Abstract
Naturally occurring polyreactive anti-DNA mAbs derived from a nonimmunized (NZB × NZW)F1 mouse with spontaneous lupus erythematosus penetrated and accumulated in the nuclei of a variety of cultured cells. These mAbs and their F(ab′)2 and Fab′ fragments, covalently coupled to fluorescein, peroxidase, or a 15-mer polynucleotide, also translocated to the cell nuclei. A 30-amino acid peptide corresponding to the combined sequences of the complementary-determining regions 2 and 3 of the heavy chain variable region of one mAb was able to penetrate into the cytoplasm and nucleus of cells of several lines. This peptide recognized DNA and was strongly polyreactive. Streptavidin-peroxidase conjugates complexed with the N-biotinylated peptide were rapidly translocated into cells. Similarly, peroxidase or anti-peroxidase polyclonal antibodies covalently coupled to the N-cysteinylated peptide through an heterobifunctional maleimide cross-linker were also rapidly internalized and frequently accumulated in nuclei. The peptide carrying 19 lysine residues at its N-terminal was highly effective in transfecting 3T3 cells with a plasmid containing the luciferase gene. Thus, penetrating mAbs and derived peptides are versatile vectors for the intracellular delivery of proteins and genes.
A long time ago, it was reported that human IgG from systemic lupus erythematosus patients with high titers directed against nuclear ribonucleoproteins and/or DNA were able to penetrate into living cells and to reach the nucleus (1). More recent studies of murine anti-DNA autoantibodies confirmed these observations and disclosed that different penetrating antibodies exhibited diverse behaviors and characteristics (2–7). In this study, we prepared several penetrating IgG anti-DNA mAbs from the spleen of a (NZB × NZW)F1 lupus mouse and examined their specificities and their abilities to act as vectors of haptens, proteins, polynucleotides, and plasmids.
MATERIALS AND METHODS
Mice and Cell Lines.
(NZB × NZW)F1 hybrids and BALB/c mice were bred in the Institut Pasteur animal facilities. Cells used were from different species and from various tissues as follows: PtK2 (Potoroo kidney fibroblasts) or CCL-39 (hamster lung), 3T3 (mouse embryo fibroblasts), and HEp-2 (human larynx carcinoma). All cells were from the American Type Culture Collection and were cultured in RPMI 1640 medium (or in DMEM for CCL-39) containing 10% heat-inactivated calf serum and supplemented with l-glutamine, sodium pyruvate, nonessential amino acids, and antibiotics (complete culture medium) at 37°C in a humidified atmosphere of 5% CO2/95% air.
mAbs.
Spleen cells from a 9-month-old nonimmunized (NZB × NZW)F1 mouse were fused with P3.X63Ag8 myeloma cells by the method of Köhler and Milstein (8), and hybridomas were selected in hypoxanthine/azaserine medium. Supernatants were tested by ELISA on double-stranded (ds) DNA-coated plates with β-galactosidase-labeled anti-Fcγ conjugate prepared from sheep antiserum (9). Isotypes were determined by using anti-IgG1-, -IgG2a-, -IgG2b-, and -IgG3-alkaline phosphatase conjugates (Southern Biotechnology Associates, Birmingham, AL). Anti-DNA-positive hybridomas were cloned and expanded, and cell culture supernatants were tested for the ability of their IgG to penetrate into living cells.
Penetration of Antibodies into Cells.
Cell monolayers were obtained by seeding 2–5 × 104 cells in 0.5 ml of complete medium on round microscope coverslips deposited in 24-well tissue culture plates. One to 2 days after culture initiation, the medium was replaced by undiluted hybridoma-positive supernatants or purified mAbs diluted in complete medium, and cultures were allowed to proceed for 2–24 h. Cells were washed with PBS, either fixed for 15 min in ethanol at −20°C and dried or fixed in 2% p-formaldehyde in 0.1 M phosphate buffer (pH 7.4) for 10 min, washed with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 2 min. Intracellular mAb was visualized by incubating fixed cells with horseradish peroxidase (PO)-coupled anti-mouse IgG Fab (10 μg/ml) prepared from sheep antiserum (9) or fluorescein isothiocyanate (FITC)-conjugated rabbit anti-mouse IgG antibodies (Southern Biotechnology Associates) (5 μg/ml) for 60 min followed by several washes in PBS. PO deposits were visualized by using the metal-enhanced diaminobenzidine substrate kit (ME-DAB; Pierce). To evaluate the amounts of mAb internalized after various incubation times, cells were trypsinized, washed, counted, and lyzed in 0.1 M Tris⋅HCl (pH 8) containing 1% Nonidet P-40. The amount of mAb in lysates was evaluated by ELISA using anti-IgG2a-alkaline phosphatase conjugate. Enzyme activity was determined by using p-nitrophenyl phosphate (Sigma).
Purification, Fragmentation, and Labeling of mAbs.
IgG mAbs were isolated by using a protein A-Sepharose column (10). The reactivities of purified mAbs with dsDNA and other antigens were tested by ELISA as described (11). Their avidities for dsDNA were estimated by using an inhibition assay (12). F(ab′)2 fragments of mAbs, prepared by pepsin digestion, were further reduced with 0.01 M cysteine to yield Fab′ fragments (13). IgG mAbs, F(ab′)2, and Fab′ fragments were coupled to FITC by published procedures (14). They were also conjugated to PO by using the two-step glutaraldehyde procedure (9). The reaction mixture was passed through a Con A-Sepharose column to eliminate unconjugated IgG, F(ab′)2 or Fab′, and the PO conjugates were eluted with 0.1 M α-methyl α-d-mannoside in PBS.
The fluoresceinated polynucleotide used was composed of 15 nucleotides with a fluorescein at the 5′ end and a free -NH2 group at the 3′ end (fluorescein-TATCTAGTCATTACT-NH2; Unité de Chimie Organique, Institut Pasteur, Paris). F(ab′)2 or Fab′ fragments of mAb J20.8 were activated and coupled to this fluorescein-polynucleotide with p-benzoquinone at a ratio of 4 nucleotides per mol of F(ab′)2 or Fab′ (9).
Gene Transfer Using mAbs.
The plasmid pHu Vim 830-T/t contained DNA encoding the simian virus 40 large tumor antigen under the control of the human vimentin promoter (15). For the transfection assays, 2 μg of plasmid were incubated with 20 μg of mAb in 20–30 μl of PBS for 15 min at room temperature, and the volume was adjusted with complete culture medium to a final volume of 0.5 ml. Controls were 2 μg of plasmid or 20 μg of mAb in 0.5 ml of complete culture medium. The reaction mixtures were then added to wells of 24-well plates that had been seeded the day before with 5 × 104 cells. After 18 h of incubation, the reaction mixtures were replaced by complete culture medium and the efficacy of gene transfer was evaluated immunocytochemically 2–5 days later. The cells were washed, fixed with methanol/acetone at −20°C for 10 min, incubated for 60 min with mAb to tumor antigen (a gift from P. Vicart, Institut Pasteur) coupled to PO, and finally visualized with ME-DAB.
Sequencing of mAbs.
Total cellular RNA was extracted from hybridoma cells by the guanidine thiocyanate technique (16). mRNA was converted into cDNA with a reverse transcriptase kit (Life Technologies) and used as a template for PCR amplification using Taq DNA polymerase (Boehringer, Mannheim) according to the manufacturer’s protocol. The amplification was performed with the primer of IgG2a (5′-GTTCTGACTAGTGGGCACTCTGGGCT) and four heavy chain variable region (VH) primers (5′-GAGGTTCAGCTCGAGCAGTCTGGGGC, 5′-GAGGTGAAGCTCGAGGAATCTGGAGG, 5′-GAAGTGCAGCTCGAGGAGTCTGGGG, and 5′-GAGGTTCAGCTCGAGCAGTCTGGAGC). PCR products were purified by using Geneclean kit (Bio 101). Chemical sequencing was carried out by Genome Express (Grenoble, France). Nucleotide sequences were analyzed by using the GenBank and EMBL databases, maintained at Institut Pasteur (Unité d’Informatique Scientifique), using the GCG sequence analysis software (17) and amino acid sequences were deduced.
Penetrating and Binding Capacities of Peptides.
Peptides corresponding to VH regions of mAb F4.1 that participate in antigen binding were prepared. Biotinylated peptides P1, P2, and P3 containing, respectively, complementary-determining region 2 (CDR2), 3 CDR3, and CDR2 plus CDR3 sequences were synthesized by solid-phase chemistry (Neosystem, Isochem, Strasbourg, France). Their sequences are reported in Table 1. Cells were incubated for 1–18 h with the biotinylated peptides in complete culture medium at concentrations from 0.1 to 20 μg/ml, washed with PBS, fixed with ethanol, incubated with streptavidin-PO (5 μg/ml) for 1 h, washed again, and exposed to ME-DAB. To examine whether the biotinylated peptides were able to transport macromolecules into cells, complexes with streptavidin-PO were prepared at various peptide/streptavidin ratios. Biotinylated peptides and streptavidin-PO conjugates in 20 μl of PBS were allowed to react for 15 min. The reaction mixtures were then diluted in complete culture medium to achieve a peptide concentration of 6–24 μg/ml and added to the cells for 1–18 h. The cells were then washed, fixed with ethanol, and incubated with the ME-DAB for PO.
Table 1.
Sequence of the peptides derived from F4.1 mAb
| Peptide | Region | Sequence |
|---|---|---|
| P1 | CDR2 | VAYISRGGVSTYYSDTVKGRFT |
| P2 | CDR3 | ARQKYNKRAMDY |
| P3 | CDR2–CDR3 | VAYISRGGVSTYYSDTVKGRFTRQKYNKRA |
To further examine the possibility of using P3 as vector for the transport of proteins, PO or immunoadsorbent-isolated sheep anti-PO antibodies were directly coupled to P3 and tested. To this end, P3 carrying a cysteine at the N-terminal was synthesized (Altergen, Schiltigheim, France) and coupled to the protein through the SH group by using succinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (Pierce) by the manufacturer’s instructions. Cells were incubated for 2–3 h with various concentrations (15–60 μg/ml) of these conjugates. Cells incubated with P3 coupled to PO were fixed and exposed to ME-DAB, whereas those incubated with P3 coupled to anti-PO antibodies were fixed, then incubated for 60 min with a PO solution (10 μg/ml), washed, and visualized as above.
To evaluate the amounts of PO internalized after incubation with the various preparations, cells were trypsinized, washed, counted, and lyzed. Enzyme activity was determined in cell lysates by using o-phenylenediamine and H2O2.
The reactivities of biotinylated P1, P2, and P3 with various antigens were examined by ELISA using streptavidin-alkaline phosphate and p-nitrophenyl phosphate as the substrate.
Gene Transfer Using P3.
P3 carrying 19 lysine residues at its N-terminal (K19-P3) was synthesized (Altergen). For transfection assays, plasmid pCMVL containing the luciferase reporter gene under the control of a cytomegalovirus promoter (a gift from U. Hazan, Institut Cochin de Génétique Moleculaire, Paris) was used. Triplicates of 3 μg of plasmid were incubated with either 18 μg of K19-P3 or 18 μg of 19-residue poly(l-lysine) (Mr = 1,000–4,000; Sigma) or 9 μl [10 equivalents of polyethylenimine polymer (PEI) nitrogen per DNA phosphate] of 800-kDa PEI (Fluka) in 30 μl of 0.15 M NaCl for 30 min at room temperature. The reaction mixtures were then added to wells of 24-well culture plates seeded the day before with 5 × 104 3T3 cells. After 5 h, the medium was replaced by fresh culture medium. After 24 h, the luciferase activity was determined by using the Luciferase Assay Reagent (Promega). The protein concentration was measured by using the Bio-Rad protein assay (Bio-Rad) and BSA as the standard. Values are expressed as luciferase activity in relative light units (RLU) per mg of protein.
RESULTS
Translocation of Anti-DNA mAbs.
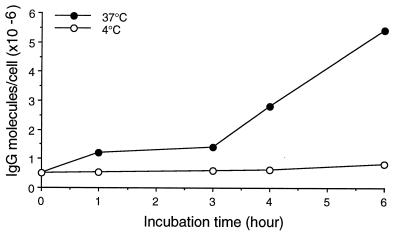
Eight IgG2a mAbs reacting with DNA were purified and tested by ELISA on various antigens (Table 2). Most of them were polyreactive, but two (A2.1 and H9.3) recognized only dsDNA. Polyreactive, but not monoreactive, anti-DNA mAbs were able to penetrate into all the living cells listed in Materials and Methods. Apparently, there was no correlation between the avidity of a mAb for DNA and its penetration capacity (Table 2). Hybridomas producing three mAbs (J20.8, F4.1, and F14.6) were bulk-cultured, and mAbs were purified and further analyzed. The purified mAbs were previously found, using phage-display peptide libraries, to recognize different peptides (18). These mAbs, used at concentrations from 10 to 50 μg/ml, penetrated all cell lines examined in a time-dependent manner. Each mAb exhibited different kinetic for intracellular localizations; F(ab′)2 and Fab′ fragments internalized like intact IgG molecules. At the above concentrations, compared with controls, no increase of cell death was noted as assessed by trypan blue dye exclusion. No change in cell metabolism was observed by using either the tetrazolium salt procedure or [3H]thymidine incorporation (data not shown). However, F4.1 translocation, which occurs at 37°C but not at 4°C (Fig. 1), was often accompanied by morphological changes and some dead cells were seen.
Table 2.
Reactivities assessed by ELISA of penetrating and nonpenetrating mAbs with a panel of antigens
| mAbs | dsDNA | ssDNA | Histone | DNase | Actin | Myosin | Tubulin | Ars | Ph-Ox | TNP | Kd (×10−9) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C2.4 (+) | 0.070* | 3.5 | 5 | 10 | 5 | 5.6 | 3 | 1.3 | 1.2 | 5 | 90 |
| C12.24 (+) | 0.022 | 10 | 5 | 10 | 5 | 5 | Neg. | 1.2 | 3 | Neg. | 80 |
| F4.1 (+) | 0.040 | 2 | 5 | 15 | 1.8 | 5 | 6 | 1.2 | 3 | 20 | 90 |
| J20.8 (+) | 0.066 | 60 | 16 | 30 | 16 | 2.5 | 3.5 | 1.8 | 5 | 30 | 350 |
| G14 (+) | 0.066 | 1.6 | 6 | 5 | 6 | 1.5 | 2 | 0.4 | 1 | 4 | 770 |
| F14.6 (+) | 0.060 | 0.8 | 0.6 | 10 | 0.6 | 0.2 | 1.8 | 0.5 | 0.45 | 3 | 150 |
| A2.1 (−) | 0.066 | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. | Neg. | 1,000 |
| H9.3 (−) | 0.140 | Neg. | 30 | Neg. | 30 | Neg. | Neg. | Neg. | Neg. | Neg. | 1,500 |
+, Penetrating mAb; −, nonpenetrating mAb; Neg., negative; ssDNA, single-stranded DNA; Ars: arsonate; Ph-Ox, phenyloxazolone; TNP, trinitrophenyl. All haptens were coupled to ovalbumin. The Kd values were calculated by using an inhibition assay (see Ref. 12).
Reactivity is expressed as the mAb concentration in μg/ml giving an OD414 of 0.200 in 2 h.
Figure 1.
Kinetic of penetration of mAb F4.1 (50 μg/ml) into 3T3 cells cultured at 37°C or 4°C. At indicated time, cells were trypsinized, counted, washed, and lyzed, and IgG2a mAb in lysates was quantified by ELISA by reference to a standard curve established with F4.1 mAb.
Intracellular Targeting of Haptens and Macromolecules by Anti-DNA mAbs.
FITC-conjugated mAbs, F(ab′)2, and Fab′ fragments penetrated cells as well as unlabeled molecules did. Confocal microscopy analysis of fluorescent cell preparations incubated for 4 h showed primarily a nuclear localization (data not shown).
The intracellular penetration of PO-conjugated IgG mAbs, F(ab′)2, and Fab′ fragments was examined on PtK2 and HEp-2 cells incubated for 16 h with the conjugates. PO-F(ab′)2 and PO-Fab′ generated intense signals mostly localized in the nucleus but sometimes also in the cytoplasm (Fig. 2A). After a 2-h incubation of PtK2 with Fab′-PO conjugate, 1–2 × 106 molecules of conjugate per cell were found to be internalized. No PO deposits were detected when cell preparations were incubated under the same conditions with nonpenetrating F(ab′)2 mAbs coupled to PO, free PO, or unlabeled penetrating F(ab′)2 mAbs plus PO.
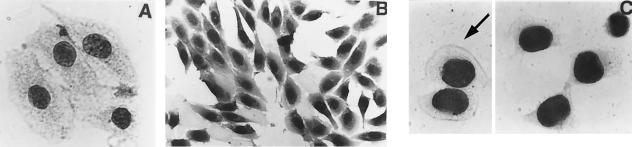
Figure 2.
Transfer of various macromolecules by mAbs or their fragments or by a derived peptide. (A) HEp-2 cells cultured overnight with J20.8 F(ab′)2 fragments coupled to PO (50 μg/ml). (B) PtK2 cells cultured with biotinylated P3 (20 μg/ml) for 1 h and after fixation incubated with streptavidin-PO (5 μg/ml). (C) Biotinylated P3 (1.4 μg) was incubated with streptavidin-PO (10 μg) in 35 μl of PBS for 20 min at room temperature, diluted to 250 μl with complete medium, and added to PtK2 cell culture for 2 h. After fixation, PO was detected with ME-DAB. Arrow shows a dividing cell.
When either FITC- or PO-coupled antibodies were translocated, the percentage of positive cells varied from 5 to 100%.
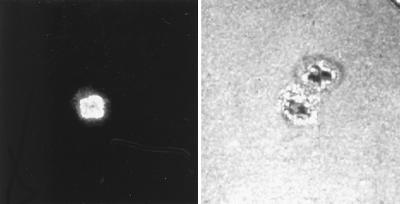
To evaluate the intracellular penetration of Fab′ conjugated to fluorescein-polynucleotide, 5 × 105 thymocytes or spleen cells from a 6-week-old BALB/c mouse were incubated in 250 μl of complete medium containing 13 μg of Fab′ conjugated to fluorescein-polynucleotide. After 3 h, cells were washed and fixed with 1% formaldehyde. Examined by flow cytometry, 4.5% of thymocytes and 15% of spleen cells were labeled. Analysis by confocal microscopy revealed that the fluorescein was localized in the nucleus (Fig. 3).
Figure 3.
Intranuclear translocation of a fluorescent nucleotide coupled to J20.8 Fab in mouse thymocytes. Cells were cultured for 2 h in the presence of the conjugated nucleotide (50 μg/ml), washed, fixed, and examined under confocal microscope. (Left) Fluorescence. (Right) Phase contrast.
The ability of mAbs J20.8, F4.1, and F14.6 to transport the plasmid pHu Vim 830-T/t were examined. Plasmid/mAb ratios were determined that avoided plasmid immunoprecipitation but allowed penetration into the nucleus. Although all three mAbs were able to transport the gene, wide variability was noted from one experiment to another, and rarely more than 0.1% of the cells expressed the transfected gene (data not shown).
Intracellular Targeting of VH Peptides of Anti-DNA mAbs.
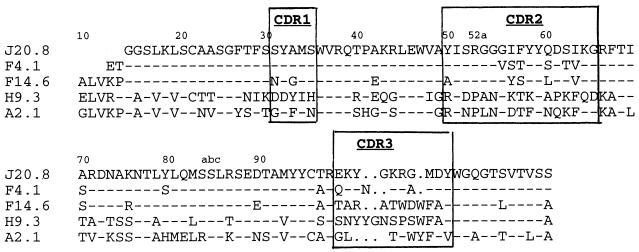
The nucleotide sequences of heavy and light chains of three penetrating and two nonpenetrating anti-DNA mAbs were determined and their amino acid sequences were deduced (Fig. 4). The penetrating mAbs (J20.8, F4.1, and F14.6) possessed highly homologous VH CDR2 sequences, including one arginine and one lysine residue, and distinct CDR3 regions, all containing arginine, a property typical of anti-DNA antibodies (19). The two nonpenetrating mAbs (H9.3 and A2.1) exhibited no CDR2 sequence homology with the penetrating mAbs. The CDR3 of one of them (A2.1) had two arginines, and the other one did not include arginine or lysine residues.
Figure 4.
Amino acid sequences of the VH genes of various mAbs deduced from their nucleotide sequences. Dashes indicate identity with the J20.8 mAb sequence; blanks indicate either that the VH does not have an amino acid at that position or a sequence was not obtained for that position.
The biotinylated peptides P1, P2, and P3 shown in Table 1 and corresponding to CDR2 and or CDR3 sequences of the penetrating mAb F4.1 were prepared and tested immunocytochemically for their capacities to penetrate into PtK2, HEp2, and CCL-39 cells. No penetration was detected in the three cell lines with P1, at all concentrations examined. With P2 at 20 μg/ml, a weak but definite labeling in the cytoplasm of most of the cells was observed. With P3 at 5–20 μg/ml, cytoplasmic positivity was noted in most of the cells, and intense nuclear labeling of PtK2 cells was also seen (Fig. 2B). Maximal penetration seemed to be achieved by 1 h, because no intensification was observed up to 5 h, after which time label intensity seemed to decrease.
To compare the reactivities of biotinylated P1, P2, and P3 and F4.1 with various antigens used in the present study (see Table 2), parallel ELISAs were performed. Of the three peptides, only P3 reacted strongly with DNA and various other antigens, although less intensely than F4.1. P2 bound weakly to a few antigens but never with DNA. P1 was always negative.
To determine whether the peptides could be used as vectors to deliver macromolecules intracellularly, complexes were prepared with biotinylated peptides and streptavidin-PO conjugate at various ratios and tested on PtK2, HEp-2, and CCL-39 cells. Complexes prepared with 1.4 μg of biotinylated P2 or P3 and 10 μg of streptavidin-PO gave the most satisfactory and reproducible results. After 1 h of incubation, P2 complexes generated a weak cytoplasmic positivity. Biotinylated P3 and streptavidin-PO complexes generated intense nuclear signals after 1 h (Fig. 2C) that were still visible 6 h later. A less intense but strong signal was noted in the cytoplasm and nuclei of almost all cells incubated for 1 h with either PO at 12–50 μg/ml or anti-PO antibodies directly coupled to P3 at 12–50 μg/ml. Measurable quantities of internalized enzymes were noted only when P3 was used as the vector. Incubation of PtK2 cells for 2 h with P3 complexed to streptavidin-PO or covalently linked to PO resulted, respectively, in the internalization of 2 × 105 to 8 × 106 molecules of streptavidin-PO and 5 × 107 to 5 × 108 of PO per cell. Values obtained with P1 and P2 were within the background level.
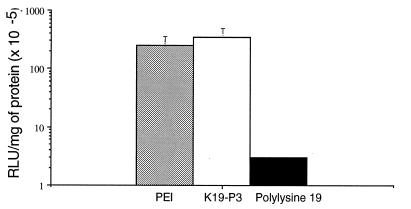
Transfection of cells with pCMVL-K19-P3 complexes resulted in a mean luciferase expression of 5 × 107 ± 1 × 107 RLU/mg of protein, values higher (two to three orders of magnitude) than those obtained with complexes of 19-residue polylysine alone (Fig. 5). The efficacy of transfection was of the same order of magnitude as that obtained by using the PEI procedure (20).
Figure 5.
Luciferase expression in 3T3 cells transfected with the pCMVL gene. Cells were incubated with 3 μg of plasmid complexed with 18 μg of K19-P3 (open bar), 10 equivalents of PEI nitrogen per DNA phosphate (shaded bar), or 18 μg of 19-residue polylysine (solid bar). Experiments were done in triplicate and the results show the average ± SEM of five experiments.
DISCUSSION
The main goal of this study was to examine the possibility of using anti-DNA mAbs, obtained from a 9-month-old (NZB × NZW)F1 mouse, as vectors for the intracellular transport of haptens and macromolecules. Many hybridoma clones indeed secreted penetrating IgG anti-DNA mAbs. We analyzed several of them of the IgG2a isotype and studied in greater detail three of them (J20.8, F4.1, and F14.6). These antibodies and their (Fab′)2 and Fab′ fragments could penetrate into various types of living cells, enter the nucleus, and accumulate there. Under the conditions used, such penetration did not appreciably alter cell viability or metabolism. Each mAb exhibited distinct kinetics of intracellular transit for a given cell line. These results confirm data previously reported for other anti-DNA antibodies (1–7). Remarkably, we found that, among the various anti-DNA antibodies from the lupus mouse, only polyreactive antibodies could penetrate into the cells. Because these polyreactive antibodies seem to possess conserved CDR1 and CDR2 and similar CDR3 sequences, the penetrating antibodies appear to represent clones of natural polyreactive autoantibodies that escape normal control and expand in lupus disease. Polyreactive anti-DNA antibodies, compared with monoreactive ones, should possess increased capabilities to bind to cell membranes. Indeed, flow cytometry analysis of our polyreactive anti-DNA mAbs showed that most, but not all, bound to various degrees to the surface of mouse spleen cells in suspension (data not shown). This finding is consistent with the results of previous work with other penetrating autoantibodies (1, 5–7), suggesting that internalization of anti-DNA mAbs is mediated by their binding to cell surface structures.
Penetrating anti-DNA mAbs and/or their F(ab′)2 or Fab′ fragments were effective vectors for the intracellular transport of haptens and macromolecules covalently linked to them. Thus fluorescein (Mr = 389), a 15-mer oligonucleotide (Mr = 5,000), and PO (Mr = 40,000) could be transported through the cytoplasm and into the nuclei of the various cell types examined. Furthermore, incubation of PtK2 cells with complexes formed between penetrating anti-DNA mAbs and a plasmid coding for the tumor antigen of simian virus 40 resulted in the nuclear expression of the tumor antigen but in no more than 0.1% of the cells. Transfection supplies evidence, independent of immunohistochemical observations, for anti-DNA mAb-mediated internalization of nucleic acids. In these experiments, the percentages of cells containing internalized mAbs detected immunocytochemically ranged, as for penetrating mAbs alone, from 5 to 100%. It will be of special interest to determine whether the limiting factor in these transfections is misrouting of most internalized plasmids or strong hindrance to the internalization of most complexes, arising from the structural alteration imposed on mAbs by plasmid binding. The observation made with site-directed mutants of a penetrating anti-DNA mAb (5), that the same residues required for binding DNA are necessary for penetration of this mAb, would agree with the latter hypothesis.
Our initial working hypothesis was that penetrating anti-DNA mAbs carry common peptides involved in their translocating abilities, which, if proven true, could be used as more effective vectors than whole antibodies. To explore this possibility, we determined the nucleotide sequences of the heavy and light chains of three penetrating mAbs. Highly homologous VH CDR2 sequences were noted. The synthetic peptide P1, corresponding to the CDR2 of F4.1, was unable to translocate into cells or to bind to antigens. In contrast, peptide P2, which corresponds to the VH CDR3 and, even more so, peptide P3 (P1 linked to P2) gave positive results. Only P3 was found by ELISA to be highly polyreactive and to bind strongly with DNA and various self and nonself antigens. Although weak binding to a few antigens, but never with DNA, was noted with P2, only P3 readily and abundantly penetrated into living cells, but small amounts of intracellular P2 could always be detected. These results suggest that the penetrating ability of F4.1 anti-DNA mAbs relies on CDR3 but is only manifested when CDR2 is associated with it. Further studies are needed to determine whether efficient penetration of CDR3 requires its linking to this specific CDR2 to achieve anti-DNA binding and increased auto- and polyreactivity or whether its association with an unrelated CDR2 would be sufficient.
Only P3 was found to be an effective vector, capable of translocating in a short time across plasma and nuclear membranes up to 5 × 108 protein macromolecules (from 40 to 200 kDa) to which it was linked covalently or not. However, although P3 at high concentrations was able to condense plasmids, it was ineffective in gene delivery. The addition of 19 lysine residues to P3 (K19-P3), to achieve DNA condensation (21), indeed generated a highly effective vector for gene transfer and expression. K19-P3 transfection does not necessitate the particular conditions (absence of serum or presence of lysotropic agents such as chloroquine) required in most of the reported transfection protocols, although its efficacy in transferring luciferase gene in 3T3 cells was of the same order of magnitude as that found with PEI, which is considered to be one of the most effective agents (20). Presumably, K19-P3 derives its very high DNA transfection efficiency from both the penetrating ability of P3 and the known properties of the added polylysine cationic peptide (21).
Several proteins or polypeptides have been proposed as vectors for the delivery of macromolecules into living cells. Among these vectors, those involving receptor-mediated mechanisms have been successful in gene delivery (22) but were reported only occasionally to be effective in internalizing substantial amounts of protein (23). In contrast, toxins and proteins from pathogenic agents, capable of translocating across the membrane and to reach the cytoplasm, have been relatively efficient in protein delivery (24–26) but of limited applicability in gene delivery. The anti-DNA mAbs studied herein delivered both proteins and genes into cells and analysis of this system indicated that P3 should be a simple versatile vector of macromolecules.
Until now, to the best of our knowledge, the only vector reported to possess somewhat similar capabilities is the Drosophila Antennapedia homeodomain protein and derived peptides (27, 28). Like penetrating mAbs, the Antennapedia protein binds to DNA. The peptide vectors derived from this protein also contain a relatively high number of positively charged amino acids (lysine and arginine), as do most proteins with nuclear localization signals (29, 30). The third α-helix of the Antennapedia protein seems to play an important role in the translocation capacity of this protein. Remarkably, an α-helix may be also involved in the high translocating ability of P3. Indeed, structural predictions by analysis software indicate that association of incomplete structures present on P1 and P2 creates a complete α-helix on P3 (M. Delepierre and N. Wolff, personal communication), which appears to confer to this peptide its translocating ability. Despite these similarities, the performances of the two translocation systems are different and may be so because they exploit markedly different molecular mechanisms. F4.1 anti-DNA mAb is internalized through a process that appears to be energy dependent, whereas Antennapedia protein operates at 4°C. F4.1-derived P3 translocates high molecular weight proteins, whereas Antennapedia peptide translocates polypeptides up to 50 amino acids. Polypeptide vectors with similar properties may thus cross membranes and follow distinct intracellular pathways, imposed, among others, by both the fine specificity of ligand-recognizing sites and by their global conformation and charges. Should this postulate prove true, by combining the CDR2 and CDR3 of different penetrating anti-DNA mAbs exhibiting different mimotopes (18), peptide vectors possessing distinct properties could be created that would be more adapted to one situation than to another.
Murine anti-DNA mAbs able to penetrate into cells under in vitro conditions were also effective when injected into mice and localized in cells of various tissues (3). Therefore, one would expect that penetrating anti-DNA antibodies and derived peptides could be used for in vivo studies. Indeed, in preliminary experiments, we have found that mice immunized with P3–protein complexes produced higher amounts of antibodies than mice immunized with the protein alone.
Acknowledgments
We thank Dr. J.-C. Mazié (Institut Pasteur) for his interest in this project and Dr. P. Lafaye (Institut Pasteur) for his help in gene sequencing. We are grateful to Dr. P. Vicart, Institut Pasteur, for providing the pHu Vim 830-T/t plasmid and the hybridoma secreting anti-tumor antigen antibody, and to Dr. U. Hazan (Institut Cochin de Génétique Moleculaire) for the gift of the pCMVL plasmid. A. Avrameas received a fellowship from Centre Européen de Bioprospective, Rouen, France.
ABBREVIATIONS
- CDR
complementary-determining region
- dsDNA
double-stranded DNA
- ME-DAB
metal-enhanced diaminobenzidine
- FITC
fluorescein isothiocyanate
- VH
Ig heavy chain variable region
- PO
horseradish peroxidase
- PEI
polyethylenimine
- RLU
relative light units
Footnotes
References
- 1.Alarcon-Segovia D S, Ruiz-Argüelles A, Fishbein E. Nature (London) 1978;271:67–69. doi: 10.1038/271067a0. [DOI] [PubMed] [Google Scholar]
- 2.Ma J, Chapman G V, Chen S-L, Melick G, Penny R, Breit S N. Clin Exp Immunol. 1991;84:83–91. doi: 10.1111/j.1365-2249.1991.tb08128.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanase K, Smith R M, Cisman B, Foster M H, Peachey L D, Jarett L, Madaio M P. Lab Invest. 1994;71:52–60. [PubMed] [Google Scholar]
- 4.Koren E, Koscec M, Wolfson-Reichlin M, Ebling F M, Tsao B, Hahn B H, Reichlin M. J Immunol. 1995;154:4857–4864. [PubMed] [Google Scholar]
- 5.Zack D J, Stempniak M, Wong A L, Taylor C, Weisbart H. J Immunol. 1996;157:2082–2088. [PubMed] [Google Scholar]
- 6.Alarcon-Segovia D S, Llorente L, Ruiz-Argüelles A. J Autoimmun. 1996;9:295–300. doi: 10.1006/jaut.1996.0038. [DOI] [PubMed] [Google Scholar]
- 7.Yanase K, Smith R M, Pucetti A, Jarett L, Madaio M P. J Clin Invest. 1997;100:25–31. doi: 10.1172/JCI119517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Köhler G, Milstein C. Eur J Immunol. 1976;6:511–514. doi: 10.1002/eji.1830060713. [DOI] [PubMed] [Google Scholar]
- 9.Avrameas, S., Ternynck, T. & Guesdon, J.-L. (1978) Scand. J. Immunol. 8, Suppl. 7, 7–23.
- 10.Ey P L, Prowse S J, Jenkin C R. Immunochemistry. 1978;15:429–436. doi: 10.1016/0161-5890(78)90070-6. [DOI] [PubMed] [Google Scholar]
- 11.Guilbert B, Dighiero G, Avrameas S. J Immunol. 1982;128:2779–2787. [PubMed] [Google Scholar]
- 12.Friguet B, Chaffotte A F, Djavadi-Ohaniance L, Goldberg M E. J Immunol Methods. 1985;77:305–319. doi: 10.1016/0022-1759(85)90044-4. [DOI] [PubMed] [Google Scholar]
- 13.Nisonoff A, Wissler F C, Lipman L N, Wornley D L. Arch Biochem Biophys. 1960;89:230–244. doi: 10.1016/0003-9861(60)90049-7. [DOI] [PubMed] [Google Scholar]
- 14.Goding J W. J Immunol Methods. 1976;13:215–226. doi: 10.1016/0022-1759(76)90068-5. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz B, Vicart P, Delouis C, Paulin D. Biol Cell. 1991;73:7–14. doi: 10.1016/0248-4900(91)90003-6. [DOI] [PubMed] [Google Scholar]
- 16.Thomas P S. Proc Natl Acad Sci USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devereux J. The GCG Sequence Analysis Software Package. Version 6.0. Madison, WI: Genetics Computer Group; 1989. [Google Scholar]
- 18.Sibille P, Ternynck T, Nato F, Buttin G, Strosberg D, Avrameas A. Eur J Immunol. 1997;27:1221–1228. doi: 10.1002/eji.1830270525. [DOI] [PubMed] [Google Scholar]
- 19.Radic M Z, Weigert M. Annu Rev Immunol. 1994;12:487–520. doi: 10.1146/annurev.iy.12.040194.002415. [DOI] [PubMed] [Google Scholar]
- 20.Boussif O, Lezoualc’h F, Zanta M A, Mergny M D, Scherman D, Demeneix B, Behr J-P. Proc Natl Acad Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wadhwa M S, Collard W T, Adami R C, McKenzie D L, Rice K G. Bioconjugate Chem. 1997;8:81–88. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]
- 22.Perales J C, Ferkol T, Molas M, Hanson R W. Eur J Biochem. 1994;226:255–266. doi: 10.1111/j.1432-1033.1994.tb20049.x. [DOI] [PubMed] [Google Scholar]
- 23.Leamon C P, Low P S. Proc Natl Acad Sci USA. 1991;88:5572–5576. doi: 10.1073/pnas.88.13.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenmark H, Moskaug J O, Madhus I H, Sandvig K, Olsnes J. J Cell Biol. 1991;113:1025–1032. doi: 10.1083/jcb.113.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prior T I, FitzGerald D J, Pastan I. Biochemistry. 1992;31:53555–3559. doi: 10.1021/bi00129a001. [DOI] [PubMed] [Google Scholar]
- 26.Fawell S, Seery J, Daikh Y, Moore C, Chen L L, Pepinsky B, Barsoum J. Proc Natl Acad Sci USA. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derossi D, Joliot A H, Chassaing G, Prochiantz A. J Biol Chem. 1994;269:10444–10450. [PubMed] [Google Scholar]
- 28.Schutze-Redelmeier M-P, Gournier H, Garcia-Pons F, Moussa M, Joliot A H, Volovitch M, Prochiantz A, Lemonnier F A. J Immunol. 1996;157:650–655. [PubMed] [Google Scholar]
- 29.Richardson W D, Roberts B L, Smith A E. Cell. 1986;44:77–85. doi: 10.1016/0092-8674(86)90486-1. [DOI] [PubMed] [Google Scholar]
- 30.Wychoski C, Benichou D, Girard M. EMBO J. 1986;5:2569–2576. doi: 10.1002/j.1460-2075.1986.tb04536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]