Abstract
Helicobacter pylori VacA, a pore-forming toxin secreted by an autotransporter pathway, causes multiple alterations in human cells, contributes to the pathogenesis of peptic ulcer disease and gastric cancer, and is a candidate antigen for inclusion in an H. pylori vaccine. Here, we present a 2.4-Å crystal structure of the VacA p55 domain, which has an important role in mediating VacA binding to host cells. The structure is predominantly a right-handed parallel β-helix, a feature that is characteristic of autotransporter passenger domains but unique among known bacterial protein toxins. Notable features of VacA p55 include disruptions in β-sheet contacts that result in five β-helix subdomains and a C-terminal domain that contains a disulfide bond. Analysis of VacA protein sequences from unrelated H. pylori strains, including m1 and m2 forms of VacA, allows us to identify structural features of the VacA surface that may be important for interactions with host receptors. Docking of the p55 structure into a 19-Å cryo-EM map of a VacA dodecamer allows us to propose a model for how VacA monomers assemble into oligomeric structures capable of membrane channel formation.
Keywords: bacterial toxin, β-helix, VacA
Helicobacter pylori is a Gram-negative bacterium that chronically infects the stomachs of >50% of the human population. H. pylori infection is a significant risk factor for the development of peptic ulcer disease, gastric adenocarcinoma, and gastric lymphoma (1, 2). One of the important virulence factors expressed by this organism is VacA, a secreted toxin named for its capacity to induce extensive vacuolation in the cytoplasm of mammalian cells (3–5). VacA is translated as a 140-kDa protoxin that undergoes N- and C-terminal cleavage during the secretion process to yield a mature 88-kDa toxin, p88 (6–8). Secretion is thought to occur through a Type Va or autotransporter pathway in which an N-terminal signal sequence directs the protein to the periplasm, and a C-terminal β-barrel domain facilitates transport of the p88 “passenger” domain across the outer membrane (8, 9) (Fig. 1a). VacA is able to intoxicate multiple types of human cells to yield a variety of cellular effects (4, 5). In addition to causing cell vacuolation, VacA can cause depolarization of membrane potential, alteration of mitochondrial membrane permeability, apoptosis, detachment of cells from the basement membrane, activation of mitogen-activated protein kinases, inhibition of antigen presentation, and inhibition of T cell activation and proliferation (4, 5). Although the mechanisms by which these processes occur are not fully understood, many of these effects depend on the capacity of VacA to form anion-selective channels in host-cell membranes (4, 5).
Fig. 1.
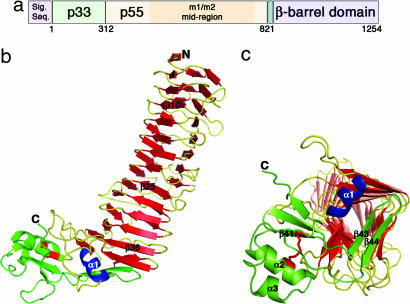
VacA structure. (a) The vacA gene encodes a 140-kDa protoxin. The mature 88-kDa VacA toxin contains two domains, designated p33 and p55. The midregion sequence that defines type m1 and m2 forms of VacA is located within p55. (b) The VacA p55 fragment adopts a β-helix structure that is composed of three parallel β-sheets (red) connected by loops of varying length and structure (yellow). The α-helix in blue (α1) is contained within one of these loops but is highlighted in blue to show how it caps the end of the β-helix. The C-terminal domain (green) has a mixture of α/β secondary structure elements and contains a disulfide bond (red), not previously observed in an autotransporter passenger domain structure. A few of the secondary structural elements are labeled to serve as landmarks and correlate to the sequences depicted in SI Fig. 7. (c) The C terminus of the β-helix is capped by a β-hairpin from the C-terminal domain (green) and the α1 α-helix (blue) located in one of the long β-helix loops. This view represents a rotation of the molecule in b by ≈90° into the plane of the page.
The first step in host-cell intoxication is binding of the toxin to the cell surface. Multiple cell surface receptors for VacA have been identified, including RPTPα, RPTPβ, and various lipids (10–13). VacA can then insert into the plasma membrane to form channels (14) or can undergo endocytosis and eventually localize with late endocytic compartments or mitochondria (15, 16). VacA-induced vacuoles correspond to swollen late endosomes, which are presumed to arise as a consequence of anion flux through VacA channels in the membranes of these compartments (5, 17).
VacA oligomerization is thought to precede membrane-channel formation (18). VacA can assemble into a variety of water-soluble oligomeric structures, including single-layered hexamers and heptamers and double-layered structures consisting of 12 or 14 subunits (19–24). The structures resemble “flowers” in which a central ring is surrounded by peripheral “petals.” Although the highest-resolution images are 19-Å cryo-EM maps of VacA dodecamers (24), atomic-force microscopy and electrophysiological studies suggest that membrane-associated VacA channels are single-layered structures (13, 22).
Two domains of VacA, p33 and p55, have been identified based on partial proteolysis of p88 into fragments of 33 and 55 kDa, respectively (7) (Fig. 1a). When expressed independently and then mixed, p33 and p55 can physically interact and reconstitute vacuolating toxin activity (25–27). The N-terminal p33 domain (residues 1–311) contains a hydrophobic sequence (residues 6–27) involved in pore formation (18, 28), whereas the p55 domain (residues 312–821) contains one or more cell-binding domains (29–31). When expressed intracellularly, the minimum portion of VacA required for cell-vacuolating activity comprises the entire p33 domain and ≈110 aa from the N terminus of p55 (25).
Analysis of H. pylori strains isolated from unrelated humans indicates a high level of genetic diversity among vacA alleles (32–35). Although frequent recombination events have eliminated phylogenetic structure from the 5′ region of vacA alleles (32, 33), phylogenetic analysis of sequences from a vacA midregion (located within p55, Fig. 1a) indicates the existence of two large families of sequences, termed m1 and m2 (34, 35). Within this ≈281-aa midregion (roughly corresponding to amino acids D455 to V735 in the VacA sequence of H. pylori strain 60190), the amino acid sequences of types m1 and m2 VacA proteins are only ≈55% identical. Differences in cell-type specificity have been noted for types m1 and m2 VacA proteins, a phenomenon that may result from binding of m1 and m2 VacA proteins to different cell-surface receptors (36–39). With the rare exception of a few m1/m2 chimeras, there has been little evidence of recombination between m1 and m2 vacA alleles within the vacA midregion, and, therefore, a phylogenetic distinction between m1 and m2 VacA sequences has remained intact (35). This distinction is the basis for a widely used typing scheme for H. pylori isolates (34, 35) and is relevant clinically because m1 strains are associated with gastric cancer (40). Despite a wealth of sequence information, little is known about the structural and functional correlates of VacA sequence diversity. To gain insights into the structural features of VacA that contribute to its cell-binding properties and to better understand the structural correlates of VacA sequence variation, we set out to determine the structure of the VacA p55 domain.
Results and Discussion
Crystal Structure of the VacA p55 Domain.
The crystal structure of the VacA p55 domain was determined to a minimum Bragg spacing of dmin = 2.4 Å by using experimental phases from multiple isomorphous replacement and anomalous scattering [see Methods and supporting information (SI) Table 1]. The p55 structure is predominantly a right-handed parallel β-helix (residues 355–735) but has a small globular domain at the C terminus (residues 736–811) with mixed α/β secondary structure elements (Fig. 1b). The structure resembles a sock in which the C-terminal domain curves from the heel and extends to the tip of the foot. The β-helical “calf” is 65 Å long with widths of 25–31 Å, whereas the C-terminal “foot” is ≈17 × 24 × 43 Å. In the crystal, pairs of p55 molecules meet at their N-terminal strands about a crystallographic 2-fold axis (SI Fig. 5). This packing is consistent with the elution of p55 as a dimer from gel filtration columns (see Methods) and a previous EM study indicating that a p55 domain secreted by H. pylori formed dimers (29).
Comparison of the VacA p55 Structure with Structures of Other Autotransporter Passenger Domains.
Crystal structures have been determined for two other type Va autotransporter passenger domains: pertactin (an adhesin from Bordetella pertussis) and Hbp (a hemoglobin protease from Escherichia coli). Pertactin (41), Hbp (42), and VacA p55 do not share sequence similarities, but all contain a β-helix fold. The β-helix fold is composed of multiple ≈25-aa quasirepeats, each of which forms a single coil of the helix. Conservation of the β-helix fold among autotransporter passenger domains suggests that this structural feature is required for efficient secretion across the outer membrane and folding (43). A C-terminal β-helix cap may be important as a nucleus and/or chaperone for secretion and folding (44). The end of the VacA p55 β-helix is capped by a β-hairpin, similar to that observed in the structure of pertactin (41). There is also a short α-helix in this region of VacA that appears to be unique among β-helix structures (Fig. 1c).
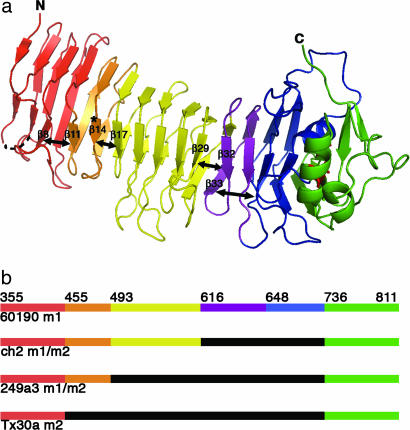
A notable difference between VacA p55 and the two other autotransporter passenger domain structures is the presence of a disulfide in the p55 C-terminal subdomain (Fig. 1c). The low cysteine content observed in autotransporter sequences is consistent with a model in which passenger domains translocate across the outer membrane in an unfolded conformation. Nevertheless, a number of autotransporter sequences have a single, closely spaced pair of cysteines near the C terminus of their passenger domains (45). In VacA, these cysteines are either 11 or 13 aa apart. Mutation of either of these cysteines to serine results in a decrease in toxin secretion but has no effect on the vacuolating activity of the toxin (ref. 45 and M.S.M., unpublished results). Here, we show experimentally that these cysteines form a disulfide and that they are positioned to buttress both the β-helix cap and a conserved C-terminal pocket (discussed below). Another feature of p55 that differs from other autotransporter passenger domains is the presence of multiple kinks that disrupt what would otherwise be continuous β-sheets. We have divided the VacA p55 β-helix into five subdomains to reflect these disruptions (Fig. 2a) and note that the divisions correlate with predicted sites of homologous recombination (as discussed further below).
Fig. 2.
The VacA p55 structure reveals marked disruptions in β-sheet contacts. (a) Breaks in the β-sheet contacts (depicted with double-headed arrows) result in subdomains (SD) within the β-helix: SD-1 (red, 355–454), SD-2 (orange, 455–492), SD-3 (yellow, 493–615), SD-4 (purple, 616–647), and SD-5 (blue, 648–735). The C-terminal domain and disulfide bond are colored green and red, respectively, and the site of the m2 23-aa insertion is indicated by an asterisk. (b) This schematic shows prototype m1 and m2 VacA proteins (strains 60190 and Tx30a, respectively) and two naturally occurring m1m2 chimeras. The breaks in β-sheet contacts (a) correlate with sites of homologous recombination (see also SI Fig. 7). In this schematic the midregion that distinguishes type m1 and type m2 sequences is colored orange/yellow/purple/blue (m1) or black (m2).
Sequence Variation Among VacA Proteins.
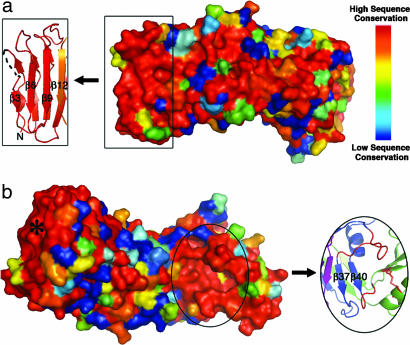
In an effort to understand how genetic differences between m1 and m2 vacA alleles relate to structural and functional differences in the proteins they encode, we have examined VacA sequence polymorphisms in the context of the p55 structure. We identified sequences for 62 m1 and 27 m2 VacA proteins in GenBank that were complete in the p55 region. In addition, we identified three m1/m2 chimeric VacA proteins for which sites of recombination between m1 and m2 sequences were easily recognizable. We aligned these sequences in various combinations using the program ClustalW (46) and mapped the sequence similarity scores to the three-dimensional m1 p55 structure using ESPript (47). The majority of residues pointing into the interior of the β-helix are either strictly or highly conserved, consistent with the idea that mutation of buried residues would result in a detrimental loss of structure and, therefore, function. The surface-exposed residues are also fairly conserved when exclusively m1 or exclusively m2 sequences are analyzed (SI Fig. 6). However, when m1 and m2 sequences are analyzed together, the surface-exposed residues are highly variable (Fig. 3). There are only two regions of the surface with notable sequence conservation. One is located at the N terminus (Fig. 3a) and will be discussed in the next section with respect to protein oligomerization. The second conserved surface is located at the C terminus of the β-helix in a cavity formed by two loops (residues 668–678 and 730–734) and the disulfide-linked C-terminal domain (Fig. 3b). The strict sequence conservation, the fact that many binding sites are located in clefts or cavities, and the fact that this is the only cavity observed in the VacA p55 structure suggest that this area may represent a receptor-binding site that is shared by m1 and m2 forms of the toxin.
Fig. 3.
The VacA p55 structure has two patches of conserved residues. (a) The 62 m1, 27 m2, and 3 m1/m2 chimeric VacA sequences were aligned and scored with a Risler matrix according to the extent of sequence variation. Scores were displayed on the p55 structure surface with a color ramp (red, orange, yellow, green, light blue, dark blue) in which strictly conserved residues are colored red, and the most variable residues are colored dark blue. One conserved region is at the N terminus of the protein (boxed) and correlates to the β3, β6, and β9 strands of the β-helix. We propose that this surface is important for VacA oligomerization. (b) Rotation of the molecule in view a by ≈180° around the long axis of the protein reveals the second conserved region of the VacA p55 structure. This surface is located in a pocket at the C terminus of the β-helix (circled) and may represent a common receptor-binding site for all m1 and m2 VacA proteins. The pocket is formed by two long β-helix loops (red) and the disulfide bond (also in red) of the C-terminal domain. Although the asterisked region is also highly conserved, this correlates to the N terminus of the β-helix that we predict to be buried when p33 is present.
Because multiple surface-exposed regions are highly divergent, it is difficult to identify a single region in the p55 structure that can explain the different binding properties of m1 and m2 VacA proteins (Fig. 3). The biggest difference between m1 and m2 sequences is the presence of a 23-aa insertion in m2 sequences (Fig. 2a, asterisk; and SI Fig. 7). The insertion is notable in that its sequence is an imperfect repeat of the sequence located immediately upstream, and the length approaches the average length of a single β-helix coil (≈25 aa) (SI Fig. 7). The 23-aa m2 insertion could therefore result in an additional β-helix coil similar in structure to the coil that precedes it. Based on an analysis of engineered m1/m2 chimeras, a region important for m1-specific binding to HeLa cells has been mapped to residues 460–496 in the m1 sequence (37). The m2 23-aa insertion is located within this region, but an engineered chimera in which these 23 m2 amino acids were inserted into an m1 sequence did not result in a loss of HeLa cell toxicity (37). Excluding the m2 insertion, there are only 10 amino acids in the 460–496 m1 sequence that consistently differ between the m1 and m2 forms (SI Fig. 7). The position of these residues in the context of the 23-aa m2 insertion is impossible to predict, and, therefore, an understanding of structural features that give rise to VacA cell-type specificity may need to await analysis of an m2 p55 structure.
Analysis of the VacA sequence alignments in the context of the m1 p55 structure reveals that disruptions of β-sheet contacts within the β-helix correlate with sites of homologous recombination events. For example, VacA residue D455 (strain 60190) represents the beginning of the midregion that defines m1 and m2 strains (Fig. 2b and SI Fig. 7) and follows a kink in the structure that disrupts β-sheet contacts between β8 and β11 (Fig. 2a and SI Fig. 7). This structural disruption and the associated sequence transition are illustrated with a change from red to orange (Fig. 2 and SI Fig. 7). Other examples of a correlation between VacA subdomains and recombination events are evident based on analysis of rare naturally occurring m1/m2 chimeras, which have m1 character in the N-terminal portion of the VacA midregion but m2 character in the C-terminal portion. The transition point from m1 to m2 in the chimeric VacA from strain 249a3 is at D493, whereas the transition point for the ch2 and v225 strains of VacA is at N616 (Fig. 2b and SI Fig. 7). As illustrated in Fig. 2a, these sequence transitions correlate with disruptions in β-sheet contacts between β14 and β17 and between β29 and β32, respectively. The final subdomain begins at residue G648. Although we did not identify a sequence with evidence of recombination at this site, the subdomain (blue) has longer loops than other subdomains and does not make β-sheet contacts with β33 from the previous subdomain (Fig. 2a).
In summary, the polymorphisms that differentiate m1 and m2 forms correspond almost entirely to surface-exposed residues. Diversification in these surface-exposed regions of VacA may have been driven by immune selective pressures. Recombination occurs commonly in H. pylori (32, 33), but m1/m2 chimeras are only rarely identified (35). This suggests that H. pylori strains with intact m1 or m2 VacA sequences have favorable functional properties and, thus, have a selective advantage compared with strains containing chimeric m1/m2 sequences (36–38). The rare naturally occurring m1/m2 chimeras shown in Fig. 2b and SI Fig. 7 probably arose as the result of recombination events that maintained favorable structural and functional properties in the encoded β-helix structures.
Assembly of VacA into Oligomeric Structures.
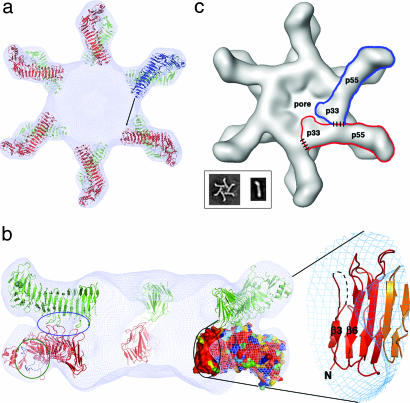
To understand the p55 structure in the context of VacA oligomers, we have docked p55 into the 19-Å cryo-EM map of the wild-type dodecamer (24) using the program COLORES (Fig. 4a) (48). The elongated shape of the β-helix and the curve of the C-terminal foot allow for unambiguous placement of 12 p55 subunits into the petal-like features of the map. The docking suggests that contacts between the p55 components of the two hexameric layers are mediated by three loops and that the N-terminal end of the p55 β-helix extends into the central density of the map (Fig. 4b). The conserved pocket that we propose as a potential common receptor binding site for m1 and m2 forms of VacA is located on the sides of the petals and would be fully accessible to cell-surface receptors when VacA is assembled in either a single- or double-layered oligomeric structure (Fig. 4b).
Fig. 4.
Docking the p55 crystal structure into a 19-Å cryo-EM map of the VacA dodecamer results in a model for oligomerization. (a) Twelve p55 subunits are shown docked into a 19-Å cryo-EM map of a VacA dodecamer (24). An arrow is shown to indicate the space that the blue molecule will occupy if p33 extends the β-helix structure of p55. (b) The view in a is rotated by 90°, such that the blue molecule moves toward the reader and is now located on the bottom of the dodecamer. The blue molecule is not visible in this side view, however, because the view has been sliced so that only the “back” of the structure is visible. On the left, a blue circle highlights three loops that mediate p55–p55 contacts between the two layers. Circled in green are the two loops that line the conserved pocket that we propose as a common receptor-binding site. This pocket is located on the side of the molecule and would therefore be accessible in both single-layered and bilayered forms of the toxin. The black circle contains the other conserved surface in p55 (also shown in Fig. 3a). This surface protrudes into the central ring of the VacA oligomer and would be accessible to the blue molecule in a for contacts that could mediate oligomerization within a hexameric or heptameric plane. We have zoomed in on the secondary structures that contribute to this surface. (c) We propose an oligomerization model in which p33 interacts with the N-terminal portion of p55 from the neighboring subunit. Regions of contact between p33 and p55 are depicted with dashed lines. (Inset) EM images of a VacA hexamer and a VacA monomer (24). The shape of a VacA hexamer (Inset) is similar to the shape of a single layer within the dodecamer (24). The rod-like shape of the p88 VacA monomer (Inset) supports a model in which the β-helix observed in p55 will extend into p33.
We hypothesize that a large portion of p33 will adopt and extend the β-helix fold observed in p55. This idea is supported by two observations. First, an extended β-helix structure is consistent with the shape of a p88 monomer obtained by EM (Fig. 4c Inset) (24). Second, the program BetaWrapPro identifies the stretch of amino acids between residues 120 and 249 as a five-coil β-helix and predicts additional β-helix strands in the region between 252 and 288 (49). BetaWrapPro uses profile wrapping for prediction and comparative modeling of β-helices and has been shown to identify the β-helix motif with high sensitivity and selectivity (49).
Based on our prediction that a large portion of p33 will extend the β-helix fold observed in p55, we suggest a model in which VacA oligomerization is mediated by contacts between p33 and the N terminus of p55 from a neighboring subunit (Fig. 4c). Specifically, one would predict the p55 oligomerization surface to contain the β-strands β3 and β6. The surface-exposed residues of β3 and β6 are accessible within the central density of the EM map and are strictly conserved among the 92 m1 and m2 sequences we surveyed, suggesting selective pressure to preserve this surface (Fig. 4b). An important functional role of this region is also supported by biochemical data indicating that VacA residues 1–422 can induce vacuolation in cultured cells when expressed intracellularly, but p33, p55, and VacA residues 1–394 cannot (25, 50). Finally, this model is supported by a yeast two-hybrid experiment in which p33 and residues 313–478 of p55 were shown to interact (27). The oligomerization surface may also contain parts of the 312–354 sequence that was not visible in this structure. Much of this region is strictly conserved among the m1 and m2 VacA sequences (SI Fig. 7).
p33 (residues 1–311) contains a putative α-helical pore-forming domain at its N terminus (28, 51). Comparison of the VacA wild-type and Δ6–27 dodecamer structures by cryo-EM suggests that the N-terminal domain of p33 is located in the central density of the structure (24). Secondary structure prediction analyses suggest that p33 has α-helical structural elements between residues 1 and 71. Short β-strands are then predicted to begin at residue 87 and extend through a region (residues 120–288) that is predicted to have a β-helical structure, based on BetaWrapPro (49). We therefore anticipate that a single subunit of VacA will adopt a roughly symmetric shape within the context of the oligomer and that residues in the N-terminal region of p33 (likely C-terminal to the pore-forming region) would be positioned to mediate oligomerization with the N-terminal region of a neighboring p55 subunit (Fig. 4c).
A protective H. pylori vaccine would potentially reduce the incidence of H. pylori infection and the serious complications of peptic ulcer disease and gastric adenocarcinoma (52), but the high level of diversity among H. pylori strains and the high frequency of genetic recombination among strains are likely to present significant challenges to vaccine development. VacA is a candidate antigen because immunization with VacA confers protective immunity in a mouse model of H. pylori infection (52, 53). We show here that two VacA surfaces are strictly conserved among all surveyed m1 and m2 sequences and propose that these surfaces are under selective pressure to be preserved to mediate receptor-binding and oligomerization functions. Efforts to selectively target these regions could result in progress toward the development of a vaccine that confers protection against multiple strains of H. pylori.
Methods
Expression and Purification of p55.
The p55 domain of VacA from H. pylori strain 60190 (a type m1 form of VacA; GenBank accession no. Q48245) with an N-terminal hexahistidine tag was expressed as described (26) except that E. coli BL21(DE3) cells were used. Harvested cells were resuspended in lysis buffer (50 mM potassium phosphate, 300 mM NaCl, 20 mM imidazole at pH 8.0) and lysed by three passages through a pressure dispersion homogenizer at 20,000 psi. Cell lysates were centrifuged at 48,000 × g for 20 min at 4°C. Native p55 was purified from the supernatant by Ni-affinity, ion exchange, and gel filtration chromatography. The sizing column retention time suggested a molecular mass of ≈130 kDa. We hypothesize that this result reflects the presence of an elongated p55 dimer. Seleno-methionine p55 was produced from E. coli BL834(DE3) in a minimal medium containing 40 mg/liter l-seleno-methionine and purified by using methods similar to those used for the native protein.
Crystallization and Preparation of Heavy-Atom Derivatives.
The native and seleno-methionine p55 were concentrated in a 100 mM NaCl, 50 mM potassium phosphate buffer and crystallized at 21°C by the hanging-drop vapor-diffusion method in which protein and precipitant were mixed in a 1:1 ratio. Crystals grew from starting protein concentrations of 5.5 mg/ml, and reservoirs containing 18–23% PEG1500. Heavy-atom derivatives were prepared by soaking crystals in 50 mM potassium tetrabromoplatinate (IV) (K2PtBr6) for 3 d, 5 mM 1,4-diacetoxymercuri-2,3-dimethoxybutane for 24 h, or 5 mM gold chloride (HAuCl4) for 24 h. Crystals were mounted on cryo loops, sequentially soaked in cryoprotectant solutions containing 25% PEG1500, and 5%, 10%, and 15% glycerol, and flash-cooled in liquid nitrogen.
Structure Determination and Refinement.
X-ray data were collected from single crystals at 100 K on beamline ID-22 at the Advanced Photon Source (Argonne, IL) by using a MAR300 image plate detector. All of the diffraction data were indexed, integrated, scaled, and merged with HKL2000 (SI Table 1) (54). Native and derivative crystals were space group P3221 and had unit cell dimensions of a = b = 57 Å, and c = 257 Å. Phases were determined by MIRAS using a native protein crystal and seleno-methionine, platinum, mercury, and gold derivatives (SI Table 1). Heavy-atom positions were located and refined with the autoSHARP program suite (55). The structure was subjected to iterative rounds of model building in O (56) and refinement in CNS (57) before applying TLS refinement (58) in REFMAC (59). The refined model (Rcryst = 18.1%, Rfree = 22.2%) consists of amino acids 355–366, 377–811, and 206 water molecules. SDS/PAGE and N-terminal sequence analysis of dissolved crystals indicated that proteolysis had occurred at the N terminus of p55 during crystallization, thus explaining the absence of residues 312–354 and 367–376 from the crystal structure. Proteolysis was prevented by the addition of protease inhibitors, but crystals did not form under these conditions.
Supplementary Material
Acknowledgments
We thank Catherine El-Bez and Jacques Dubochet for providing the cryo-EM map. X-ray data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory (Argonne, IL). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. This work was also supported by National Institutes of Health Grant R01 AI39657 (to T.L.C.), the Medical Research Service of the Department of Veterans Affairs (T.L.C.), the Sartain-Lanier Family Foundation and an American Cancer Society Institutional Research Grant IRG-58-009-48 (to D.B.L.), and the T32 GM 08320 Molecular Biophysics Training Program (K.A.G.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2QV3).
This article contains supporting information online at www.pnas.org/cgi/content/full/0707447104/DC1.
References
- 1.Marshall BJ, Warren JR. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Suerbaum S, Michetti P. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 3.Cover TL, Blaser MJ. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 4.Cover TL, Blanke SR. Nat Rev Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 5.de Bernard M, Cappon A, Del Giudice G, Rappuoli R, Montecucco C. Int J Med Microbiol. 2004;293:589–597. doi: 10.1078/1438-4221-00299. [DOI] [PubMed] [Google Scholar]
- 6.Cover TL, Tummuru MK, Cao P, Thompson SA, Blaser MJ. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 7.Telford JL, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z, et al. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitt W, Haas R. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 9.Fischer W, Buhrdorf R, Gerland E, Haas R. Infect Immun. 2001;69:6769–6775. doi: 10.1128/IAI.69.11.6769-6775.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yahiro K, Niidome T, Kimura M, Hatakeyama T, Aoyagi H, Kurazono H, Imagawa K, Wada A, Moss J, Hirayama T. J Biol Chem. 1999;274:36693–36699. doi: 10.1074/jbc.274.51.36693. [DOI] [PubMed] [Google Scholar]
- 11.Yahiro K, Wada A, Nakayama M, Kimura T, Ogushi K, Niidome T, Aoyagi H, Yoshino K, Yonezawa K, Moss J, et al. J Biol Chem. 2003;278:19183–19189. doi: 10.1074/jbc.M300117200. [DOI] [PubMed] [Google Scholar]
- 12.Roche N, Ilver D, Angstrom J, Barone S, Telford JL, Teneberg S. Microbes Infect. 2007;9:605–614. doi: 10.1016/j.micinf.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 13.Iwamoto H, Czajkowsky DM, Cover TL, Szabo G, Shao Z. FEBS Lett. 1999;450:101–104. doi: 10.1016/s0014-5793(99)00474-3. [DOI] [PubMed] [Google Scholar]
- 14.Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, Rappuoli R, Montecucco C, Papini E, Zoratti M. EMBO J. 1999;18:5517–5527. doi: 10.1093/emboj/18.20.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galmiche A, Rassow J, Doye A, Cagnol S, Chambard JC, Contamin S, de Thillot V, Just I, Ricci V, Solcia E, et al. EMBO J. 2000;19:6361–6370. doi: 10.1093/emboj/19.23.6361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauthier NC, Monzo P, Gonzalez T, Doye A, Oldani A, Gounon P, Ricci V, Cormont M, Boquet P. J Cell Biol. 2007;177:343–354. doi: 10.1083/jcb.200609061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Genisset C, Puhar A, Calore F, de Bernard M, Dell'antone P, Montecucco C. Cell Microbiol. 2007;9:1481–1490. doi: 10.1111/j.1462-5822.2006.00886.x. [DOI] [PubMed] [Google Scholar]
- 18.Vinion-Dubiel AD, McClain MS, Czajkowsky DM, Iwamoto H, Ye D, Cao P, Schraw W, Szabo G, Blanke SR, Shao Z, et al. J Biol Chem. 1999;274:37736–37742. doi: 10.1074/jbc.274.53.37736. [DOI] [PubMed] [Google Scholar]
- 19.Cover TL, Hanson PI, Heuser JE. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanzavecchia S, Bellon PL, Lupetti P, Dallai R, Rappuoli R, Telford JL. J Struct Biol. 1998;121:9–18. doi: 10.1006/jsbi.1997.3941. [DOI] [PubMed] [Google Scholar]
- 21.Lupetti P, Heuser JE, Manetti R, Massari P, Lanzavecchia S, Bellon PL, Dallai R, Rappuoli R, Telford JL. J Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Czajkowsky DM, Iwamoto H, Cover TL, Shao Z. Proc Natl Acad Sci USA. 1999;96:2001–2006. doi: 10.1073/pnas.96.5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adrian M, Cover TL, Dubochet J, Heuser JE. J Mol Biol. 2002;318:121–133. doi: 10.1016/S0022-2836(02)00047-5. [DOI] [PubMed] [Google Scholar]
- 24.El-Bez C, Adrian M, Dubochet J, Cover TL. J Struct Biol. 2005;151:215–228. doi: 10.1016/j.jsb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Ye D, Willhite DC, Blanke SR. J Biol Chem. 1999;274:9277–9282. doi: 10.1074/jbc.274.14.9277. [DOI] [PubMed] [Google Scholar]
- 26.Torres VJ, Ivie SE, McClain MS, Cover TL. J Biol Chem. 2005;280:21107–21114. doi: 10.1074/jbc.M501042200. [DOI] [PubMed] [Google Scholar]
- 27.Torres VJ, McClain MS, Cover TL. J Biol Chem. 2004;279:2324–2331. doi: 10.1074/jbc.M310159200. [DOI] [PubMed] [Google Scholar]
- 28.McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, Shao Z, Cover TL. J Biol Chem. 2003;278:12101–12108. doi: 10.1074/jbc.M212595200. [DOI] [PubMed] [Google Scholar]
- 29.Reyrat JM, Lanzavecchia S, Lupetti P, de Bernard M, Pagliaccia C, Pelicic V, Charrel M, Ulivieri C, Norais N, Ji X, et al. J Mol Biol. 1999;290:459–470. doi: 10.1006/jmbi.1999.2877. [DOI] [PubMed] [Google Scholar]
- 30.Garner JA, Cover TL. Infect Immun. 1996;64:4197–4203. doi: 10.1128/iai.64.10.4197-4203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang HJ, Wang WC. Biochem Biophys Res Commun. 2000;278:449–454. doi: 10.1006/bbrc.2000.3820. [DOI] [PubMed] [Google Scholar]
- 32.Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, Dyrek I, Achtman M. Proc Natl Acad Sci USA. 1998;95:12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottke MU, Fallone CA, Barkun AN, Vogt K, Loo V, Trautmann M, Tong JZ, Nguyen TN, Fainsilber T, Hahn HH, et al. J Infect Dis. 2000;181:1674–1681. doi: 10.1086/315425. [DOI] [PubMed] [Google Scholar]
- 34.Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 35.Atherton JC, Sharp PM, Cover TL, Gonzalez-Valencia G, Peek RM, Jr, Thompson SA, Hawkey CJ, Blaser MJ. Curr Microbiol. 1999;39:211–218. doi: 10.1007/s002849900447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pagliaccia C, de Bernard M, Lupetti P, Ji X, Burroni D, Cover TL, Papini E, Rappuoli R, Telford JL, Reyrat JM. Proc Natl Acad Sci USA. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji X, Fernandez T, Burroni D, Pagliaccia C, Atherton JC, Reyrat JM, Rappuoli R, Telford JL. Infect Immun. 2000;68:3754–3757. doi: 10.1128/iai.68.6.3754-3757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skibinski DA, Genisset C, Barone S, Telford JL. Infect Immun. 2006;74:49–55. doi: 10.1128/IAI.74.1.49-55.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang WC, Wang HJ, Kuo CH. Biochemistry. 2001;40:11887–11896. doi: 10.1021/bi010065u. [DOI] [PubMed] [Google Scholar]
- 40.Figueiredo C, Machado JC, Pharoah P, Seruca R, Sousa S, Carvalho R, Capelinha AF, Quint W, Caldas C, van Doorn LJ, et al. J Natl Cancer Inst. 2002;94:1680–1687. doi: 10.1093/jnci/94.22.1680. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Charles IG, Fairweather NF, Isaacs NW. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- 42.Otto BR, Sijbrandi R, Luirink J, Oudega B, Heddle JG, Mizutani K, Park SY, Tame JR. J Biol Chem. 2005;280:17339–17345. doi: 10.1074/jbc.M412885200. [DOI] [PubMed] [Google Scholar]
- 43.Junker M, Schuster CC, McDonnell AV, Sorg KA, Finn MC, Berger B, Clark PL. Proc Natl Acad Sci USA. 2006;103:4918–4923. doi: 10.1073/pnas.0507923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver DC, Huang G, Nodel E, Pleasance S, Fernandez RC. Mol Microbiol. 2003;47:1367–1383. doi: 10.1046/j.1365-2958.2003.03377.x. [DOI] [PubMed] [Google Scholar]
- 45.Letley DP, Rhead JL, Bishop K, Atherton JC. Microbiology. 2006;152:1319–1325. doi: 10.1099/mic.0.28548-0. [DOI] [PubMed] [Google Scholar]
- 46.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gouet P, Courcelle E, Stuart DI, Metoz F. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 48.Wriggers W, Milligan RA, McCammon JA. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 49.McDonnell AV, Menke M, Palmer N, King J, Cowen L, Berger B. Proteins. 2006;63:976–985. doi: 10.1002/prot.20942. [DOI] [PubMed] [Google Scholar]
- 50.Ye D, Blanke SR. Mol Microbiol. 2002;43:1243–1253. doi: 10.1046/j.1365-2958.2002.02818.x. [DOI] [PubMed] [Google Scholar]
- 51.Kim S, Chamberlain AK, Bowie JU. Proc Natl Acad Sci USA. 2004;101:5988–5991. doi: 10.1073/pnas.0308694101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruggiero P, Peppoloni S, Rappuoli R, Del Giudice G. Microbes Infect. 2003;5:749–756. doi: 10.1016/s1286-4579(03)00125-4. [DOI] [PubMed] [Google Scholar]
- 53.Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. Science. 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- 54.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 55.de la Fortelle E, Bricogne G. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 56.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 57.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 58.Winn MD, Isupov MN, Murshudov GN. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 59.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.