Abstract
The features which contribute to the activity of the single-stranded origin of the Bacillus plasmid pBAA1 were investigated. This origin is contained on a DNA fragment greater than 116 but less than 191 bases in size. There is the potential to form three stem-loop structures within this fragment. Comparison of the sequence of this origin from pBAA1 with the sequence of a homologous fragment from the Bacillus thuringiensis plasmid pGI2 indicates that both the structure and the relative positioning of the predicted stem-loops are important for origin activity. Deletion analysis suggests that it is the structure of stem-loop III which is important, because it can be replaced by a nonrelated dyad element without significant loss of origin activity. Three sequence motifs are conserved between the origins from pBAA1 and pGI2. Mutation of motif 1 leads to attenuation of single-stranded origin activity. A second motif (motif 3) shares significant homology with a group of single-strand initiation (ssi) sites found on plasmids isolated from Escherichia coli, suggesting that it also contributes to single-stranded origin activity. Our results also indicate that RNA polymerase is utilized to synthesize the RNA primer at the pBAA1 single-stranded origin and that this origin can function in both Bacillus subtilis and Staphylococcus aureus.
Full text
PDF

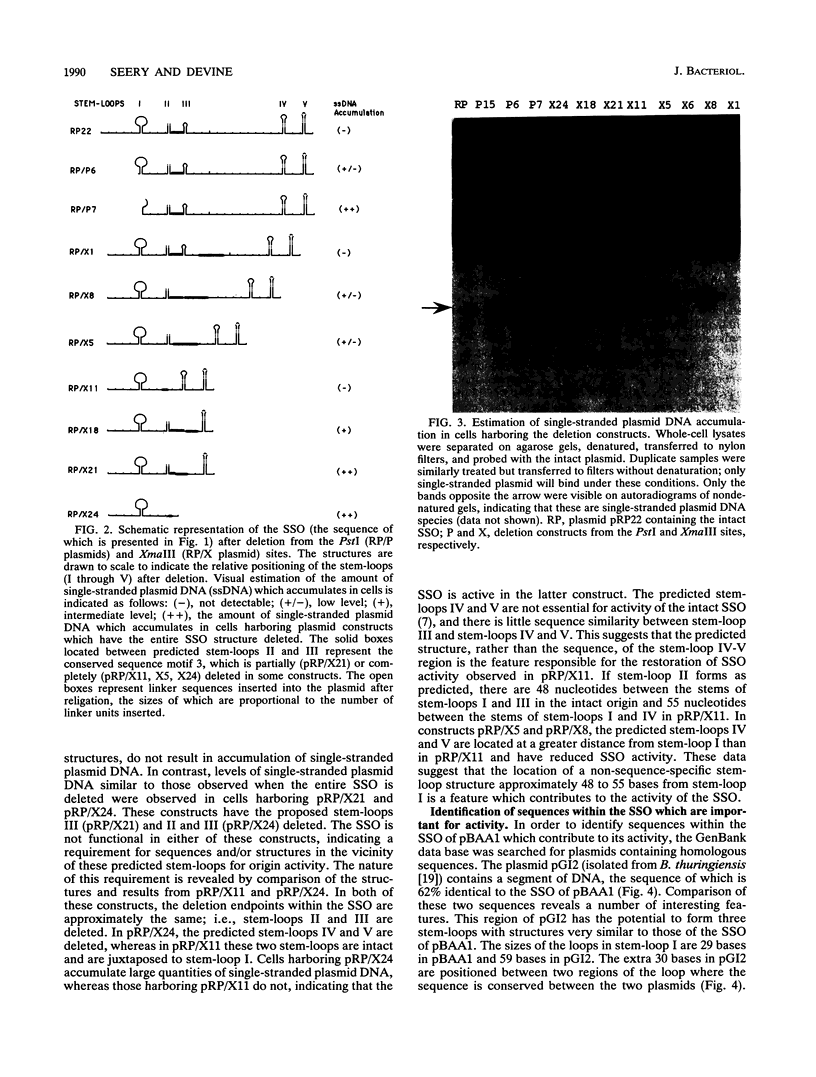




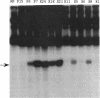
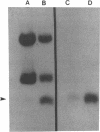
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Kornberg A. Unique primed start of phage phi X174 DNA replication and mobility of the primosome in a direction opposite chain synthesis. Proc Natl Acad Sci U S A. 1981 Jan;78(1):69–73. doi: 10.1073/pnas.78.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai K., Low R. L., Kornberg A. Movement and site selection for priming by the primosome in phage phi X174 DNA replication. Proc Natl Acad Sci U S A. 1981 Feb;78(2):707–711. doi: 10.1073/pnas.78.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas P. D. DNA replication of single-stranded Escherichia coli DNA phages. Biochim Biophys Acta. 1985 Jun 24;825(2):111–139. doi: 10.1016/0167-4781(85)90096-x. [DOI] [PubMed] [Google Scholar]
- Baas P. D., Jansz H. S. Single-stranded DNA phage origins. Curr Top Microbiol Immunol. 1988;136:31–70. doi: 10.1007/978-3-642-73115-0_3. [DOI] [PubMed] [Google Scholar]
- Boe L., Gros M. F., te Riele H., Ehrlich S. D., Gruss A. Replication origins of single-stranded-DNA plasmid pUB110. J Bacteriol. 1989 Jun;171(6):3366–3372. doi: 10.1128/jb.171.6.3366-3372.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine K. M., Hogan S. T., Higgins D. G., McConnell D. J. Replication and segregational stability of Bacillus plasmid pBAA1. J Bacteriol. 1989 Feb;171(2):1166–1172. doi: 10.1128/jb.171.2.1166-1172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J. H., Marians K. J. The interaction of Escherichia coli replication factor Y with complementary strand origins of DNA replication. Contact points revealed by DNase footprinting and protection from methylation. J Biol Chem. 1984 Feb 25;259(4):2594–2601. [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiasa H., Sakai H., Tanaka K., Honda Y., Komano T., Godson G. N. Mutational analysis of the primer RNA template region in the replication origin (oric) of bacteriophage G4: priming signal recognition by Escherichia coli primase. Gene. 1989 Dec 7;84(1):9–16. doi: 10.1016/0378-1119(89)90133-9. [DOI] [PubMed] [Google Scholar]
- Hiasa H., Tanaka K., Sakai H., Yoshida K., Honda Y., Komano T., Godson G. N. Distinct functional contributions of three potential secondary structures in the phage G4 origin of complementary DNA strand synthesis. Gene. 1989 Dec 7;84(1):17–22. doi: 10.1016/0378-1119(89)90134-0. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Gouy M. Interfacing similarity search software with the sequence retrieval system ACNUC. Comput Appl Biosci. 1987 Sep;3(3):239–241. doi: 10.1093/bioinformatics/3.3.239. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989 Apr;5(2):151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- Kim M. H., Hines J. C., Ray D. S. Viable deletions of the M13 complementary strand origin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6784–6788. doi: 10.1073/pnas.78.11.6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert P. F., Kawashima E., Reznikoff W. S. Secondary structure at the bacteriophage G4 origin of complementary-strand DNA synthesis: in vivo requirements. Gene. 1987;53(2-3):257–264. doi: 10.1016/0378-1119(87)90014-x. [DOI] [PubMed] [Google Scholar]
- Lee M. S., Marians K. J. The Escherichia coli primosome can translocate actively in either direction along a DNA strand. J Biol Chem. 1989 Aug 25;264(24):14531–14542. [PubMed] [Google Scholar]
- Mahillon J., Seurinck J. Complete nucleotide sequence of pGI2, a Bacillus thuringiensis plasmid containing Tn4430. Nucleic Acids Res. 1988 Dec 23;16(24):11827–11828. doi: 10.1093/nar/16.24.11827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai H., Nomura N., Arai K. The ABC-primosome. A novel priming system employing dnaA, dnaB, dnaC, and primase on a hairpin containing a dnaA box sequence. J Biol Chem. 1990 Sep 5;265(25):15134–15144. [PubMed] [Google Scholar]
- Masai H., Nomura N., Kubota Y., Arai K. Roles of phi X174 type primosome- and G4 type primase-dependent primings in initiation of lagging and leading strand syntheses of DNA replication. J Biol Chem. 1990 Sep 5;265(25):15124–15133. [PubMed] [Google Scholar]
- Nomura N., Masai H., Inuzuka M., Miyazaki C., Ohtsubo E., Itoh T., Sasamoto S., Matsui M., Ishizaki R., Arai K. Identification of eleven single-strand initiation sequences (ssi) for priming of DNA replication in the F, R6K, R100 and ColE2 plasmids. Gene. 1991 Dec 1;108(1):15–22. doi: 10.1016/0378-1119(91)90482-q. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ryder T. B., Maeda Y., Armstrong K., Ohtsubo E. DNA replication of the resistance plasmid R100 and its control. Adv Biophys. 1986;21:115–133. doi: 10.1016/0065-227x(86)90018-3. [DOI] [PubMed] [Google Scholar]
- Ray D. S., Hines J. C., Kim M. H., Imber R., Nomura N. M13 vectors for selective cloning of sequences specifying initiation of DNA synthesis on single-stranded templates. Gene. 1982 Jun;18(3):231–238. doi: 10.1016/0378-1119(82)90160-3. [DOI] [PubMed] [Google Scholar]
- Sakai H., Komano T., Godson G. N. Replication origin (oric) on the complementary DNA strand of Escherichia coli phage G4: biological properties of mutants. Gene. 1987;53(2-3):265–273. doi: 10.1016/0378-1119(87)90015-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul D., Spiers A. J., McAnulty J., Gibbs M. G., Bergquist P. L., Hill D. F. Nucleotide sequence and replication characteristics of RepFIB, a basic replicon of IncF plasmids. J Bacteriol. 1989 May;171(5):2697–2707. doi: 10.1128/jb.171.5.2697-2707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer G., Som T., Itoh T., Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983 Jan;32(1):119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., Helinski D. R. DNA segments of the IncX plasmid R485 determining replication and incompatibility with plasmid R6K. Plasmid. 1985 Nov;14(3):245–254. doi: 10.1016/0147-619x(85)90008-3. [DOI] [PubMed] [Google Scholar]
- Stuitje A. R., Weisbeek P. J., Meijer M. Initiation signals for complementary strand DNA synthesis in the region of the replication origin of the Escherichia coli chromosome. Nucleic Acids Res. 1984 Apr 11;12(7):3321–3332. doi: 10.1093/nar/12.7.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viret J. F., Alonso J. C. A DNA sequence outside the pUB110 minimal replicon is required for normal replication in Bacillus subtilis. Nucleic Acids Res. 1988 May 25;16(10):4389–4406. doi: 10.1093/nar/16.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]