cbic.200500076. [DOI] [PubMed] [Google Scholar]
- 50.Selenko P, Serber Z, Gade B, Ruderman J, Wagner G. Proc Natl Acad Sci USA. 2006;103:11904–11909. doi: 10.1073/pnas.0604667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grobler JA, Wang M, Pike ACW, Brew K. J Biol Chem. 1994;269:5106–5114. [PubMed] [Google Scholar]
- 52.Cornilescu G, Marquardt JL, Ottiger M, Bax A. J Am Chem Soc. 1998;120:6836–6837. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.