Summary
Escherichia coli (E. coli) single stranded (ss)DNA binding (SSB) protein binds ssDNA in multiple binding modes and regulates many DNA processes via protein-protein interactions. Here, we present direct evidence for fluctuations between the two major modes of SSB binding, (SSB)35 and (SSB)65 formed on (dT)70, with rates of interconversion on time scales that vary as much as 200-fold for a mere 4-fold change in NaCl concentration. Such remarkable electrostatic effects allow only one of the two modes to be significantly populated outside a narrow range of salt concentration, providing a context for precise control of SSB function in cellular processes via SSB expression levels and interactions with other proteins. Deletion of the acidic C-terminus of SSB, the site of binding of several proteins involved in DNA metabolism, does not affect the strong salt dependence, but shifts the equilibrium towards the highly cooperative (SSB)35 mode, suggesting that interactions of proteins with the C-terminus may regulate the binding mode transition and vice versa. Single molecule analysis further revealed a novel low abundance binding configuration and provides a direct demonstration that the SSB-ssDNA complex is a finely tuned assembly in dynamic equilibrium among several well-defined structural and functional states.
Keywords: DNA-Protein Interactions, Single-stranded DNA Binding Protein, Binding Modes, Replication, FRET, single molecule, kinetics
Introduction
The presence of extensive amounts of single stranded DNA (ssDNA) in the cell is often a sign of trouble as ssDNA is an obligate intermediate in recombination-mediated DNA repair. Single stranded DNA binding (SSB) proteins are essential in all organisms and bind selectively to ssDNA independent of sequence1; 2. They also modulate the functions of many DNA processing enzymes either via protein-protein interactions or by controlling accessibility to ssDNA1; 2; 3; 4; 5. While it is now well established that SSB proteins can display a multiplicity of modes of interaction with DNA5; 6, the dynamic nature of the transitions among these modes is yet to be explored. We have therefore examined the dynamic interconversion among these SSB/DNA complexes at the single molecule level.
The Escherichia coli SSB protein forms a stable homotetramer, with each of its 19kD subunits possessing an oligonucleotide/oligosaccharide binding (OB) fold5; 7; 8; 9, hence the tetramer has four potential ssDNA binding sites. The SSB tetramer can bind long ssDNA in a variety of binding modes depending on solution conditions, especially salt concentration and type5; 10; 11; 12; 13; 14. At low monovalent salt concentrations (< 10 mM NaCl) and high protein to DNA ratios, an SSB tetramer binds to ssDNA with high inter-tetramer cooperativity using only two out of four subunits on average, and occluding ~35 nucleotides (nt) ((SSB)35 mode)15. However, at higher salt concentrations (> 0.2 M NaCl), an SSB tetramer binds to ssDNA using all four subunits with approximately 65 nts wrapped around the tetramer to form the low cooperativity (SSB)65 mode5; 8.
A model for SSB bound to ssDNA in its (SSB)65 binding mode has been suggested based on the x-ray crystallographic structure of a tetramer of SSBc (a C-terminal truncation of 42 residues) bound to two molecules of (dC)358. In this model, ~65 nt of ssDNA interact with all four SSB subunits with a topology approximating the stitching around a baseball (Fig. 1a), hence the entry and exit sites of a ~65 nt ssDNA are in close proximity. A more speculative structural model for the (SSB)35 binding mode was also proposed as shown in Figure 1b8. The transition between these binding modes is reversible13 but little information is currently available about the dynamics of interconversion between these binding modes.
Figure 1. E. coli SSB binding modes.
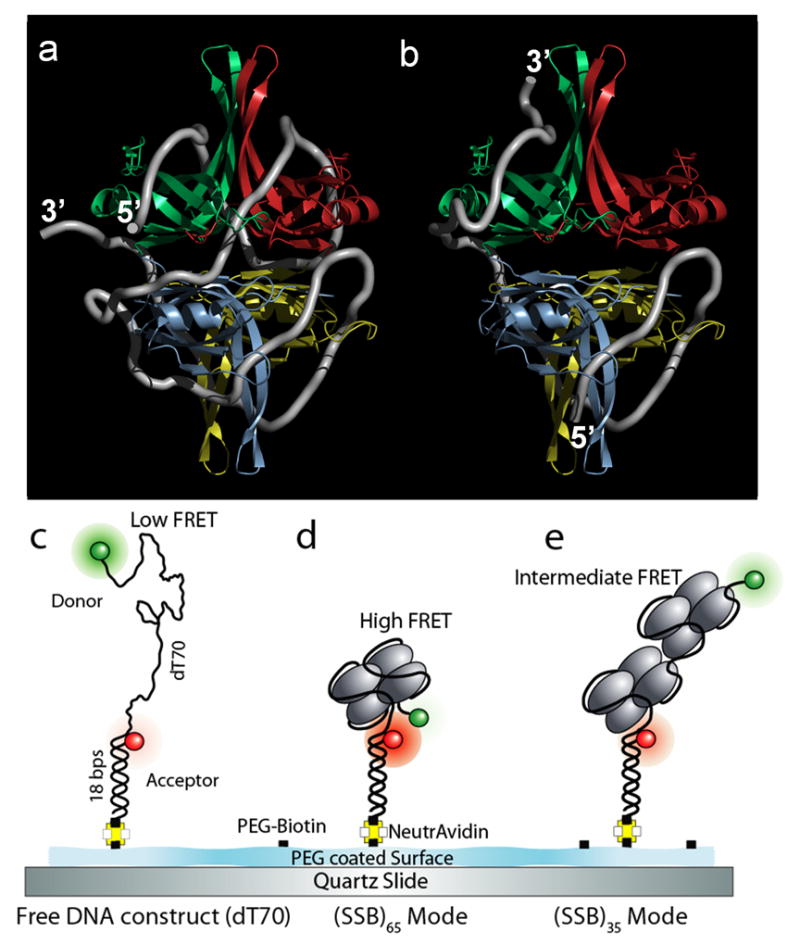
(a) Model of the (SSB)65 binding mode derived from the crystal structure of a chymotrypsin truncated SSB tetramer (missing 42 C-terminal residues) bound to two (dC)35 oligonucleotides8. Single stranded (ss)DNA (70 nucleotides; white tube) wraps around the SSB tetramer (subunits colored individually; C-terminus not shown) binding to all four subunits.
(b) Model of the (SSB)35 binding mode. ~35 nucleotides (white tube) are bound to approximately two subunits on average8. Figures 1a and 1b were generated using PyMol54.
(c–e) Schematic design for the single molecule FRET experiment for SSB
(c) Partial duplex DNA, (dT)70 immobilized on ‘biocompatible’ PEG coated quartz surface has low FRET.
(d) Binding of a single SSB tetramer in (SSB)65 binding mode results in high FRET.
(e) Binding of another SSB (in (SSB)35 binding mode) will result in an intermediate FRET state.
In this report, we use single molecule fluorescence resonance energy transfer (smFRET) techniques16; 17 to examine directly the dynamics of the spontaneous structural changes between the two SSB-DNA binding modes using ssDNA that accommodates only one tetramer bound in the (SSB)65 mode or two tetramers in the (SSB)35 mode. The results agree well with stopped-flow ensemble studies and further reveal a new binding configuration that we term (SSB)35b that can be formed from the (SSB)35 mode via a major rearrangement of the protein-DNA complex without protein dissociation. Deletion of 42 amino acids from the C-termini of SSB shifts the equilibrium to favor the (SSB)35 mode, suggesting that interaction of SSB with other replication/recombination proteins via its C-termini can potentially be regulated by these binding modes and vice versa.
Results
DNA constructs for monitoring SSB binding mode transitions
For single molecule experiments, we used a surface-tethered partial duplex DNA labeled with a donor (Cy3) and an acceptor (Cy5) so that FRET between them reports on the conformations of the intervening 70 nt of ssDNA (Fig. 1c, referred to hereafter as “(dT)70”). While the DNA alone has low FRET due to a large time-averaged distance between the two fluorophores18, ssDNA wrapping around an SSB tetramer in the (SSB)65 mode should result in very high FRET (Fig. 1d)19. Upon binding two tetramers in the (SSB)35 mode, an intermediate FRET signal is expected due to a lower degree of compaction (Fig. 1e). To demonstrate this in bulk solution, we performed an equilibrium titration of Cy5-(T)68-Cy3-T-3′ (referred to as “(dT)68”) with SSB at 80 mM NaCl. Figure 2 shows the results of a titration of “(dT)68” with SSB plotted as the normalized Cy5 fluorescence as a function of the ratio of the total [SSB] to total [(dT)68]. Upon addition of SSB, the Cy5 fluorescence increases linearly up to a ratio of 1 SSB tetramer per “(dT)68” indicating stoichiometric formation of the fully wrapped (SSB)65 complex (1:1 molar complex) characterized by high FRET indicating that Cy3 and Cy5 are in close proximity. Upon further addition of SSB, there is a more gradual decrease in Cy5 fluorescence reflecting formation of the (SSB)35 complex with two SSB tetramers bound and characterized by an intermediate FRET value. Binding of the first SSB tetramer occurs with very high affinity (stoichiometrically), whereas the apparent affinity of the second SSB tetramer is substantially lower. Although the second SSB tetramer binds with high cooperativity in the (SSB)35 mode, this apparent negative cooperativity is due to the very high affinity of a single tetramer in the (SSB)65 mode15. The binding isotherm in Fig. 2 was fit well by a model in which the DNA can bind two SSB tetramers (see Supplementary Materials) characterized by two step-wise binding constants, k1=5×1010 M−1 and k2=(3.3±0.6)×107 M−1, reflecting binding of the first and the second SSB tetramers to DNA. The data in Figure 2 show that the DNA binds two SSB tetramers at saturation, with each SSB bound in the (SSB)35 binding mode.
Figure 2. SSB concentration dependent changes in binding modes observed in bulk solution.
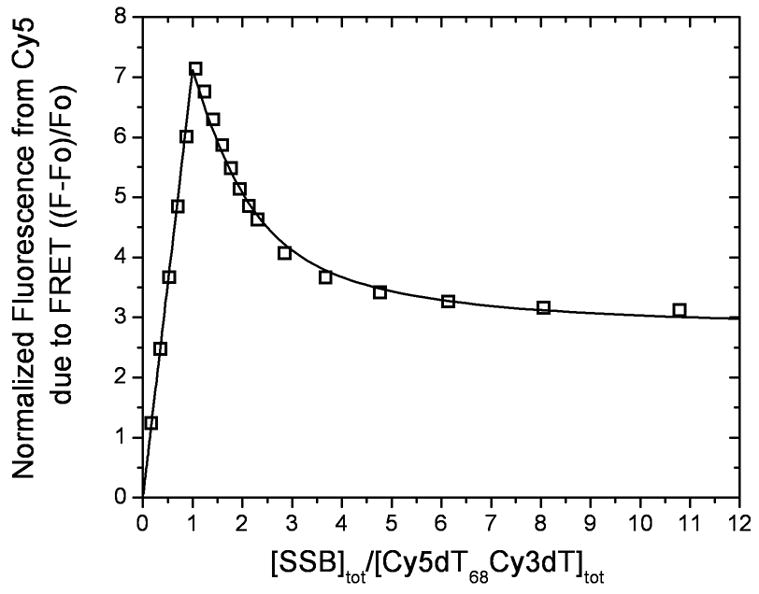
Results of an equilibrium titration of Cy5(dT)68Cy3dT-3′ (0.1μM) with SSB (buffer T, 80 mM NaCl, 25°C) plotted as normalized Cy5 fluorescence (Fn=(F−F0)/F0) versus ratio of total protein to DNA concentrations (where F0 is fluorescence intensity of DNA alone and F is the fluorescence measured at each point in the titration). The biphasic character of the binding isotherm indicates that two types of complexes can form, the first having one and the second having two tetramers bound and characterized by high and intermediate FRET values ((SSB)65 and (SSB)35, respectively), respectively. The solid line represents the best fit to the data with k1=5×1010 M−1 (minimum estimate) and k2=(3.3±0.6)×107 M−1 and two additional parameters F1=7.1±0.1 and F2= 2.7±0.1, reflecting the maximum Cy5 fluorescence observed for one and two tetramers bound, respectively.
Single molecule studies show a salt-dependent distribution of binding modes
We first tested whether the two major SSB binding modes can be detected as two distinct FRET states at the single molecule level (Fig. 3a and b). In the presence of 10 nM SSB tetramer, at low NaCl concentrations (< 50 mM), we observed a single population with low apparent FRET efficiency (Eapp ~ 0.2) calculated as the acceptor intensity divided by the total intensity. This low FRET species at 10 nM SSB is easily distinguishable from DNA alone (Eapp ~ 0.1) and from the donor only population (Eapp ~0) (FRET histograms in Fig. 3a). Upon increasing the [NaCl], the low FRET species disappeared concomitantly with the increase of a high FRET species (Eapp ~ 0.75). Since higher salt concentrations favor the (SSB)65 mode13, we tentatively assigned the high and low FRET states to the (SSB)65 and (SSB)35 modes, respectively.
Figure 3. Salt and SSB dependent changes in SSB binding modes.
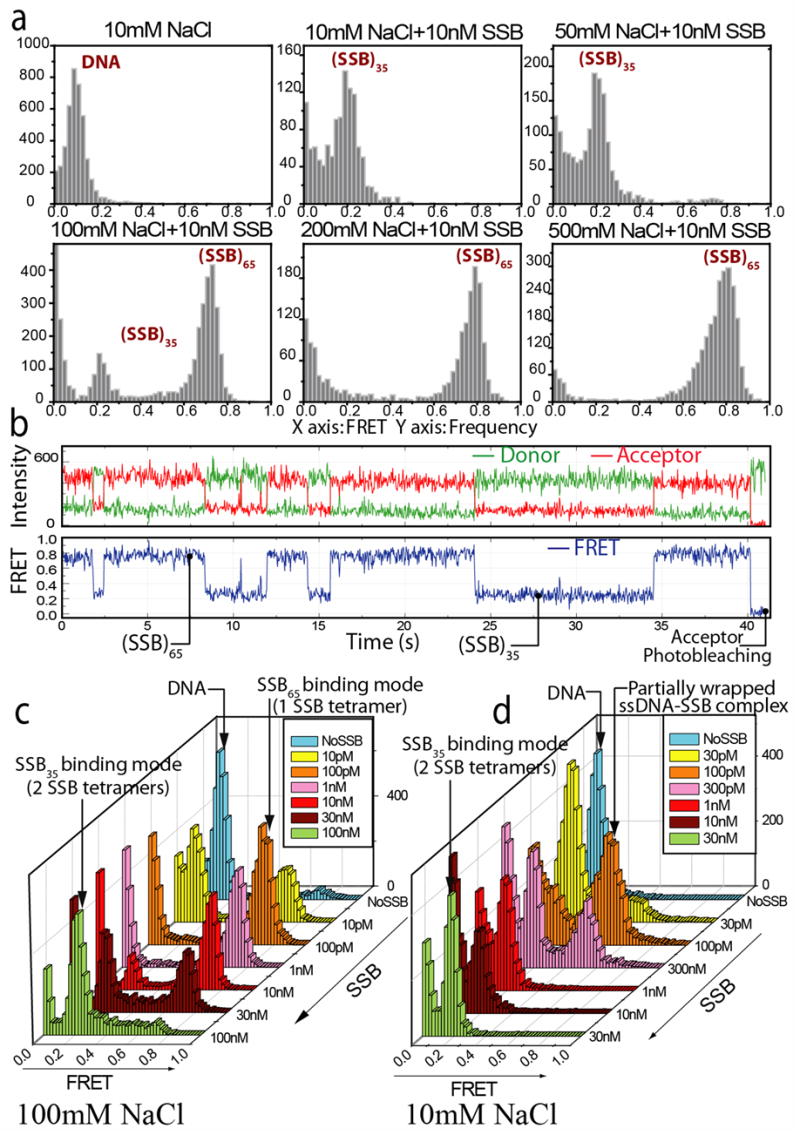
(a) Histograms representing the distributions of average FRET values for single molecules of (dT)70. The FRET peak for DNA only (E ~ 0.1) can be distinguished from a protein bound state (E ~ 0.2) at low salt corresponding to the (SSB)35 binding mode. Increasing salt concentration results in conversion to a high FRET state representing predominantly the (SSB)65 binding mode. Kinetic analysis of the transitions reveals the presence of another minor binding configuration, termed (SSB)35b, within the high FRET population (see text).
(b) Donor and Acceptor Intensity and FRET time record of a single DNA molecule (100 mM NaCl and 10 nM SSB) shows distinct transitions between low and high FRET states, corresponding to the (SSB)35 and (SSB)65 binding modes, respectively.
(c) FRET histograms as a function of SSB concentration at 100 mM NaCl. (SSB)65 binding mode (high FRET) is favored at low SSB concentrations, but is replaced by (SSB)35 binding mode (E ~ 0.2) at higher SSB concentration.
(d) FRET histograms as a function of SSB concentration at 10 mM NaCl. Formation of a 1:1 complex is still favored at low [SSB], although with a somewhat lower FRET efficiency, Eapp ~ 0.6, (due to suggested rapid partial unwrapping of the labeled DNA from the SSB), although lower SSB concentrations (> 300 pM) are required to induce the transition to the (SSB)35 mode (low FRET) with two tetramers bound to DNA
We evaluated whether the high FRET values that we observe are consistent with the expectations based on the model of the (SSB)65 complex inferred from the SSB-DNA crystal structure8. The apparent FRET efficiency, Eapp, should provide a good measure of the true FRET efficiency because the correction factor, γ, which accounts for the differences in quantum yield and detection efficiency between the donor and the acceptor is 1.03 ± 0.02 (details in Materials and Methods) for NaCl concentrations ranging from 100mM to 500mM20. FRET values increased from 0.73 at 100 mM NaCl to 0.80 at 500 mM NaCl (Fig. 3a), probably reflecting some compaction of the extra bases not in contact with the protein at the higher salt concentrations18. Based on a R0 value of 60 Å for the Cy3-Cy5 FRET pair18, the calculated inter-dye distance for the high FRET state is 47–51 Å. From the structure-based model of a (dT)70 bound to an SSBc tetramer, we estimate an average distance between the first and seventieth base to be 33 Å. This agreement is reasonably good considering that the dyes on the (dT)70 in the single molecule experiments are placed on two different strands of the partial duplex via flexible linkers, and suggests that DNA wrapping in the (SSB)65 mode indeed induces the DNA ends to come together as inferred from the crystal structure.
The midpoint of the NaCl-induced transition between the low and high FRET states ranged from 80–130 mM NaCl for the SSB concentrations (10–200 nM) used here. We also observed a transition from the low to high FRET state induced by increasing [MgCl2], with a midpoint of ~2 mM MgCl2 at 10 nM SSB (Supplementary Fig. S1). Previous ensemble studies of SSB binding to poly(dT) reported midpoints of ~20 mM NaCl (or 0.06 mM MgCl2) and ~160mM NaCl (or 64 mM MgCl2) for the (SSB)35→(SSB)56 and (SSB)56→(SSB)65 transitions, respectively5; 10; 11. Direct comparison of these midpoints is difficult because the (SSB)56 mode is presumably only formed on long ssDNA, such as poly(dT), and appears to be absent in our experiments with (dT)70. However, the salt concentration ranges over which the binding mode transitions occur, are in general agreement suggesting that the smFRET measurements of DNA tethered on a surface recapitulate the major features of SSB-DNA complexes in bulk solution.
Fig. 3b shows representative time traces of the donor and acceptor intensities and Eapp from a single DNA-SSB complex which exhibits clear fluctuations between discrete states with Eapp ~ 0.2 and 0.75 (100 mM NaCl, 10 nM SSB). Similar two-state transitions were observed for a range of SSB and NaCl concentrations indicating the clear existence of two distinct SSB-(dT)70 binding modes which interconvert on a time scale ranging from sub-seconds to a minute as we discuss below.
Effects of SSB concentration
Previous ensemble studies15 have shown a SSB concentration dependent transition from a state with one SSB tetramer bound to (dA)70 ((SSB)65 mode) to a state with two tetramers bound to (dA)70 ((SSB)35 mode). An ensemble titration of “(dT)68” with SSB at 80mM NaCl also shows this transition (Fig. 2). Therefore, to further test our FRET state assignments in the single molecule experiments with (dT)70, we examined the effect of SSB concentration at three NaCl concentrations. At 500mM NaCl, a single FRET population in the high FRET state (Eapp ~ 0.75) was observed at all protein concentrations (10 pM–100 nM) (data not shown). SSB binding was very tight such that even at 100 pM SSB, we did not observe unliganded ssDNA (that could be detected as molecules with Eapp ~ 0.1). At 100mM NaCl, the high FRET state (Eapp ~ 0.73) of (dT)70 was first formed at low SSB concentrations, followed by a population shift to the low FRET state (~0.2) at higher SSB concentrations (Fig. 3c), indicating that the low FRET state of (dT)70 has more SSB bound. This is consistent with our assignment that the high and low FRET states have one and two tetramers bound, respectively.
Even at the lowest NaCl concentration examined (10 mM), which favors the (SSB)35 mode, we first observe binding of a single tetramer (at 100 pM SSB), followed by binding of a second SSB tetramer resulting in formation of a low FRET state as the SSB concentration is increased (Fig. 3d). The single tetramer-(dT)70 complex formed at 10 mM NaCl displays Eapp ~ 0.6, which is significantly lower than the values observed at higher salts (100 and 500 mM NaCl). We suggest that this reduction in FRET at 10 mM NaCl may reflect the possibility that a fully wrapped (SSB)65 complex is in dynamic equilibrium with partially unwrapped forms due to the significant negative cooperativity for ssDNA binding to the third and fourth SSB subunits that occurs at low [NaCl]21; 22; 23. Our preliminary investigations indicate that similar complexes formed at low [NaCl] are indeed undergoing fast fluctuations (DNA unwrapping and rewrapping) that are fully averaged during the FRET histogram binning time (300 ms) used here (R. Roy, unpublished data).
Single molecule transition rates
Two-state time trajectories (Fig. 3b, 30 ms time resolution) were analyzed (details in Materials and Methods) to yield transition rates for a range of solution conditions (50–200 mM NaCl; 10–200 nM SSB). The integrated dwell time histograms were fit to single or bi-exponential functions. For the high FRET state, single exponential fits were adequate at high salt ([NaCl] > 100 mM), but at lower salt (50–100 mM NaCl) the data were better fit by a bi-exponential function (Fig. 4a), suggesting that two different species share the same high FRET value. The fast component (with lifetimes < 180 ms independent of SSB concentration) is detectable only at low salt (its amplitude drops sharply with respect to the amplitude of the slow component with increasing [NaCl]; Fig 4b) and is due to a short-lived species we term (SSB35b as will be discussed later. The slow component has a lifetime which is inversely proportional to [SSB] and therefore its inverse represents the observed rate constant for the (SSB)65→(SSB)35 transition, k65→35,obs,sm, which involves binding of an additional SSB tetramer. The observed rate constants at higher salt concentrations (NaCl > 100 mM) were determined from single exponential fits. The values of k65→35,obs,sm increased sharply with decreasing [NaCl] (Fig. 4c and d), consistent with low salt favoring the (SSB)35 mode13. The slope of a plot of k65→35,obs,sm vs. [SSB] (Fig. 4d) yields the apparent bimolecular rate constant, k65→35,sm which ranges from 106 to 2×107 M−1s−1 depending on the [NaCl] (Table I).
Figure 4. Dependence of transition kinetics between different binding modes on NaCl and SSB concentrations (from single molecule experiments).
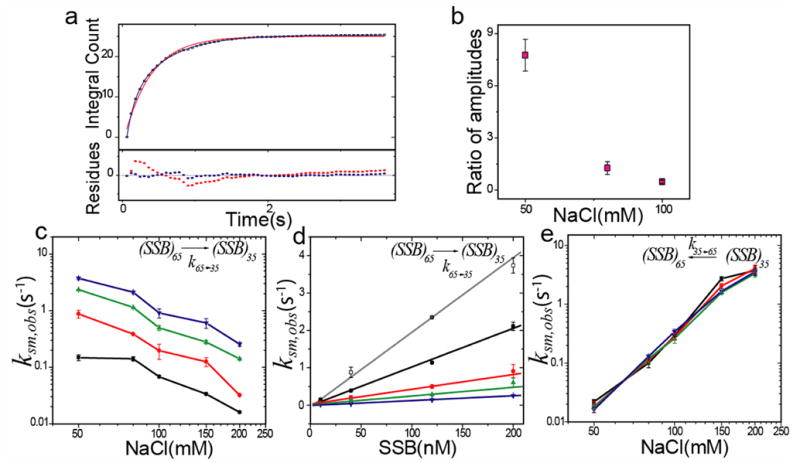
(a) Integrated dwell time distribution in high FRET state for (50 mM NaCl and 10 nM SSB) fit to single (red solid line) and double (blue solid line) exponentials. Residuals of the corresponding fits are shown in the lower panel.
(b) Ratio of amplitudes of fast to slow component (panel a) as a function of [NaCl]. Amplitude of faster component from the double-exponential fits decreases dramatically with increasing salt concentration such that only single exponential fits are needed above 100 mM NaCl.
(c) Rates of transition from the (SSB)65 to the (SSB)35 mode, k65→35,obs,sm, as a function of [NaCl] at different [SSB] (black squares- 10 nM SSB, red circles- 40 nM SSB, green triangles- 120 nM SSB, blue inverted triangles- 200 nM SSB).
(d) Dependence of observed rates, k65→35,obs,sm, on [SSB] at various [NaCl] (white, black, red, green and blue represent 50, 80, 100, 150, 200 mM NaCl, respectively). The slopes of the linear least squares fits (shown in grey, black, red, green and blue) yield the bimolecular rate constant for the transition at specified [NaCl] (tabulated in Table I).
(e) Rate of transition from (SSB)35 → (SSB)65 binding mode, k35→65,obs,sm increases with increasing [NaCl], but is independent of [SSB] (symbols and colors are similar to those in panel c).
Table I.
Kinetic parameters for transitions between (SSB)35 and (SSB)65 binding modes for wild type SSB
| [NaCl] | k35→65,sf, s−1 (intercept, stopped flow) | k35→65,sm, s−1 (average, single molecule) | k65→35,sf, M−1s−1 (slope, stopped flow) | k65→35,sm, M−1s−1 (slope, single molecule) |
|---|---|---|---|---|
| 0.04 M | <0.08 | - | (6.96±0.34)×106 | - |
| 0.05 M | - | 0.018±0.01 | - | (2.0±0.02)×107 |
| 0.06 M | <0.08 | - | (4.73±0.17)×106 | - |
| 0.08 M | 0.08±0.02 | 0.11±0.01 | (3.42±0.16)×106 | (1.04±0.06)×107 |
| 0.1 M | 0.31±0.02 | 0.29±0.02 | (2.53±0.17)×106 | (3.96±0.4)×106 |
| 0.15 M | 1.69±0.04 | 2.0±0.26 | (0.77±0.32)×106 | (2.33±0.17)×106 |
| 0.2 M | 4.47±0.02a | 3.68±0.15 | - | (1.29±0.09)×106 |
| 0.3 M | 14.8±0.05a | - | - | - |
| 0.5 M | 46.78±0.22a | - | - | - |
| 0.8 M | 90.66±0.54a | - | - | - |
| 1.0 M | 104.9±0.70a | - | - | - |
values of k35→65,sf are calculated as an average of ksf,obs, at different protein-DNA ratios, since there is no dependence on [SSB] at these [NaCl] (see Figure 5b)
Dwell times for the low FRET state were well described by single exponential fits and yielded the observed rate klow→high. We obtained the actual rate constant for the (SSB)35→(SSB)65 transition (k35→65,obs,sm) by correcting for the contributions from the alternative path of depopulating (SSB)35 to form (SSB)35b (see Materials and Methods). Remarkably, k35→65,obs,sm increased 200 fold for a mere four-fold increase in [NaCl] (Fig. 4e). The value of k35→65,obs,sm does not depend on SSB concentration as expected for a dissociation reaction and the average value of k35→65,sm, is shown in Table I. This represents the first determination of the kinetic parameters for this SSB binding mode transition.
Stopped-flow studies of the (SSB)35–(SSB)65 binding mode transition initiated by salt-jumps
To compare with the single molecule studies, we also performed stopped-flow kinetic studies of the transition between the (SSB)35 and (SSB)65 modes in bulk solution (buffer T, pH 8.1, 25°C). The (SSB)35 complex was pre-formed at low salt concentration (no added NaCl with SSB in molar excess over (dT)68) and the transition to the (SSB)65 mode was initiated by the addition of buffer containing a series of higher [NaCl]. Example time courses (20 nM Cy5(dT)68Cy3dT and 40 nM SSB) are shown in Figure 5a and are all well described by a single exponential increase in Cy5 fluorescence yielding observed rate constants, ksf,obs. Figure 5b shows a plot of logksf,obs vs. log[NaCl] determined for a variety of SSB to ssDNA concentration ratios indicating that ksf,obs is very sensitive to [NaCl]. For example, for the starting concentrations used for the time courses shown in Figure 5a, ksf,obs increases by almost 5000-fold (light green line in Fig. 5b) as the [NaCl] is increased from below 20 mM to 1 M.
Figure 5. Stopped-flow kinetics of the (SSB)35 to (SSB)65 binding mode transition initiated by salt-jumps.
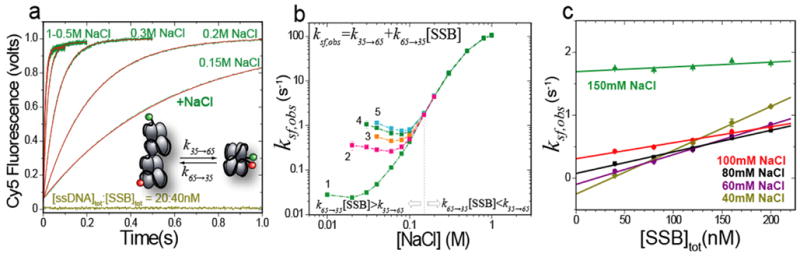
(a) Cy5 FRET time courses (green) obtained upon addition of buffer T with increasing [NaCl] to a premixed (Cy5-T68-Cy3-T)-SSB complex (20 nM: 40 nM final ratio) in the stopped-flow in buffer T with no salt at 25°C. Each time course shows an increase in FRET signal reflecting formation of (SSB)65 mode. The fits of the traces to single exponential functions are shown in red.
(b) Observed rates (ksf,obs) determined from the single exponential fits each time course are plotted as a function of [NaCl] for different initial ratios of [Cy5-T68-Cy3-T]tot/[SSB]tot : (1) 20:40 nM, (2) 20:80 nM, (3) 20:120 nM, (4) 20:160 nM and (5) 20:200 nM.
(c) Dependences of ksf,obs on [SSB] obtained from the data in panel (b) for [NaCl] < 0.2 M. The slope and intercept of the linear fits yield the bimolecular rate constant of (SSB)65→(SSB)35 (k65→35,sf) and rate constant for (SSB)35→(SSB)65 (k35→65,sf), respectively (see Table I). For [NaCl] ≥ 0.2 M ksf,obs is independent on [SSB] (see Table I).
The value of ksf,obs is independent of SSB concentration at high salt (above 0.15 M NaCl) but displays a linear dependence on [SSB] at lower salt concentrations (Fig. 5c). The slopes of the plots in Fig. 5c decrease with increasing [NaCl], whereas the intercepts increase (also tabulated in Table I). A simple one-step kinetic mechanism, as shown in the inset to Figure 5a, can explain the data, where k35→65,sf and k65→35,sf are the rate constants for the (SSB)35→(SSB)65 and (SSB)65→(SSB)35 transitions, respectively. Based on this mechanism, the observed rate, ksf,obs, is given by equation (1).
| (1) |
Therefore, the individual rate constants, k35→65,sf and k65→35,sf can be obtained from the intercept and slope of the linear dependences of ksf,obs on [SSB] shown in Fig. 5c (Table I). It is clear from Figures 5b and c that the first term in equation (1) dominates at [NaCl] > 0.15 M where the (SSB)65 mode is favored. As the [NaCl] concentration is decreased, k35→65,sf also decreases and the second term in equation (1) becomes significant.
The rates for the (SSB)35→ (SSB)65 transition determined from the single molecule and stopped-flow measurements agree very well as shown in Table I (compare k35→65,sm and k35→65,sf). The bimolecular rate constants determined for the (SSB)65→ (SSB)35 transition are also comparable. For example, k65→35,sm which varies by a factor of ten upon increasing the [NaCl] from 50 mM to 200 mM was only 1.5–3 fold higher than k65→35,sf. This small difference may be due to the fact that the ssDNA used in the single molecule studies is longer, by 2 nucleotides, than that used in the stopped-flow studies, which should facilitate the binding of the second tetramer (each with an occluded site size of ~35 nt). A similar enhancement (~2 fold) in rate is observed for SSB binding to (dT)68 vs. (dT)66 in stopped-flow measurements (A. G. Kozlov, unpublished results).
C-terminal truncated SSB and electrostatic effects
The C-terminus of E. coli SSB protein is highly acidic2; 24, a feature shared by many other SSB proteins, such as phage T4 gene 32 protein (gp32)25; 26 and phage T7 gene 2.5 protein (gp2.5) 1,27; 28. The C-terminal domain of T4 gp32 has previously been shown to modulate DNA binding via a salt dependent structural rearrangement 25; 26; 29; 30. Both for E. coli SSB 2; 5; 31; 32; 33; 34; 35 and the T7 gp2.5 28; 36; 37, the C-terminal domain is the site for interactions with other proteins involved in DNA processing. Since the DNA wrapping topology (Fig. 1a) was derived from a crystal structure of ssDNA bound to a 42 amino acid C-terminal truncation of SSB (termed SSBc)8 and the DNA was very poorly resolved in a crystal structure of the full length SSB38, we examined the binding mode transitions for ssDNA binding to SSBc, to determine whether deletion of the C-termini influences the salt-dependent dynamics of these complexes. Importantly, deletion of the 42 amino acids from each SSB subunit does not affect either the stability of the SSB tetramer or its high salt occluded site size (A. G. Kozlov, unpublished experiments).
Upon performing single molecule FRET experiments with the SSBc tetramer, we still observed transitions between two FRET states with identical FRET values as observed for the full length SSB tetramer except that the equilibrium is shifted in favor of the (SSBc)35 mode (data not shown). Only the lowest salt concentration tested (100mM NaCl) displayed any bi-exponential behavior in the high FRET state dwell time distribution, and for that case the slower component was used to estimate the rate constant for the (SSBc)65→(SSBc)35 transition. Both the forward and reverse rate constants had similar dependences on NaCl concentration as was observed for full length SSB (Fig. 6a–c). However, there is a two-fold decrease in the rate for the (SSBc)35→(SSBc)65 transition and a ~ two-fold increase in the bimolecular rate constant for the (SSBc)65→(SSBc)35 transition compared to full length SSB (Tables I and II). The net result is a four-fold change in the equilibrium constant for the transition, K65→35 ≡ k65→35,sm/k35→65,sm (Fig. 6d), indicating that deletion of the C-termini favors the (SSB)35 mode.
Figure 6. Kinetics of binding mode transition for SSBc (C-terminal truncated SSB).
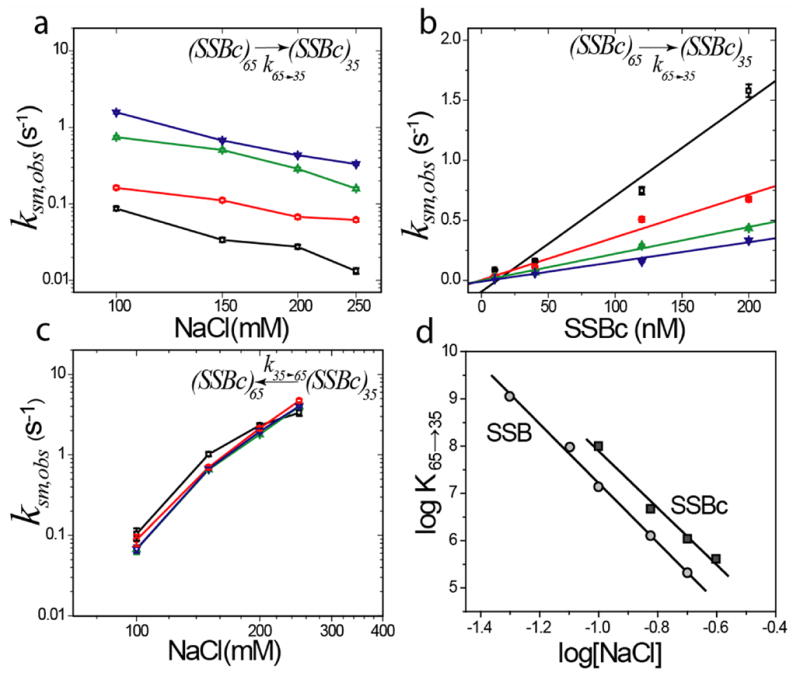
(a) Observed rate of transition from (SSBc)65 → (SSBc)35 mode, k65→35,obs,sm decreases with increasing [NaCl] but increases with [SSBc]. (Black squares, red circles, green triangles and blue inverted triangles represent 10, 40, 120 and 200 nM SSBc respectively)
(b) Slope of k65→35,obs,sm plotted as a function of SSBc concentration yields a bimolecular rate constant which is ~2 fold higher than for wild type SSB for all NaCl concentrations (see Tables I & II).
(c) Observed rate for (SSBc)35 → (SSBc)65 binding mode transition, k35→65,obs,sm increases with [NaCl] albeit 2 fold slower than for wild type SSB for the same salt concentrations. There is no change in k35→65,obs,sm with [SSBc] ([SSBc] color coding same as in panel a).
(d) Dependences of the equilibrium constant, K65→35 (calculated as k65→35/k35→65 from single molecule data) on [NaCl] (log-log plot) for SSB (circles) and SSBc (squares). The slopes, −6.3±0.2 and −6.0±0.5 for SSB and SSBc, respectively, indicate a net release of ~6 ions.
Table II.
Kinetic parameters for transitions between (SSBc)35 and (SSBc)65 binding modes for truncated SSB (SSBc) from single molecule experiments
| [NaCl] | k35→65,sm, s−1 (average) | k65→35,sm, M−1s−1 (slope) |
|---|---|---|
| 0.1 M | 0.08±0.01 | (7.98±0.88)×106 |
| 0.15 M | 0.76±0.09 | (3.56±0.45)×106 |
| 0.2 M | 2.05±0.11 | (2.23±0.15)×106 |
| 0.25 M | 4.02±0.28 | (1.64±0.16)×106 |
From the slope of a plot of log(K65→35) vs. log[NaCl], one can estimate the net number of ions (Na+ + Cl−) released or absorbed in the transition from the (SSB)65 to the (SSB)35 modes11. Such plots are shown in Figure 6d for both the SSB and SSBc binding mode transitions. In each case, the log-log slopes are negative, with slopes of −6.3 ± 0.2 and −6.0 ± 0.5 for SSB and SSBc, indicating a net release of ~ 6 ions upon binding the second SSB tetramer to form the (SSB)35 mode. Overall, the results suggest that the DNA wrapping topology deduced from the ssDNA-SSBc structures is likely to be the same for SSBc and the full length protein. However, the C-terminus does exert a significant influence on the equilibrium between the two binding modes. Since the C-terminus of SSB is the region involved in interactions with other accessory proteins, it is possible that the binding of these proteins to the C-terminus could alter either the equilibrium and/or dynamics of the transition between the SSB binding modes.
(SSB)35b, a novel (SSB)35 complex
The results presented above indicate evidence for two distinct SSB-DNA species that co-exist within the high FRET state, but that have markedly different lifetimes. We assigned the longer-lived species to the (SSB)65 mode and the shorter-lived species to a new binding configuration termed (SSB)35b since it exhibits the same stoichiometry as in the (SSB)35 mode. The rate constant for the transition from the (SSB)35b to the (SSB)35 mode, k35b→35, was observed to be independent of [SSB] (open squares in Fig. 7a) and only slightly dependent on [NaCl]. The rate constant for the reverse step, k35→35b, calculated as described in Materials and Methods (open circles in Fig. 7a), is independent of [NaCl] and nearly 100-fold smaller in magnitude than k35b→35.
Figure 7. Kinetics of the (SSB)35b↔ (SSB)35 transition.
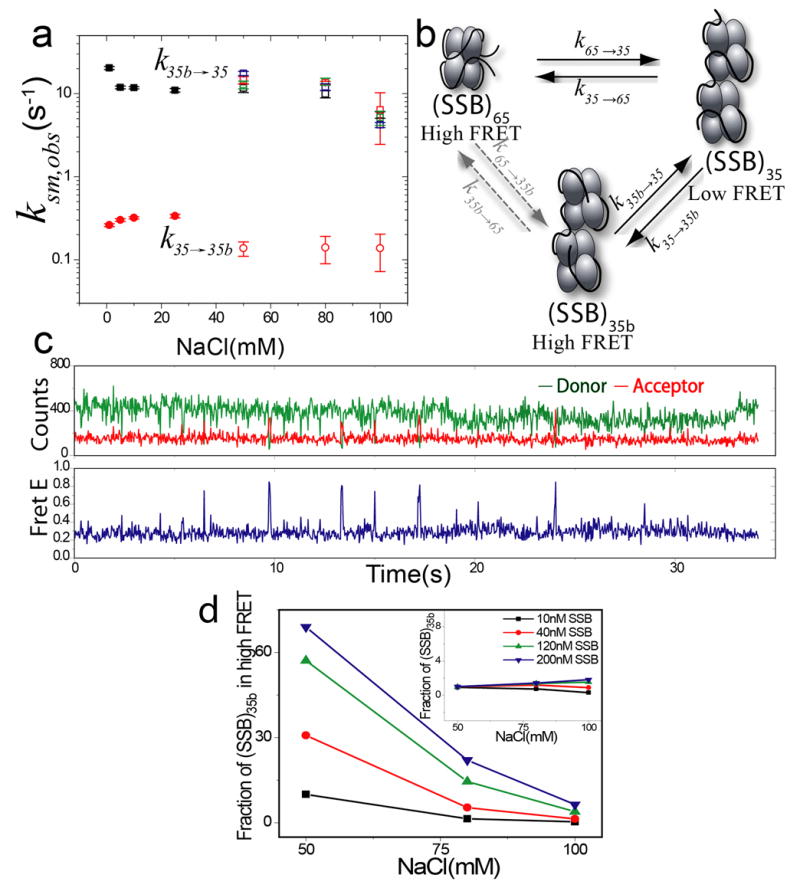
(a) Rate constants for the (SSB)35↔ (SSB)35b transition k35→35b (~0.3s−1; solid red circles) and k35b→35 (20–12s−1 solid black squares) determined for isolated (SSB)35 complexes ([SSB]:[dT]70=2:1, no SSB in solution) at low [NaCl] (1–25 mM) from the analysis of dwell times of the traces as shown in panel (c). These rates are close to those estimated from experiments performed in the presence of free SSB in solution at intermediate [NaCl] (50–100mM), k35→35b (~ 0.1–0.2 s−1; open red circles) and k35b→35 (~ 15–6 s−1, open squares; different colors represent different SSB concentrations – 10 nM (black), 40 nM (red), 120 nM SSB (green) and 200 nM (blue)).
(b) Kinetic scheme showing interconversion among the SSB binding modes on (dT)70 (see text for details).
(c) Fluorescence Intensity and FRET time record of single DNA complexed with 2 SSB tetramers in 10 mM NaCl in the absence of free SSB show brief excursions to a high FRET state indicating that the (SSB)35 binding mode can undergo a transition to a high FRET state without dissociation of SSB from the ssDNA.
(d) Fraction of (SSB)35b in the high FRET state as a function of NaCl and SSB concentrations (10 nM (black ), 40 nM (red), 120 nM SSB (green) and 200 nM (blue)). This is a minor species in the population except at low salt (50mM) and high protein concentrations (> 40 nM SSB). Insert: Fraction of (SSB)35b in the total equilibrium population is always < 2%. Colors for SSB concentrations are as in main panel.
We hypothesize that (SSB)35b represents a new configuration of SSB binding to ssDNA consisting of two tetramers bound per (dT)70 as in (SSB)35, but which differs in that the two ends of (dT)70 are in much closer proximity. If this is the case, the (SSB)35b complex should be able to be formed directly from the (SSB)35 mode without any change in the extent of SSB binding to the ssDNA (Fig. 7b). Indeed, when we form the (SSB)35 mode at low [NaCl] and remove free SSB from solution, an isolated complex in the (SSB)35 mode displays transient excursions to the high FRET state (Fig. 7c).
Next, we compared the kinetic rate constants for the transition between the (SSB)35b and (SSB)35 modes determined from the two sets of experiments described above. The values of k35b→35 obtained from analysis of the time-dependent fluctuations in the isolated (SSB)35 complexes display only a moderate salt dependence over the range from 1 mM to 25 mM NaCl (Fig. 7a, filled black squares). These values approach the values of k35b→35 determined from the fast phase of the bi-exponential decay in the presence of free SSB in the range from 50 – 100 mM NaCl (Fig. 7a, open squares). Furthermore, the values of k35→35b, calculated from analysis of the isolated (SSB)35 complexes (solid circles in Fig. 7a) were similar to the values of k35→35b estimated at higher salt concentrations in the presence of free SSB (open circles in Fig. 7a).
Using these rate constants, we estimated the equilibrium population of (SSB)35b for the range of solution conditions in this study (details in Materials and Methods). Figure 7d shows the relative population of (SSB)35b within the high FRET state which increases with increasing protein concentration and decreasing [NaCl]. However, the fraction of (SSB)35b in the total population of SSB-DNA complexes at equilibrium is less than 2% under all solution conditions examined (Fig. 7d inset).
On the basis of the arguments presented above, we use the kinetic scheme depicted in Figure 7b to describe the transitions among these three conformations. The (SSB)35 binding mode with 2 SSB tetramers bound per (dT)70 can undergo transitions either to the (SSB)65 mode upon dissociation of one tetramer or to the (SSB)35b configuration via a conformational rearrangement without SSB dissociation. Although the kinetic scheme in Figure 7b indicates a direct pathway for interconversion between the two high FRET binding modes, (SSB)65 and (SSB)35b (dashed arrows in Fig. 7b), the rate constants indicate that the preferred pathway for this interconversion would proceed through the (SSB)35 binding mode.
Discussion
In this study, we have used single molecule and stopped-flow FRET methods to determine, for the first time, the dynamics of the spontaneous transitions between the major SSB/DNA binding modes. Importantly, our single molecule data show in a most direct way that SSB binds to ssDNA in well-defined structural states that reflect these different binding modes that may regulate the functional activity of SSB in vivo5. The experiments described here lay the foundation for further single molecule investigations of the central role of this key protein in DNA replication and recombination processes.
Kinetic analysis of single molecule FRET data revealed evidence that three distinct complexes can form on a 70 nt ssDNA: (SSB)35 with two SSB tetramers, (SSB)65 with one tetramer, and (SSB)35b, a newly uncovered minority species with two tetramers bound. Furthermore, we have established the reaction pathways connecting these species (Fig. 7b). The rate constants of the transitions between the (SSB)35 and (SSB)65 modes are extremely sensitive to salt concentration. For example, k65→35 decreases 10–16 fold with a four-fold increase in [NaCl] (from 50 mM to 200 mM). Even more remarkably, k35→65 varies by 200-fold over the same range of salt concentration. As a consequence, outside of a narrow range of salt concentrations, only one or the other of the two modes is populated significantly. We have not performed a kinetic analysis as a function of Mg2+, but equilibrium data (Fig. S1) suggest that a similar dichotomy holds for Mg2+ around its physiologically relevant concentrations (which most likely lies close to the higher end of the reported range of 6 μM to 10mM39; 40; 41). In fact, the 35 to 56 transition can also be induced at micromolar concentrations of the polyamine, spermidine42. Therefore, charged species present in the cell or a change in the expression level of SSB can easily shift the delicate balance between the two modes providing the possibility for significant regulatory consequences.
We observe that the (SSB)35 complex can also undergo a conformational transformation to form a previously undetected, low abundance ternary complex (2SSB: DNA) which we term (SSB)35b. This minor complex was revealed only through kinetic analysis of the single molecule data and is not populated sufficiently to be detected in ensemble experiments. Extremely fast dynamics of DNA wrapping/unwrapping occurring in the microsecond time scale have been observed within the (SSB)65 complex43. In the (SSB)65 complex, SSB contacts ssDNA via all four subunits and therefore should be capable of remaining bound to DNA even if a long segment of the DNA is unraveled. Here, we provide evidence that the (SSB)35 binding conformation is also dynamically capable of sampling alternate topologies without protein dissociation, even though the SSB is bound to ~35 nts using only two subunits. These results suggest that small scale DNA wrapping/unwrapping dynamics might occur in this mode with implications for how SSB tetramers might undergo direct transfer between single strands of DNA44.
Deletion of the negatively charged C-terminus of SSB shifts the equilibrium constant for the (SSB)65 to (SSB)35 transition by a factor of four in favor of the (SSB)35 mode. This raises the possibility that the binding of SSB to replication/recombination proteins, such as the χ subunit of DNA polymerase III34; 45, exonuclease I31, PriA33 and RecO46, which occurs primarily via the C-termini of SSB may also influence the equilibrium between SSB-ssDNA binding modes. The (SSB)35 mode has been proposed to function during DNA replication by virtue of its high cooperativity5; 14, and our observation with SSBc suggests the interesting possibility that binding of the χ subunit of DNA polymerase III holoenzyme to the C-terminus of SSB35 may stabilize the (SSB)35 mode and induce the formation of long SSB clusters. More generally, other proteins may modulate SSB function by controlling its DNA binding mode. Future studies will test this model of reciprocity where the DNA binding mode of SSB is influenced by its interaction with other proteins via the C-terminus and vice versa.
The existence of DNA binding proteins that can bind in multiple, salt-dependent binding modes is more prevalent than generally thought. In fact, any DNA binding protein possessing multiple DNA binding sites has the potential for binding DNA in multiple binding modes. Such salt dependent binding mode transitions have been observed for ssDNA binding of yeast RPA (Replication Protein A, the eukaryotic SSB protein, which possesses 4–5 OB-folds)6, as well as for hnRNP (heterogeneous nuclear Ribonucleoprotein) binding to pre-mRNA47. The strong dependence of the stabilities and rates of interconversion between the SSB-ssDNA binding modes on salt and SSB concentration emphasizes the need to consider explicitly the different properties of its different binding modes5; 11,14 when interpreting experiments that include SSB as a component.
Materials and Methods
Oligodeoxyribonucleotides and SSB proteins
For single molecule experiments, a partial duplex with a 71 base long ssDNA 3′ tail was generated by annealing strands 5′-Cy5-GCCTCGCTGCCGTCGCCA-Biotin-3′ and 5′-TGGCGACGGCAGCGAGGC(T)70-Cy3-T-3′ so that the FRET (Cy3-Cy5) pairs are 70 bases apart (“(dT)70”). Stopped-flow studies and equilibrium titrations were done with 5′-Cy5-(T)68-Cy3-T-3′ with concentration determined as described earlier19. The oligodeoxynucleotides were synthesized using standard β-cyanoethyl phosphoramidite chemistry using an ABI model 391 automated DNA synthesizer (Applied Biosystems, Foster City, CA). Biotin was incorporated using BiotinTEG CPG (all modification reagents were from Glen Research, Sterling, VA). E. coli SSB and SSBc proteins (>99% homogeneity) were purified and concentrations determined spectrophotometrically as described48.
Single-molecule TIR spectroscopy and analysis
All single molecule measurements were performed at 23±1°C in 10mM Tris (pH 8.0), 0.1mM Na3EDTA, 0.1mg/ml BSA (NEB, Ipswich, MA), oxygen scavenging system49; 50 (0.5% w/v glucose, 1.5mM Trolox51, 165U/ml glucose oxidase and 2170U/ml catalase (Roche, Indianapolis, IN)) and indicated amounts of NaCl and SSB. Trolox concentration was determined spectrophotometrically using an extinction coefficient ε290= 2350±100 M−1cm−1.
A wide-field prism-type total internal reflection (TIR) microscope (IX 70, Olympus) was used to image single immobilized (<50pM) DNA on NeutrAvidin (Pierce, Rockford, IL) treated biotin-PEG (PEG-MW 5,000, Nektar Therapeutics, Huntsville, AL) quartz surface50. Donor (Cy3) was excited with an Nd:YAG laser (532nm, 50mW; Reno, NV). Donor and Acceptor (Cy5) fluorescence was collected using water immersion objective (60x, 1.2 numerical aperture, Olympus). Laser scatter was rejected using a long pass filter at 550nm (Chroma, Rockingham, VT). Cy3 and Cy5 fluorescence was split using a dichroic mirror at 630nm (Chroma) and imaged side by side on an electron multiplying charge coupled device (CCD) camera (iXon DV 887-BI, Andor Technology, CT). Mapping calibration between the donor and acceptor channels is achieved with the help of immobilized fluorescent beads (0.2μm, 540/560nm, Molecular Probes). Fluorescence data as successive frames of images were acquired with custom Visual C++ (Microsoft) routines and IDL (Research Systems Inc.) programs were used to obtain donor and acceptor intensity time traces with 30ms integration time.
After correcting for the leakage and background in both channels, apparent FRET efficiency was calculated as Eapp = IA/(IA + ID) where IA and ID represent acceptor and donor intensity, respectively. Single-molecule FRET histograms were generated by averaging for 300ms. Gamma correction factor, γwas calculated by estimating the ratio of change in the acceptor intensity, ΔIA to change in the donor intensity, ΔID after acceptor photobleaching (γ=ΔIA/ΔID)20. Gamma factor calculated (>50 molecules) was binned and fitted to a gaussian to yield the representative gamma factor for each solution condition (data not shown). Dwell times in each of the FRET states were estimated from time traces (having at least 10 turnovers; 60–120sec long) using custom MATLAB (Mathworks) routines using a ‘thresholding’ criterion52. Histograms of dwell times in each of the FRET states were integrated and fitted to single or bi-exponential decay functions to obtain the average lifetimes and respective amplitudes. Rate constants were estimated as the inverse of the lifetimes. For rate constants ≤ 0.1s−1, time traces from multiple molecules were concatenated for dwell time analysis to prevent underestimation of the dwell times.
In the presence of two high FRET states ((SSB)65 and (SSB)35b), the observed rate for the transition from the low to the high FRET state (klow→high, obtained as the inverse of low FRET lifetime) reflects the weighted sum of k35→65,obs,sm (the observed rate constant for (SSB)35→(SSB)65) and k35→35b,obs,sm (the rate constant for (SSB)35→(SSB)35b). Hence, k35→65,obs,sm and k35→35b,obs,sm can be approximated using equation 2,
| (2a) |
| (2b) |
where Ampslow and Ampfast represent the amplitudes of the fast and slow components determined from the bi-exponential fits. The fraction of (SSB)35b state in the high FRET state and the fraction of (SSB)35b in the total population are related to the rate constants k35→35b, k35b→35, k35→65, k65→35 and [SSB] according to the expressions in equations 3a and b, respectively.
| (3a) |
| (3b) |
Stopped-flow kinetics and equilibrium fluorescence titrations
Stopped-flow data were acquired using an SX.18MV stopped-flow instrument (Applied Photophysics Ltd., Leatherhead, UK) as described19. Cy3 was excited at 515nm and sensitized Cy5 emission was collected using a 665nm long pass filter. Reactant solutions were prepared on ice and pre-incubated at 25°C for 5 mins prior to mixing. All kinetic time courses used here represent an average of 6–10 individual traces that were fitted to single exponentials. Fluorescence titrations were performed using a QM-4 spectrofluorometer (Photon Technology International, Lawrenceville, NJ) exciting Cy3 donor (515 nm) and monitoring sensitized emission from Cy5 acceptor at 664 nm applying all corrections as described53. All measurements were carried out in buffer T (10mM Tris pH 8.1, 0.1mM Na3EDTA) at 25±0.1°C. All chemicals were reagent grade and from Sigma-Aldrich (St. Louis, MO), if not stated otherwise.
Supplementary Material
Acknowledgments
RR would like to thank S. McKinney for data acquisition programs; C. Joo, I. Rasnik, S. Hohng, M. Nahas and S. Myong for experimental help and discussion. We also thank Dr. G. Waksman for providing the pdb structures for Figure 1 and T. Ho for DNA synthesis. Funding for the work was provided by NIH grants to T.H. (GM065367) and T.M.L. (GM030498). T.H. is an investigator with the Howard Hughes Medical Institute.
Abbreviations
- SSB
single stranded DNA binding
- Tris
tris(hydroxymethyl)aminomethane
- Trolox
6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid
- EDTA
ethylenediaminetetraacetic acid
- ssDNA
single-stranded DNA
- FRET
fluorescence resonance energy transfer
- TIR
Total Internal Reflection
- BSA
Bovine Serum Albumin
- PEG
poly-ethyleneglycol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–36. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 2.Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990;54:342–80. doi: 10.1128/mr.54.4.342-380.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuzhakov A, Kelman Z, O’Donnell M. Trading places on DNA--a three-point switch underlies primer handoff from primase to the replicative DNA polymerase. Cell. 1999;96:153–63. doi: 10.1016/s0092-8674(00)80968-x. [DOI] [PubMed] [Google Scholar]
- 4.Sun W, Godson GN. Structure of the Escherichia coli primase/single-strand DNA-binding protein/phage G4oric complex required for primer RNA synthesis. J Mol Biol. 1998;276:689–703. doi: 10.1006/jmbi.1997.1471. [DOI] [PubMed] [Google Scholar]
- 5.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–70. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 6.Kumaran S, Kozlov AG, Lohman TM. Saccharomyces cerevisiae replication protein A binds to single-stranded DNA in multiple salt-dependent modes. Biochemistry. 2006;45:11958–73. doi: 10.1021/bi060994r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94:6652–7. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–52. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 9.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct. 2003;32:115–33. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bujalowski W, Lohman TM. Escherichia coli single-strand binding protein forms multiple, distinct complexes with single-stranded DNA. Biochemistry. 1986;25:7799–802. doi: 10.1021/bi00372a003. [DOI] [PubMed] [Google Scholar]
- 11.Bujalowski W, Overman LB, Lohman TM. Binding mode transitions of Escherichia coli single strand binding protein-single-stranded DNA complexes. Cation, anion, pH, and binding density effects. J Biol Chem. 1988;263:4629–40. [PubMed] [Google Scholar]
- 12.Griffith JD, Harris LD, Register J., 3rd Visualization of SSB-ssDNA complexes active in the assembly of stable RecA-DNA filaments. Cold Spring Harb Symp Quant Biol. 1984;49:553–9. doi: 10.1101/sqb.1984.049.01.062. [DOI] [PubMed] [Google Scholar]
- 13.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–603. [PubMed] [Google Scholar]
- 14.Lohman TM, Bujalowski W, Overman LB. E. coli single strand binding protein: a new look at helix-destabilizing proteins. Trends Biochem Sci. 1988;13:250–5. [PubMed] [Google Scholar]
- 15.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236:106–23. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 16.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 17.Ha T. Single-molecule fluorescence methods for the study of nucleic acids. Curr Opin Struct Biol. 2001;11:287–92. doi: 10.1016/s0959-440x(00)00204-9. [DOI] [PubMed] [Google Scholar]
- 18.Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys J. 2004;86:2530–7. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozlov AG, Lohman TM. Stopped-flow studies of the kinetics of single-stranded DNA binding and wrapping around the Escherichia coli SSB tetramer. Biochemistry. 2002;41:6032–44. doi: 10.1021/bi020122z. [DOI] [PubMed] [Google Scholar]
- 20.Ha T, Ting AY, Liang J, Caldwell WB, Deniz AA, Chemla DS, Schultz PG, Weiss S. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc Natl Acad Sci U S A. 1999;96:893–8. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. I Evidence and a quantitative model. J Mol Biol. 1989;207:249–68. doi: 10.1016/0022-2836(89)90454-3. [DOI] [PubMed] [Google Scholar]
- 22.Bujalowski W, Lohman TM. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. II Salt, temperature and oligonucleotide length effects. J Mol Biol. 1989;207:269–88. doi: 10.1016/0022-2836(89)90455-5. [DOI] [PubMed] [Google Scholar]
- 23.Lohman TM, Bujalowski W. Negative cooperativity within individual tetramers of Escherichia coli single strand binding protein is responsible for the transition between the (SSB)35 and (SSB)56 DNA binding modes. Biochemistry. 1988;27:2260–5. doi: 10.1021/bi00407a002. [DOI] [PubMed] [Google Scholar]
- 24.Chase JW, L’Italien JJ, Murphy JB, Spicer EK, Williams KR. Characterization of the Escherichia coli SSB-113 mutant single-stranded DNA-binding protein. Cloning of the gene, DNA and protein sequence analysis, high pressure liquid chromatography peptide mapping, and DNA-binding studies. J Biol Chem. 1984;259:805–14. [PubMed] [Google Scholar]
- 25.Lonberg N, Kowalczykowski SC, Paul LS, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. III Binding properties of two specific proteolytic digestion products of the protein (G32P*I and G32P*III) J Mol Biol. 1981;145:123–38. doi: 10.1016/0022-2836(81)90337-5. [DOI] [PubMed] [Google Scholar]
- 26.Waidner LA, Flynn EK, Wu M, Li X, Karpel RL. Domain effects on the DNA-interactive properties of bacteriophage T4 gene 32 protein. J Biol Chem. 2001;276:2509–2516. doi: 10.1074/jbc.M007778200. [DOI] [PubMed] [Google Scholar]
- 27.Marintcheva B, Hamdan SM, Lee SJ, Richardson CC. Essential Residues in the C Terminus of the Bacteriophage T7 Gene 2.5 Single-stranded DNA-binding Protein. J Biol Chem. 2006;281:25831–40. doi: 10.1074/jbc.M604601200. [DOI] [PubMed] [Google Scholar]
- 28.Kong DC, Richardson CC. Role of the acidic carboxyl-terminal domain of the single-stranded DNA-binding protein of bacteriophage T7 in specific protein-protein interactions. J Biol Chem. 1998;273:6556–6564. doi: 10.1074/jbc.273.11.6556. [DOI] [PubMed] [Google Scholar]
- 29.Hosoda J, Moise H. Purification and physicochemical properties of limited proteolysis products of T4 helix destabilizing protein (gene 32 protein) J Biol Chem. 1978;253:7547–7558. [PubMed] [Google Scholar]
- 30.Pant K, Karpel RL, Rouzina I, Williams MC. Salt dependent binding of T4 gene 32 protein to single and double-stranded DNA: single molecule force spectroscopy measurements. J Mol Biol. 2005;349:317–30. doi: 10.1016/j.jmb.2005.03.065. [DOI] [PubMed] [Google Scholar]
- 31.Genschel J, Curth U, Urbanke C. Interaction of E. coli single-stranded DNA binding protein (SSB) with exonuclease I. The carboxy-terminus of SSB is the recognition site for the nuclease. Biol Chem. 2000;381:183–92. doi: 10.1515/BC.2000.025. [DOI] [PubMed] [Google Scholar]
- 32.Curth U, Genschel J, Urbanke C, Greipel J. In vitro and in vivo function of the C-terminus of Escherichia coli single-stranded DNA binding protein. Nucleic Acids Res. 1996;24:2706–11. doi: 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–87. doi: 10.1093/nar/gkh980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelman Z, Yuzhakov A, Andjelkovic J, O’Donnell M. Devoted to the lagging strand-the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–49. doi: 10.1093/emboj/17.8.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–40. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J Biol Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 37.He ZG, Richardson CC. Effect of single-stranded DNA-binding proteins on the helicase and primase activities of the bacteriophage T7 gene 4 protein. J Biol Chem. 2004;279:22190–22197. doi: 10.1074/jbc.M401100200. [DOI] [PubMed] [Google Scholar]
- 38.Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G. The C-terminal domain of full-length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–7. doi: 10.1110/ps.04661904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kung FC, Raymond J, Glaser DA. Metal ion content of Escherichia coli versus cell age. J Bacteriol. 1976;126:1089–95. doi: 10.1128/jb.126.3.1089-1095.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lusk JE, Williams RJ, Kennedy EP. Magnesium and the growth of Escherichia coli. J Biol Chem. 1968;243:2618–24. [PubMed] [Google Scholar]
- 41.Hurwitz C, Rosano CL. The intracellular concentration of bound and unbound magnesium ions in Escherichia coli. J Biol Chem. 1967;242:3719–22. [PubMed] [Google Scholar]
- 42.Wei TF, Bujalowski W, Lohman TM. Cooperative binding of polyamines induces the Escherichia coli single-strand binding protein-DNA binding mode transitions. Biochemistry. 1992;31:6166–74. doi: 10.1021/bi00141a029. [DOI] [PubMed] [Google Scholar]
- 43.Kuznetsov SV, Kozlov AG, Lohman TM, Ansari A. Microsecond dynamics of protein-DNA interactions: direct observation of the wrapping/unwrapping kinetics of single-stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. doi: 10.1016/j.jmb.2006.02.070. [DOI] [PubMed] [Google Scholar]
- 44.Kozlov AG, Lohman TM. Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry. 2002;41:11611–27. doi: 10.1021/bi020361m. [DOI] [PubMed] [Google Scholar]
- 45.Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single-stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. doi: 10.1093/nar/gkg498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hobbs MD, Sakai A, Cox MM. SSB limits RecOR binding onto single strand DNA. 2007 doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 47.Nadler SG, Merrill BM, Roberts WJ, Keating KM, Lisbin MJ, Barnett SF, Wilson SH, Williams KR. Interactions of the A1 heterogeneous nuclear ribonucleoprotein and its proteolytic derivative, UP1, with RNA and DNA: evidence for multiple RNA binding domains and salt-dependent binding mode transitions. Biochemistry. 1991;30:2968–76. doi: 10.1021/bi00225a034. [DOI] [PubMed] [Google Scholar]
- 48.Lohman TM, Green JM, Beyer RS. Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under lambda PL control. Biochemistry. 1986;25:21–5. doi: 10.1021/bi00349a004. [DOI] [PubMed] [Google Scholar]
- 49.Benesch RE, Benesch R. Enzymatic removal of oxygen for polarography and related methods. Science. 1953;118:447–8. doi: 10.1126/science.118.3068.447. [DOI] [PubMed] [Google Scholar]
- 50.Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 51.Rasnik I, McKinney SA, Ha T. Nonblinking and long-lasting single-molecule fluorescence imaging. Nat Methods. 2006;3:891–3. doi: 10.1038/nmeth934. [DOI] [PubMed] [Google Scholar]
- 52.McKinney SA, Declais AC, Lilley DM, Ha T. Structural dynamics of individual Holliday junctions. Nat Struct Biol. 2003;10:93–7. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 53.Lohman TM, Mascotti DP. Nonspecific ligand-DNA equilibrium binding parameters determined by fluorescence methods. Methods Enzymol. 1992;212:424–58. doi: 10.1016/0076-6879(92)12027-n. [DOI] [PubMed] [Google Scholar]
- 54.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


