Abstract
Pyrrolysine (Pyl), the 22nd natural amino acid and genetically encoded by UAG, becomes attached to its cognate tRNA by pyrrolysyl-tRNA synthetase (PylRS). We have determined three crystal structures of the Methanosarcina mazei PylRS complexed with either AMP–PNP, Pyl–AMP plus pyrophosphate, or the Pyl analogue N-ε-[(cylopentyloxy)carbonyl]-l-lysine plus ATP. The structures reveal that PylRS utilizes a deep hydrophobic pocket for recognition of the Pyl side chain. A comparison of these structures with previously determined class II tRNA synthetase complexes illustrates that different substrate specificities derive from changes in a small number of residues that form the substrate side-chain-binding pocket. The knowledge of these structures allowed the placement of PylRS in the aminoacyl-tRNA synthetase (aaRS) tree as the last known synthetase that evolved for genetic code expansion, as well as the finding that Pyl arose before the last universal common ancestral state. The PylRS structure provides an excellent framework for designing new aaRSs with altered amino acid specificity.
Keywords: aminoacyl-tRNA synthetase, evolution, pyrrolysine
The aminoacyl-tRNA synthetases (aaRSs) arose early in evolution as key enzymes involved in the faithful transmission of genetic information (1). They catalyze the formation of all cognate aminoacyl-tRNAs, the substrates for ribosomal protein synthesis. Recently, some unique aaRSs were discovered that expand the code to noncanonical amino acids (2). Phosphoseryl-tRNA synthase (SepRS) attaches O-phosphoserine to tRNACys during the formation of Cys-tRNA in methanogens (3, 4). Most excitingly, pyrrolysine (Pyl), the 22nd amino acid that is cotranslationally inserted in response to an inframe UAG codon, is charged to its unique amber suppressor tRNA (5) by pyrrolysyl-tRNA synthetase (PylRS) (6, 7). Pyl is thought to be an essential active site residue in methylamine methyltransferases in the Methanosarcina mazei (8).
The tRNA synthetases fall into two unrelated classes based on the topology of their ATP-binding domain. When compared with how other amino acids associate with their cognate class II aaRSs, phosphoserine is bound by SepRS in an unusual conformation (9). Because Pyl is significantly larger than the canonical 20 amino acids, knowledge of the PylRS structure allows an inquiry of how a class II aaRS evolved to accommodate this unusual amino acid. Based on genome analyses and biochemical data, it appears that PylRS is the last known aaRS that can be identified in the sequence databases.
Not only does the structure of PylRS represent the final major chapter in the aaRS structural repertoire, but it also grants insight into the evolution of the genetic code. The codon catalog of 20 canonical amino acids appears to predate the class I and class II aaRSs (10), which means that the aaRSs displaced some now extinct aminoacylation system. The development of the universal genetic code was indeed an ancient event in the history of life because the aaRSs had already evolved their modern specificities by the time of the last universal common ancestral state (LUCAS) (11, 12). How Pyl was included in the set of genetically encoded amino acids has remained an open evolutionary question. Because phylogeny based on 3D protein structures allows reconstruction of distant evolutionary events (12), knowledge of the PylRS structure can delineate the position of this enzyme in a tRNA synthetase family tree and can suggest how Pyl was added to the genetic code.
Here we present structures of the catalytic domain of the M. mazei PylRS, as well as the results of a phylogenetic analysis of the subclass IIc aaRSs.
Results
We present three crystal structures of PylRS in complex with AMP–PNP, ATP and the Pyl analogue N-ε-[(cylopentyloxy)carbonyl]-l-lysine (Cyc), and Pyl–AMP with pyrophosphate. These structures allow us to describe in detail the organization of the enzyme-binding pocket for the Pyl side chain that elucidates the principles governing Pyl recognition by PylRS.
Structure Determination of the Enzyme.
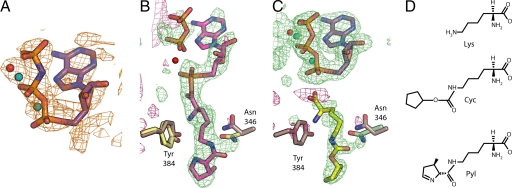
Crystals of the substrate complexes with the PylRS were grown by using modifications (see Materials and Methods) of a published procedure (13). The crystal structure of PylRS and the nonhydrolyzable ATP analogue, AMP–PNP, was determined by using a trimethyl lead acetate derivative to produce experimental phases derived from single isomorphous replacement with anomalous scattering. After cycles of solvent flattening and phase calculation, the experimental map was of sufficient quality to initiate model building (Fig. 1A). The final model has been refined at 1.8 Å resolution and has an Rfactor of 18.1% and an Rfree of 20.1% (Table 1).
Fig. 1.
PylRS substrates. (A) The solvent-flattened, experimentally phased map calculated to 2.5 Å resolution and contoured at 2σ for the PylRS structure bound to AMP–PNP. The final refined model of AMP–PNP is shown as sticks and the magnesium ions as blue spheres. (B and C) Unbiased Fo-Fc maps calculated with experimental amplitudes from data collected from PylRS crystals that were soaked with either ATP and Pyl to 2.2 Å resolution (B) or ATP and Cyc to 1.9 Å (C). Calculated amplitudes were generated from a model of PylRS that lacked nucleotide. Both maps are contoured at +1.5σ (green) and −1.5σ (maroon). The position of the side chains from the original AMP–PNP complex are shown in brown, whereas the final refined positions of the side chains for the complex with Pyl–AMP are in yellow (B) or are displayed in green for the complex with Cyc (C). The final positions of the Pyl–AMP and pyrophosphate are in pink (B), and the amino acid substrate Cyc is shown in yellow (C). The final positions of two magnesium ions (blue spheres) were confirmed by anomalous difference maps from crystals that had been soaked with manganese. Based on similarity with LysRS (19), a third metal position (red sphere) was identified, but tentatively modeled as a water due to a lack of anomalous difference density. (D) Chemical diagrams of Lys, Cyc, and Pyl.
Table 1.
Data collection, phasing, and refinement statistics
| AMPPNP | AMPPNP + TMLA | ATP + Cyc | Adenylated Pyl | |
|---|---|---|---|---|
| Space group | P64 | P64 | P64 | P64 |
| Unit cell dimensions | ||||
| a = b, and c, Å | 105.12, 70.31 | 105.15, 70.82 | 105.22, 71.82 | 106.07, 70.26 |
| α, β, and γ, ° | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Resolution, Å | 50–1.8 | 50–2.5 | 40–1.9 | 40–2.1 |
| Rmerge, %†‡ | 4.6 (87.5) | 8.2 (40.3) | 7.8 (32.8) | 8.7 (>100) |
| I/σI‡ | 48.2 (2.0) | 13.8 (2.1) | 29.3 (5.2) | 26.1 (1.5) |
| Completeness, %‡ | 99.1 (95.7) | 98.4 (86.8) | 99.6 (96.5) | 100 (100) |
| Redundancy‡ | 10.6 (7.3) | 3.7 (2.5) | 10.8 (7.5) | 11.2 (11.0) |
| Rcryst, %‡§ | 18.1 (27.9) | — | 17.4 (20.1) | 16.9 (27.0) |
| Rfree, %‡§ | 20.1 (33.3) | — | 20.3 (21.7) | 21.4 (39.5) |
| rmsd bond length, Å | 0.010 | — | 0.010 | 0.012 |
| rmsd bond angle, ° | 1.357 | — | 1.403 | 1.495 |
| Phasing powerঠ| ||||
| Acentric | — | 1.28 (0.81) | — | — |
| Centric | — | 0.60 (0.65) | — | — |
| Figure of merit‡ | 0.40 (0.26) |
†Rmerge is Σ|Ij − 〈I〉|/ΣI, where Ij is the intensity of an individual reflection and 〈I〉 is the mean intensity for multiply recorded reflections.
‡The values in parentheses are for the highest resolution shell.
§Rcryst is Σ|Fo − Fc|/ΣFo, where Fo is an observed amplitude and Fc is a calculated amplitude; Rfree is the same statistic calculated over a subset of the data that has not been used for refinement.
¶Phasing power is the rms isomorphous difference divided by the rms lack of closure.
Both the preadenylated and adenylated complex structures were solved by using difference Fourier methods. Initial attempts at soaking the amino acid substrate into existing crystals of the apo complex failed to yield interpretable electron density corresponding to the substrate. Therefore, to obtain structures of the complex of PylRS and amino acid substrate, crystals grown in the presence of AMP–PNP were first soaked in a buffer containing EDTA to chelate the magnesium ions, thereby helping to remove the nucleotide from the enzyme, and were then transferred to a buffer containing ATP, magnesium, and either amino acid substrate. With this method, electron density corresponding to Cyc was easily identified in the initial Fo-Fc difference Fourier maps (Fig. 1C). For Pyl, the Fo-Fc difference Fourier maps clearly showed a break in the electron density between the α- and β-phosphates of ATP. This result indicates that the adenylation reaction occurred in the crystals, yielding a complex of PylRS with adenylated Pyl and pyrophosphate (Fig. 1B). The final model for each PylRS complex with Cyc and Pyl were refined at 1.9 Å resolution with an Rfactor of 17.4% and an Rfree of 20.3% and at 2.2 Å resolution with an Rfactor of 16.9% and an Rfree of 21.4% (Table 1).
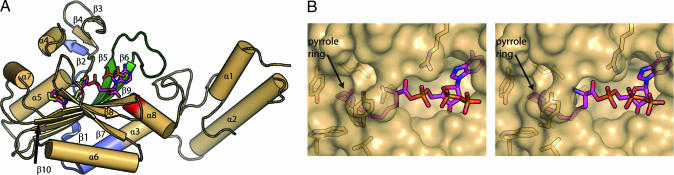
The 3D organization of the PylRS catalytic domain resembles that of other synthetases from the class II family. PylRS contains the typical core β-sheet surrounded by several long helices (14), with signature motifs 2 and 3 that recognize the nucleotide and motif 1 that mediates the dimerization interface of PylRS (15) (Fig. 2A). PylRS crystals contain one protein molecule in the asymmetric unit, and a dimer of PylRS is created by crystallographic symmetry between neighboring enzymes in the crystal lattice with the interface mediated by residues from motif 1. This truncated PylRS construct also runs as a dimer on gel filtration (data not shown). When the homotetrameric SepRS (4, 9), a close structural homologue to PylRS (see Phylogeny of Subclass IIc aaRSs), is superimposed onto a homodimer of PylRS, the dimer interface for the two enzymes is similar. A possible tetramer of the PylRS catalytic core can be generated by the superposition of a second PylRS dimer on the SepRS tetrameric core without producing any steric clashes.
Fig. 2.
Structure of PylRS. (A) A secondary structure diagram of PylRS with the amino acid analogue substrate Cyc shown as sticks (yellow) and the Pyl-AMP and pyrophosphate shown as sticks (pink). For clarity, the position of the ATP from the PylRS complex with Cyc was not shown. The conserved class II synthetase motifs 1, 2, and 3 are shown in blue, green, and red, respectively. (B) A stereo diagram of a surface representation of PylRS highlighting the enzyme's deep amino acid substrate-binding pocket. The surface of the enzyme has been made transparent to reveal the positions of the side chains that interact with the Pyl (pink), which is partially occluded by the enzyme.
Recognition of Pyl–AMP.
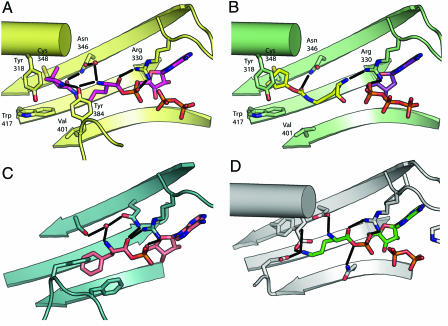
The Pyl–AMP binds in a deep hydrophobic pocket, with its position coordinated by a hydrogen-bonding network with PylRS. Analysis of the initial difference electron density maps clearly showed the position of the adenosine ring, the α-amino, Cα, and primary carbonyl of Pyl (Fig. 1B). The enzyme is highly coordinated to the substrate via three hydrogen bonds to both the α-amino and primary carbonyl (Fig. 3A). Although electron density corresponding to the hydrocarbon chain of Pyl was present, it was significantly weaker, suggesting that it adopts a less rigid conformation on binding to PylRS. The pyrrole ring is buried in a deep hydrophobic pocket of the enzyme (Tyr-384, Trp-417, Cys-348, and Val-401) (Fig. 2B). The orientation of the pyrrole ring relative to the rest of Pyl is slightly ambiguous because no electron density corresponding to the methyl substitution on the ring is present. Because the presence of the methyl substitution in the Pyl preparation was confirmed by mass spectroscopy (7), the lack of electron density corresponding to this group may be the consequence of a small rotational disorder of the pyrrole ring. The ring was modeled to place the nitrogen of the pyrrole ring within hydrogen-bonding distance to the γ-OH of Tyr-384 (Fig. 3A). The only component of Pyl that could participate in a hydrogen bond but does not is the ε-amino group.
Fig. 3.
Structures of the amino acid-binding sites of given synthetases with their amino acid substrates. Each panel is oriented similarly. (A) Side chains that interact with substrate are shown as sticks. The interactions of PylRS (yellow) with Pyl–AMP and pyrophosphate (pink) are shown. (B) The recognition of PylRS (green) with the substrate mimic Cyc (yellow) and ATP (purple) is shown. Two other synthetases are shown for comparison with PylRS. PheRS (blue) with Phe–AMP (tan) in C and LysRS (gray) with Lys–AMP (green) in D. Hydrogen bonds are indicated by solid black lines. Water molecules that mediate hydrogen bonds with substrates are shown as red spheres.
Although the structure of the backbone of PylRS is relatively unchanged between the AMP–PNP and Pyl–AMP intermediate complex structures, there are side-chain movements associated with Pyl recognition and positioning. Recognition of the substrate is carried out only by slight modifications of side-chain positions. The two significant differences are Asn-346 and Tyr-384. On binding of the intermediate, Asn-346 moves so that it can hydrogen bond to both the secondary carbonyl and through a water-mediated contact with the primary amino group of Pyl. Although Tyr-384 is only weakly ordered in the AMP–PNP complex structure, it becomes significantly more ordered in the Pyl structure and makes specific hydrogen-bonding interactions with both the pyrrole nitrogen and the α-amino group of the substrate. These two strictly conserved residues [supporting information (SI) Fig. 5] are most likely the key players in providing the specificity of PylRS for its substrate.
A Complex with an Amino Acid Analogue.
Soaking of PylRS crystals with both ATP and the Pyl analogue, Cyc, produced a ternary complex structure of the enzyme bound to both components and captured the step before adenylation (Fig. 3B). Because the crystals were subjected to a shorter incubation with Cyc than with Pyl, it is not clear whether the lack of reaction reflects a slower reaction rate or an insufficient incubation time. The position of the pentane ring through to the ε-amino group of Cyc was clear in the electron density difference Fourier maps (Fig. 1C) and allows for a hydrogen bond between the primary carbonyl of Cyc and Asn-346 (Fig. 3B). The corresponding electron density for the lysyl section of the Cyc substrate was weaker and less defined. The orientation of the α-amino, Cα, and primary carbonyl of Cyc was ambiguous from the electron density alone. However, comparisons with other synthetase complex structures and satisfaction of hydrogen-bonding interactions, specifically between the primary carbonyl of Cyc and Asn-330, guided the final modeling for this segment of the analogue.
Phylogeny of Subclass IIc aaRSs.
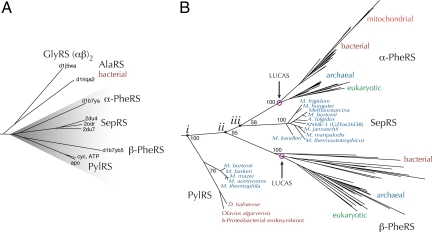
Our structure allows the assignment of PylRS to the aaRS subclass IIc. The class II aaRS family is subdivided into three phylogenetic subclasses, and here we present a structural phylogeny of subclass IIc (Fig. 4A). Interestingly, sequence-based search methods, such as Blast and Pfam, fail to provide a confident consensus regarding the relationship of PylRS to the other members of its protein family. Once thought only to include the tetrameric glycyl-tRNA synthetase and the highly divergent α- and β-subunits of phenylalanyl-tRNA synthetase (PheRS), structural similarity makes it evident that this subclass also includes alanyl-tRNA synthetase (AlaRS), SepRS, and PylRS.
Fig. 4.
Phylogenetic trees for the subclass IIc aaRSs are shown. (A) A structural phylogeny with subclass IIa and IIb aaRS structures as outgroups. (B) A sequence-based phylogeny derived from a structure-based, multiple-sequence alignment with AlaRS sequences as the outgroup. Bootstrap support is indicated for major branches. Other bootstrap values were reported previously (16).
Subclass IIc aaRSs not only present similarity in the core domain, but in their quaternary structure as well. SepRS and AlaRS are the only known homotetrameric aaRSs. Although PheRS is a heterotetramer, in which the α-chain contains the class II core catalytic domain and the β-chain contains a homologous but nonenzymatic domain, the PheRS quaternary structure is homologous to that observed in SepRS and presumably AlaRS as well. With the exception of glycyl-tRNA synthetase, the members of this group all share a homologous quaternary organization, so it is possible that PylRS is a homotetramer in vivo. Our model of the putative PylRS tetramer (described above) shows that well conserved residues mediate the tetramer interface (SI Fig. 5). A cluster of compensatory electrostatic residues are found on opposite faces of the tetramer interface in PheRS (Glu-112, Arg-115) and in SepRS (Gln-57, Arg-61). The homologous positions in PylRS (Arg-252, Asp-256) appear to contribute to the putative tetramer interface in our model.
In addition to properly placing PylRS in subclass IIc, this structure enables a more accurate structure-based alignment of PylRS, SepRS, and PheRS sequences. Protein structures can give insight into distant evolutionary history (16). Because there are many more sequences in the database than structures, the wealth of sequence information can be used to provide better phylogenetic resolution of key evolutionary events only if an accurate alignment can be generated.
Our PylRS structure, along with the recent SepRS structures, finally permits construction of a highly resolved sequence-based phylogenetic tree of these enzymes (Fig. 4B). This tree shows the expected canonical phylogenetic patterns in α- and β-PheRS, where the bacterial versions are specifically related to, but deeply separated from, the archaeal and eukaryotic sister lineages (Fig. 4B). As marked in the figure, the separate roots of the α- and β-PheRS groups represent LUCAS (11, 16). Bifurcations that occur before these points in the tree must have occurred before LUCAS (17) in the common ancestral community that gave rise to all life on Earth (18). The phylogenetic analysis implies that both SepRS and PylRS had already evolved in the ancestral community, although today they are found mostly in methanogenic archaea.
This phylogeny also suggests that SepRS is derived from α-PheRS, whereas PylRS evolved earlier, before PheRS differentiated into a heterotetramer. At this early stage, it is likely that PheRS existed in a homotetrameric form, similar to SepRS and AlaRS. There are three ancestral nodes (i, ii, and iii) that must be considered to understand how Pyl may have been added to the genetic code (Fig. 4B). At node ii, the α- and β-subunits of PheRS diverged from each other; thus, the ancestral enzyme represented by node ii was most likely a PheRS. SepRS does not code directly for an amino acid. Also, there is another known pathway for Cys-tRNACys formation, so the common ancestor of α-PheRS and SepRS (node iii) was likely responsible for Phe coding. Finally, node i, joining PheRS and PylRS, may represent an ancestral PheRS, PylRS, or perhaps a synthetase that ambiguously recognized Pyl or Phe. Because Phe coding is essential to all life, whereas Pyl coding is not, the ancestral enzyme at node i must have been a PheRS. Thus, we infer that Pyl was added to the genetic code after Phe.
Discussion
Substrate-Binding Specificity of PylRS.
The affinity of PylRS for its amino acid substrate arises significantly from hydrophobic interactions, but its specificity derives from five hydrogen bonds between PylRS and the amino acid substrate. The two interactions of Asn-346 with the secondary carbonyl and of Arg-330 with the primary carbonyl of the substrate are seen in both of the complexes with Pyl or Cyc. Although the cyclic component of both substrates is bound in a roughly equivalent position in the hydrophobic pocket of PylRS, additional interactions are required for the proper positioning of the substrate in the enzymatic active site because the electron density describing the correct substrate, Pyl, is stronger than that for Cyc.
Two residues are primarily responsible for the amino acid-specificity determining the hydrogen-bonding network between PylRS and the substrate. Of the three additional hydrogen bond interactions made between PylRS and Pyl that are not made with Cyc, the most interesting are those between the pyrrole ring and Tyr-384. The Tyr-384 interaction plays an important role in orienting Pyl through hydrogen bonds between its γ-OH and the pyrrole ring nitrogen as well as the α-amino group. Furthermore, the complex is also stabilized from the even greater binding energy gained from van der Waals contacts when Tyr-384 binds to Pyl. Tyr-384 is mobile in the structure of the complex bound to Cyc most likely because there is no hydrogen-bonding partner for this residue in Cyc. The Tyr side chain seals off the hydrophobic pocket when the correct substrate is bound, thereby completely surrounding the substrate. The other hydrogen bond interaction between the enzyme and Pyl that is absent in the complex with Cyc is a water-mediated hydrogen bond with Asn-346. Interestingly, although there is water hydrogen-bonded to Asn-346 in the Cyc complex structure, it is not correctly positioned to interact with the α-amino group of Cyc. We believe that Tyr-384 and Asn-346 are the key players in establishing substrate-specificity.
Small Changes Control Specificity.
Class II tRNA synthetases have evolved to discriminate among their amino acid substrates principally through alteration of the amino acid side chains that line the binding pocket, rather than employing changes that also affect the position of protein backbone or secondary structure elements. We compared our complex structures with those of two synthetases complexed with their cognate aminoacyl adenylates that recognize amino acid side chains with similar chemical properties to Pyl, lysyl-tRNA synthetase (LysRS) (19), and PheRS (20). For each of these enzymes, the base of the substrate-binding pocket is composed of a glycine-rich β-strand (β10, β11, and β8 in PylRS, PheRS, and LysRS), which immediately precedes Motif 3 in sequence space. The unique side-chain-specificity elements for the substrate of each synthetase build off this flat surface. This property greatly facilitates the formation of synthetases that charge novel amino acids, either by evolution, as occurred in these cases, or hopefully by intelligent design in future experiments.
The hydrophobic pocket of both PheRS and PylRS are similarly organized. The cycloalkane group of Pyl and the phenyl group of phenylalanine bind in the hydrophobic pocket of their respective synthetase (Fig. 3 A and C). In both enzymes, the interior surface of this hydrophobic pocket is formed by aromatic residues, and the pocket is sealed off by a loop containing an aromatic residue (Tyr-384–PylRS or Phe-260–PheRS). This loop also functions as a substrate-specificity element in other class II synthetases for their cognate aminoacyl substrates (16). The only difference among these relatively hydrophobic pockets is that PylRS contains aromatic residues that are able to participate in hydrogen-bonding interactions with the pyrrole ring, whereas PheRS has aromatic residues lacking potential for hydrogen-bonding interactions, consistent with its substrate's phenyl group. For both enzymes, the shape complementarity between the hydrophobic pocket of the enzyme and the substrate provides van der Waals contacts that contribute to the overall binding energy for the complex.
Despite the similar position of lysine and the lysyl moiety of Pyl in their respective enzymatic-binding pockets, the positions are coordinated by different specificity elements on each synthetase. A Cα-based superposition of substrate-bound complexes of LysRS and PylRS results in the placement of the ε-amino group of Lys–AMP within 1 Å of the ε-amino group of Pyl. However, LysRS specifically recognizes the ε-amino group of Lys through three hydrogen bonds (Fig. 3D). In contrast, every atom of Pyl that can make a hydrogen bond with the synthetase makes at least one contact with the enzyme except for the ε-amino group of Pyl. The lack of a hydrogen-bonding partner for the ε-amino group of the lysyl moiety of Pyl further discriminates against Lys binding to PylRS because the binding of the charged ε-amino group would have no hydrogen-binding partners, thereby enhancing the discrimination of PylRS against Lys.
Evolution of PylRS.
The principal evolutionary question surrounding Pyl concerns its connection to the evolution of the genetic code. Examining the relationship of PylRS to PheRS reveals that PylRS is an ancient enzyme that evolved before LUCAS. The phylogeny also indicates that, because PylRS is derived from PheRS, Pyl must have been added to the code after Phe. Supported by experimental evidence indicating that tRNA identity is older than the modern aaRSs, we previously concluded that the aaRSs evolved only after the emergence of the universal genetic code (10). Here we also note that the nongenetically encoded Sep, similarly derived from PheRS, is an addition to the coding process and not to the genetic code. The post-LUCAS advent of glutaminyl-tRNA synthetase and asparaginyl-tRNA synthetase (11), which replaced the primordial indirect coding pathway for Asn and Gln (21) in only some organisms, also reminds us that the aaRSs were more involved in adaptations to the coding process than in the establishment of the codon catalog. Unlike the canonical set of 20 amino acids, Pyl encoding is not essential for life. Thus, we find it plausible that Pyl coding did not exist until PylRS evolved from an ancient gene duplication of PheRS. These notions imply that the only aaRS that evolved to expand the codon catalog was PylRS, which remains an evolutionary remnant of early archaeal innovation to the genetic code.
Conclusion.
The structures of the three substrate complexes with PylRS presented here reveal the organization of the amino acid side-chain-binding pocket and the elements that determine the enzyme's substrate-specificity. Comparison of the PylRS structure with the structures of other tRNA synthetases demonstrates how the class II synthetase fold acts as a scaffold on which simple side-chain alterations determine the specificity for the amino acid substrate. Taken together, these results lay the foundation for future protein engineering of this PylRS enzyme to alter its amino acid-specificity. PylRS is a particularly attractive aaRS for this task as tRNAPyl directs amino acid insertion in response to UAG, a codon that is not normally present in the ORF. Also, PylRS and tRNAPyl are a perfect orthogonal pair because they have negligible cross-reactivity with other aaRSs or tRNAs (6, 22). Thus, a redesigned PylRS may become a key enzyme in studies to genetically incorporate unnatural amino acids into proteins.
Materials and Methods
General.
Pyl was chemically synthesized as described (7, 23). Cyc was purchased from Sigma–Aldrich (St. Louis, MO). The genomic DNA from M. mazei (Barker), Mah, and Kuhn (DSM 3647) was obtained from the American Type Culture Collection (Manassas, VA). Oligonucleotide synthesis and DNA sequencing were performed at the Keck Foundation Research Biotechnology Resource Laboratory (Yale University, New Haven, CT). Pymol was used to generate all figures (24).
PylRS Gene Constructs and Enzyme Purification.
The catalytic domain of PylRS (185–454) was PCR-amplified from the genomic DNA of M. mazei (MM1445) and cloned into the pET15b vector (Novagen, Madison, WI), which encodes an N-terminal hexahistidine tag. The fusion construct was expressed in Escherichia coli Codon Plus-RIL (DE3) cells (Stratagene, La Jolla, CA). Clarified lysate was loaded onto a HisTrap column (Amersham, Piscataway, NJ) and eluted with an imidazole gradient. The protein sample was then buffer-exchanged on a desalting column before being loaded onto a Heparin column (Amersham). The PylRS fraction eluted at 0.25 M NaCl and was directly loaded onto a HiLoad Superdex 200 (26/60) column equilibrated with 10 mM Hepes (pH 7.4), 0.3 M NaCl, 5 mM MgCl2, and 1 mM DTT. PylRS-containing fractions were concentrated to a concentration of 10 mg/ml. Aliquots were flash-frozen in liquid nitrogen and stored until needed for crystallization experiments.
Crystallization.
Hexagonal-shaped crystals of a complex between PylRS and AMP–PNP were grown at 16°C by vapor diffusion. A solution containing 10 mg/ml of PylRS and 10 mM AMP–PNP was mixed at a ratio of 2:1 with well solution [100 mM Tris (pH 7.0–8.0) and 8–14% PEG 2000 monomethyl ether]. Crystals appeared overnight and grew to dimensions of 300 × 300 × 150 μm. Stabilization of the crystals proceeded in a stepwise fashion. First, the crystals were transferred to a well solution supplemented with 5 mM MgCl2, 10 mM AMP–PNP, and an additional 2% PEG. Second, the crystals were transferred, stepwise, into the same buffer, but with 30% ethylene glycol. Stabilized crystals were flash-frozen in liquid propane.
Diffusion of substrates into crystals was performed on cryostabilized crystals. Initial attempts to soak amino acid substrate into existing crystals of PylRS and AMP–PNP failed. As a result, the AMP–PNP had to be removed from the crystals. Cryostabilized crystals were transferred to a cryosolution containing 5 mM EDTA, as well as the appropriate buffer, PEG, and ethylene glycol, to remove the magnesium ions and nucleotide. The crystals were incubated with EDTA overnight. Crystals were then transferred to a cryosolution supplemented with either 10 mM ATP, 5 mM MgCl2, and 50 mM Cyc for 1 h or 10 mM ATP, 5 mM MgCl2, and 10 mM Pyl overnight before flash-freezing.
Structure Solution and Refinement.
Frozen PylRS crystals diffracted x-rays at synchrotron sources to 1.8 Å resolution. Diffraction data were collected on stations X-29 at the Brookhaven National Laboratory (Upton, NY), 24ID at the Argonne Photon Source (Argonne, IL), or the Yale Center for Structural Biology home source (New Haven, CT). HKL2000 was used to process diffraction data (25). The crystals belong to space group P64, with one molecule in the asymmetric unit (unit cell dimensions: a and b, 105 Å; c, 70 Å; α and β, 90°; γ, 120°). This crystal form is essentially the same as reported previously (13). Initial phases were obtained by soaking cryoprotected crystals in a buffer supplemented with 50 mM trimethyl lead acetate for 2 h before freezing. Heavy atom sites were found, and single isomorphous replacement with anomalous scattering phases were calculated in SOLVE (26). Solvent flattening was performed in RESOLVE (27). Iterative rounds of building and refinement were performed in COOT (28) and REFMAC (29), respectively. Refinement parameter files and initial coordinates for Cyc, Pyl, and Pyl–AMP were generated by using the prordg2 server (30).
Phylogenetic Analyses.
Sequences were downloaded from the National Center for Biotechnology Information nonredundant database and the Integrated Microbial Genomes with Microbiome Samples database (31), and structures were obtained from the Protein Data Bank (32). Structure-based alignment and phylogeny, as well as sequence alignments, were completed by using the Multiseq 2.0 module in VMD 1.8.5 (33) and performed as described (16). Sequence alignments were also edited by using the CINEMA alignment editor (34). The structural similarity measure QH (35), along with the NEIGHBOR and DRAWTREE programs in PHYLIP version 3.66 (36), were used in constructing the structure-based phylogenetic tree. As detailed previously (16), the sequence-based phylogeny was generated by using a combined maximum parsimony/maximum likelihood method with the programs PAUP* (37) and PHYML version 2.4.4 (38), and bootstrap values were computed with MOLPHY (39).
Supplementary Material
Acknowledgments
We thank Sotiria Palioura for inspired discussions and Robin Evans for help with data collection. This work was supported by National Institute of General Medical Sciences Grants GM 22778 (to T.A.S.) and GM 22854 (to D.S.), Department of Energy Grant DE-FG02–98ER20311 (to D.S.), National Science Foundation Grant MCB-0645283 (to D.S.), a Feodor Lynen Postdoctoral Fellowship of the Alexander von Humboldt Stiftung (to M.E.), and a National Science Foundation postdoctoral fellowship in Biological Informatics (to P.O.).
Abbreviations
- Cyc
N-ε-[(cylopentyloxy)carbonyl]-l-lysine
- LUCAS
last universal common ancestral state
- Pyl
pyrrolysine
- PylRS
pyrrolysyl-tRNA synthetase
- aaRS
aminoacyl-tRNA synthetase
- AlaRS
alanyl-tRNA synthetase
- PheRS
phenylalanyl-tRNA synthetase
- LysRS
lysyl-tRNA synthetase
- Pyl–AMP
pyrrolysyl adenylate
- SepRS
phosphoseryl-tRNA synthase.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2Q7E, 2Q7G, and 2Q7H).
This article contains supporting information online at www.pnas.org/cgi/content/full/0704769104/DC1.
References
- 1.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ambrogelly A, Palioura S, Söll D. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 3.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, III, Ibba M, Söll D. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 4.Kamtekar S, Hohn MJ, Park HS, Schnitzbauer M, Sauerwald A, Söll D, Steitz TA. Proc Natl Acad Sci USA. 2007;104:2620–2625. doi: 10.1073/pnas.0611504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srinivasan G, James CM, Krzycki JA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 6.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 7.Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Söll D. Proc Natl Acad Sci USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krzycki JA. Curr Opin Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Fukunaga R, Yokoyama S. Nat Struct Mol Biol. 2007;14:272–279. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 10.Hohn MJ, Park HS, O'Donoghue P, Schnitzbauer M, Söll D. Proc Natl Acad Sci USA. 2006;103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woese CR, Olsen GJ, Ibba M, Söll D. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Donoghue P, Luthey-Schulten Z. Microbiol Mol Biol Rev. 2003;67:550–573. doi: 10.1128/MMBR.67.4.550-573.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagisawa T, Ishii R, Fukunaga R, Nureki O, Yokoyama S. Acta Crystallogr F Struct Biol Cryst Commun. 2006;62:1031–1033. doi: 10.1107/S1744309106036700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cusack S, Berthet-Colominas C, Härtlein M, Nassar N, Leberman R. Nature. 1990;347:249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- 15.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 16.O'Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. Proc Natl Acad Sci USA. 2005;102:19003–19008. doi: 10.1073/pnas.0509617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwabe N, Kuma K, Hasegawa M, Osawa S, Miyata T. Proc Natl Acad Sci USA. 1989;86:9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woese CR. Proc Natl Acad Sci USA. 2002;99:8742–8747. doi: 10.1073/pnas.132266999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desogus G, Todone F, Brick P, Onesti S. Biochemistry. 2000;39:8418–8425. doi: 10.1021/bi0006722. [DOI] [PubMed] [Google Scholar]
- 20.Fishman R, Ankilova V, Moor N, Safro M. Acta Crystallogr D Biol Crystallogr. 2001;57:1534–1544. doi: 10.1107/s090744490101321x. [DOI] [PubMed] [Google Scholar]
- 21.Tumbula DL, Becker HD, Chang WZ, Söll D. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 22.Ambrogelly A, Gundllapalli S, Herring S, Polycarpo C, Frauer C, Söll D. Proc Natl Acad Sci USA. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polycarpo CR, Herring S, Berube A, Wood JL, Söll D, Ambrogelly A. FEBS Lett. 2006;580:6695–6700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delano LLC. PyMOL. Palo Alto, CA: Delano LLC; 2002. [Google Scholar]
- 25.Otwinowski Z, Minor W. Macr Crystallogr. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 26.Terwilliger TC, Berendzen J. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terwilliger TC. Acta Crystallogr D Biol Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Schuttelkopf AW, van Aalten DMF. Acta Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 31.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, et al. Nucleic Acids Res. 2006;34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pettifer SR, Sinnott JR, Attwood TK. Comp Funct Genomics. 2004;5:56–60. doi: 10.1002/cfg.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Donoghue P, Luthey-Schulten Z. J Mol Biol. 2005;346:875–894. doi: 10.1016/j.jmb.2004.11.053. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 37.Swofford D. Sunderland, MA: Sinauer Associates; 2003. PAUP*. Phylogenetic Analysis Using Parsimony, version 4. [Google Scholar]
- 38.Guindon S, Gascuel O. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 39.Adachi M, Hasegawa M. Comp Sci Monogr. 1996;28:1–150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.