Abstract
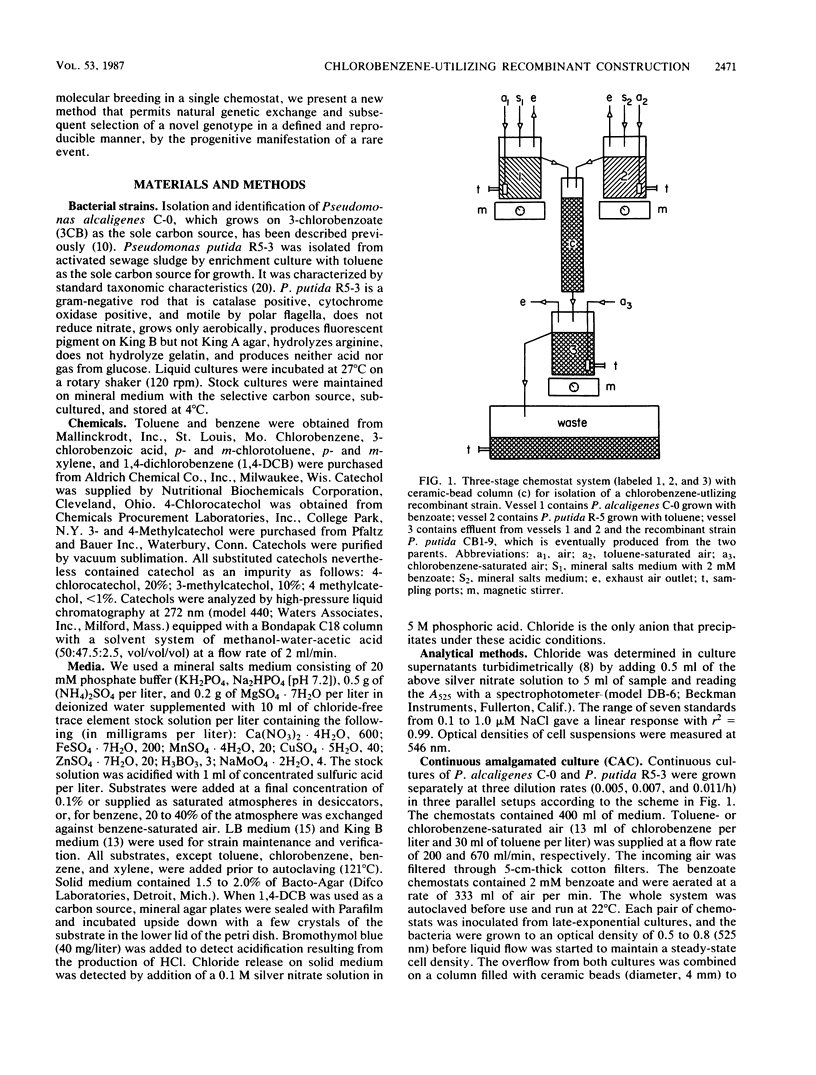
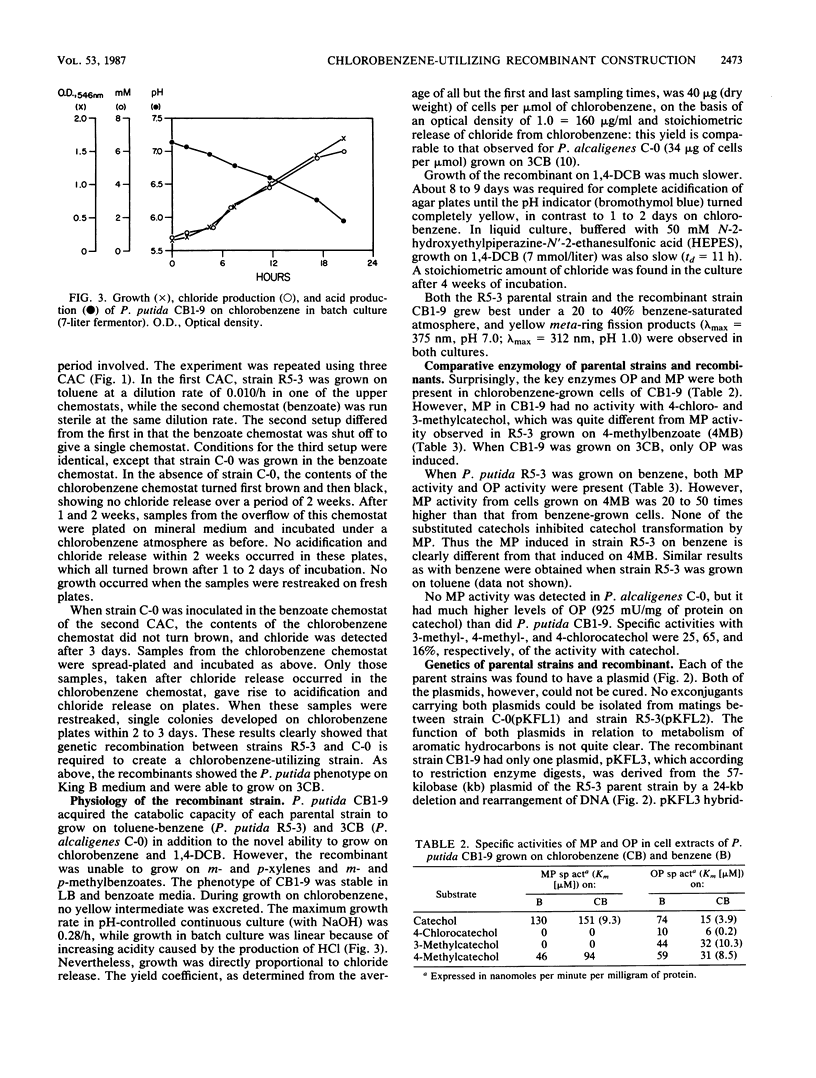
Separate continuous cultures of Pseudomonas putida R5-3, grown on toluene, and Pseudomonas alcaligenes C-O, grown on benzoate, were concentrated and continuously amalgamated on a ceramic bead column, which was subjected to a continuous stream of chlorobenzene vapors. A recombinant strain, P. putida CB1-9, was isolated in less than 1 month. P. alcaligenes C-0 grew on benzoate and 3-chlorobenzoate but not on toluene, P. putida R5-3 grew on benzoate and toluene but not on 3-chlorobenzoate, and neither strain grew on chlorobenzene or 1,4-dichlorobenzene; however, the recombinant P. putida CB1-9 grew on all of these substrates. Chlorobenzene-utilizing strains were not found in continuous cultures run at the lowest growth rate (0.05/h) or in the absence of the donor strain, P. alcaligenes C-0. Chloride was released in stoichiometric amounts when P. putida CB1-9 was grown on either chlorobenzene or 1,4-dichlorobenzene. The recombinant strain was related to P. putida R5-3, phenotypically and genetically. Restriction enzyme digests of the single 57-kilobase (kb) plasmid in R5-3 and of the single 33-kb plasmid in CB1-9 were similar, but also indicated rearrangement of plasmid DNA. Coincidental or causal to the loss of the 24-kb fragment was the observation that the recombinant--unlike its parent, R5-3--did not grow on xylenes or methylbenzoates. Although both ortho-pyrocatechase (OP) and meta-pyrocatechase (MP) were found in CB1-9 and R5-3, MP activity was 20- to 50-fold higher in R5-3 cells grown on 4-methylbenzoate than in the same cells grown on benzene.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartels I., Knackmuss H. J., Reineke W. Suicide Inactivation of Catechol 2,3-Dioxygenase from Pseudomonas putida mt-2 by 3-Halocatechols. Appl Environ Microbiol. 1984 Mar;47(3):500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chatfield L. K., Williams P. A. Naturally occurring TOL plasmids in Pseudomonas strains carry either two homologous or two nonhomologous catechol 2,3-oxygenase genes. J Bacteriol. 1986 Nov;168(2):878–885. doi: 10.1128/jb.168.2.878-885.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Two catechol 1,2-dioxygenases from a 3-chlorobenzoate-grown pseudomonad. Biochem J. 1978 Jul 15;174(1):73–84. doi: 10.1042/bj1740073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focht D. D., Alexander M. Aerobic cometabolism of DDT analogues by Hydrogenomonas sp. J Agric Food Chem. 1971 Jan-Feb;19(1):20–22. doi: 10.1021/jf60173a042. [DOI] [PubMed] [Google Scholar]
- Focht D. D., Brunner W. Kinetics of biphenyl and polychlorinated biphenyl metabolism in soil. Appl Environ Microbiol. 1985 Oct;50(4):1058–1063. doi: 10.1128/aem.50.4.1058-1063.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focht D. D., Shelton D. Growth kinetics of Pseudomonas alcaligenes C-0 relative to inoculation and 3-chlorobenzoate metabolism in soil. Appl Environ Microbiol. 1987 Aug;53(8):1846–1849. doi: 10.1128/aem.53.8.1846-1849.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kellogg S. T., Chatterjee D. K., Chakrabarty A. M. Plasmid-assisted molecular breeding: new technique for enhanced biodegradation of persistent toxic chemicals. Science. 1981 Dec 4;214(4525):1133–1135. doi: 10.1126/science.7302584. [DOI] [PubMed] [Google Scholar]
- Kilbane J. J., Chatterjee D. K., Karns J. S., Kellogg S. T., Chakrabarty A. M. Biodegradation of 2,4,5-trichlorophenoxyacetic acid by a pure culture of Pseudomonas cepacia. Appl Environ Microbiol. 1982 Jul;44(1):72–78. doi: 10.1128/aem.44.1.72-78.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klecka G. M., Gibson D. T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida by 3-chlorocatechol. Appl Environ Microbiol. 1981 May;41(5):1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol. 1984 Feb;47(2):395–402. doi: 10.1128/aem.47.2.395-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraa G., Boone M. L., Jetten M. S., van Neerven A. R., Colberg P. J., Zehnder A. J. Degradation of 1,4-dichlorobenzene by Alcaligenes sp. strain A175. Appl Environ Microbiol. 1986 Dec;52(6):1374–1381. doi: 10.1128/aem.52.6.1374-1381.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain J. C., Nishino S. F. Degradation of 1,4-dichlorobenzene by a Pseudomonas sp. Appl Environ Microbiol. 1987 May;53(5):1010–1019. doi: 10.1128/aem.53.5.1010-1019.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
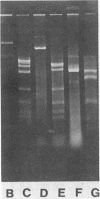
- de Bont J. A., Vorage M. J., Hartmans S., van den Tweel W. J. Microbial degradation of 1,3-dichlorobenzene. Appl Environ Microbiol. 1986 Oct;52(4):677–680. doi: 10.1128/aem.52.4.677-680.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]