Abstract
Background and purpose:
Two compounds, URB602 and URB754, have been reported in the literature to be selective inhibitors of monoacylglycerol lipase, although a recent study has questioned their ability to prevent 2-arachidonoyl hydrolysis by brain homogenates and cerebellar membranes. In the present study, the ability of these compounds to inhibit monoacylglycerol lipase and fatty acid amide hydrolase has been reinvestigated.
Experimental approach:
Homogenates and cell lines were incubated with test compounds and, thereafter, with either [3H]-2-oleoylglycerol or [3H]-anandamide. Labelled reaction products were separated from substrate using chloroform: methanol extraction.
Key results:
In cytosolic fractions from rat brain, URB602 and URB754 inhibited the hydrolysis of 2-oleoylglycerol with IC50 values of 25 and 48 μM, respectively. Anandamide hydrolysis by brain membranes was not sensitive to URB754, but was inhibited by URB602 (IC50 value 17 μM). Hydrolysis of 2-oleoylglycerol by human recombinant monoacylglycerol lipase was sensitive to URB602, but not URB754. The lack of selectivity of URB602 for 2-oleoylglycerol compared to anandamide hydrolysis was also observed for intact RBL2H3 basophilic leukaemia cells. C6 glioma expressed mRNA for monoacylglycerol lipase, and hydrolyzed 2-oleoylglycerol in a manner sensitive to inhibition by methyl arachidonoyl fluorophosphonate but not URB754 or URB597. MC3T3-E1 mouse osteoblastic cells, which did not express mRNA for monoacylglycerol lipase, hydrolyzed 2-oleoylglycerol in the presence of URB597, but the hydrolysis was less sensitive to methyl arachidonoyl fluorophosphonate than for C6 cells.
Conclusions and implications:
The data demonstrate that the compounds URB602 and URB754 do not behave as selective and/or potent inhibitors of monoacylglycerol lipase.
Keywords: 2-acylglycerol, anandamide, monoacylglycerol lipase, fatty acid amide hydrolase, URB597, URB602, URB754
Introduction
The primary enzyme involved in the metabolism of the endocannabinoid anandamide (AEA) is fatty acid amide hydrolase (FAAH), and selective inhibitors of this enzyme, such as URB597 (3′-carbamoyl-biphenyl-3-yl-cyclohexylcarbamate) have been shown to produce potentially useful effects in models of pain and inflammation (see e.g. Kathuria et al., 2003; Holt et al., 2005; Jayamanne et al., 2006 for studies with this compound). The other main endocannabinoid, 2-arachidonoylglycerol (2-AG) is also metabolized by FAAH (Goparaju et al., 1998), and microinjection of an FAAH inhibitor into the periaqueductal grey increases 2-AG levels in that region (Maione et al., 2006). However, the enzyme monoacylglycerol lipase (MAGL), which is capable of hydrolyzing both 2-AG and its oleoyl homologue 2-oleoylglycerol (2-OG) (Tornqvist and Belfrage, 1976; Dinh et al., 2002), may be a more important metabolic enzyme in the brain for this endocannabinoid (Dinh et al., 2002). This enzyme is not inhibited by URB597 (Makara et al., 2005), but is sensitive to the non-selective serine hydrolase inhibitor methyl arachidonoyl fluorophosphonate (MAFP) (Dinh et al., 2002; see also http://www.caymanchem.com/app/template/Product.vm/catalog/10005192).
Until recently, no selective inhibitors of MAGL had been described in the literature. However, in 2005, Hohmann et al. found that URB602 ([1,1′-biphenyl]-3-yl-carbamic acid, cyclohexyl ester) inhibited the hydrolysis of 2-OG by rat brain cytosolic fractions with a half-maximal inhibitory concentration (IC50) value of 28 μM, but had little effect upon FAAH, diacylglycerol lipase or cyclooxygenase-2 activity, and did not ‘significantly influence (IC50 ⩾5 μM)' the binding of the cannabinoid receptor agonist [3H]WIN55,212-2 to CB1 or CB2 cannabinoid receptors. The compound also inhibited rat brain recombinant MAGL activity with an IC50 value of 75 μM (Makara et al., 2005). In contrast, Saario et al. (2006) found that a concentration of 1 mM of URB602 did not affect the hydrolysis of 2-AG by either rat brain homogenates or rat cerebellar membrane fractions. An even larger difference was seen with respect to another compound, URB754 (6-methyl-2-[(4-methylphenyl)amino]-4H-3,1-benzoxazin-4-one), which was identified when a library of compounds was screened towards recombinant MAGL and was demonstrated to inhibit this enzyme with an IC50 value of 200 nM (Makara et al., 2005). Commercially available URB754, however, did not inhibit 2-AG metabolism in the brain homogenates and cerebella membranes at a concentration of 100 μM (Saario et al., 2006). In order to shed further light upon the pharmacology of these compounds, we have investigated their ability to prevent 2-OG hydrolysis, using membrane and cytosolic sources of the enzyme, recombinant human MAGL and intact cells. Also, the selectivity of these compounds for 2-OG compared to AEA hydrolysis has been determined.
Methods
Cell-free preparations for MAGL and FAAH assays
Cerebella from adult male Wistar rats were used. These were thawed and homogenized at 4°C in sodium phosphate buffer (50 mM, pH 8) containing 0.32 M sucrose. Homogenates were centrifuged at 100 000 g for 60 min at 4°C, and supernatants (‘cytosol fractions') were collected. The pellets were resuspended in sodium phosphate buffer (50 mM, pH 8) (‘membrane fractions'). The fractions were stored frozen in aliquots at −70°C until used for assay of either rat MAGL or FAAH, as appropriate. Data for human MAGL were obtained using recombinant enzyme purified and expressed in Escherichia Coli by co-author Labar, the protocol of which will be published elsewhere. Protein concentrations of the fractions were determined by using the method described by Harrington (1990), with bovine serum albumin (BSA) as standard. Ethical permission for the animals was obtained from the local animal ethical committee.
Preparation of cell cultures
Rat C6 glioma cells were obtained from the European Collection of Cell Cultures (Porton Down, UK) and used over a passage range of 10–34. Rat RBL2H3 basophilic leukaemia cells were obtained from the American Type Culture Collection (Manassas, VA, USA) and used over a passage range of 21–28. The murine calvarial osteoblastic cell line MC3T3-E1 was a kind gift from Dr Masayoshi Kumegawa, Meikai University School of Dentistry, Saitama, Japan, and was used at passages 12–20. Culture media were: C6 cells, F-10 Ham with 10% fetal bovine serum (FBS) and 100 units ml−1 penicillin and 100 μg ml−1 streptomycin (1% PEST); RBL2H3 cells, MEM with Earls salts, 2 mM L-glutamine, 15% FBS and 1% PEST; MC3T3-E1 cells, α-MEM with 10% FBS, 2 mM L-glutamine and antibiotics (gentamicin 2.5 μg ml−1, streptomycin 70 μg ml−1, bensylpenicillin 35 μg ml−1).
Assay of MAGL and FAAH
For cell-free preparations, MAGL and FAAH were assayed as described previously (see Vandevoorde et al., 2005), although for the experiments run in Sweden a different extraction protocol (Boldrup et al., 2004) was used for the three highest concentrations of URB602 and for FAAH; data shown as an inset to Figure 1a. For intact cells, a modification of the assay described by Jonsson et al. (2001) was used. Cells (2.5 × 105 per well) were cultured overnight in 24-well culture plates. The cells were then washed once with 500 μl of Krebs–Henseleit-Bicarbonate buffer (120 mM NaCl, 4.7 mM KCl, 2.2 mM CaCl2, 10 mM 4-(2-hydroxyethyl)-1-piperazineethyl-sulphonic acid (HEPES), 0.12 mM KH2PO4, 0.12 mM MgSO4 in milliQ deionized water, pH 7.4) containing 1% w v−1 BSA. Thereafter Krebs–Henseleit-Bicarbonate buffer was added followed by test compound, and the samples were incubated for 10–60 min at 37°C. Substrate (0.25 μM final concentration, in buffer containing fatty acid-free BSA at a final assay concentration 0.1% w v−1) was added to give an assay volume of 400 μl. The samples were incubated for the times shown in the figure legend, then the plates were placed upon ice. Methanol (400 μl) was added and the cells were collected by scraping the wells. Aliquots (400 μl) were transferred to glass tubes, chloroform (200 μl) was added, and the assays completed as described for the cell-free preparations (Vandevoorde et al., 2005). Results are expressed as percentage of controls treated with the same concentration of vehicle.
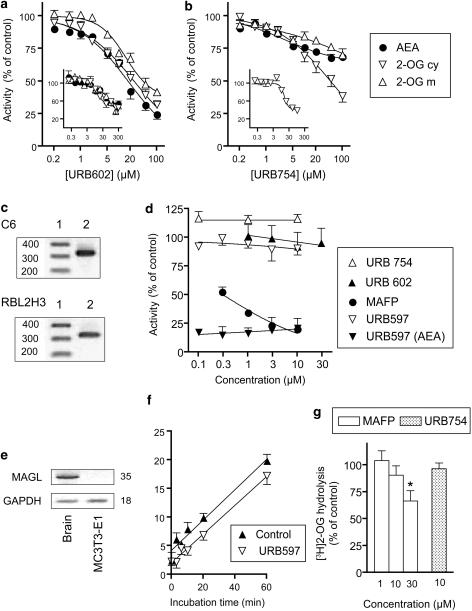
Figure 1.
(a and b) Inhibition of the hydrolysis of [3H]AEA (membrane preparations), [3H]2-OG (cytosolic [‘cy'] and URB597-treated membrane [‘m'] preparations) by (a) URB 602 and (b) URB 754. Shown are means and s.e.m. (vertical lines), n=3–6, of the activity as percentage of the corresponding vehicle controls. The experiments were performed in Belgium, and the insets show the corresponding data from experiments performed in Sweden. The substrate concentrations were 2 μM and incubation times 10 min. (c) Representative PCR analyses for MAGL from C6 (upper) and RBL2H3 (lower) cells. Lane 1: 100 bp ladder, lane 2: rat MAGL mRNA (30 PCR cycles), expected PCR fragment size was 327 bp. The identity of the PCR fragment as MAGL was confirmed by sequencing, as described in Methods section. (d) Inhibition of 0.25 μM [3H]2-OG hydrolysis in intact C6 cells by URB597, URB754, URB602 and MAFP. Compounds (10 μl in ethanol or dimethyl sulphoxide (DMSO), as appropriate) were preincubated for 10 min before the addition of substrate and incubation for a further 5 min at 37°C. Corresponding data for URB597 and 0.25 μM [3H]AEA hydrolysis are also shown in the figure. (e) Semi-quantitative PCR analysis of MAGL in MC3T3-E1 cells and mouse brain. The PCR reactions were normalized with glyceraldehyde-3-phosphate dehydrogenase, and the numbers in the figure indicate the number of PCR cycles. (f) Time course of 0.25 μM [3H]2-OG hydrolysis in the absence and presence of 1 μM URB597 by the MC3T3-E1 cells. Shown are means and s.e.m., n=3–4, of the hydrolysis for wells seeded with 2.5 × 105 cells after subtraction of the observed hydrolysis for the wells alone. (g) Effects of URB754 and MAFP on the hydrolysis of 0.25 μM [3H]2-OG. Compounds were preincubated for 10 min before the addition of [3H]2-OG and incubation for a further 20 min. URB597 (1 μM) was present during the preincubation phase (means and s.e.mean are shown, n=4–8 of the activities expressed as percentage of the corresponding vehicle controls).
It should be pointed out that the assay used, although a ‘standard' method, separates the product of enzyme action upon 2-OG on the basis of its partition into the methanol/water phase vs the chloroform phase but is not necessarily specific for MAGL. Treatment of rat brain cytosolic fractions with an antibody to MAGL reduces, but does not completely block their ability to hydrolyze 2-OG (Dinh et al., 2004), suggesting that additional enzymes may be operative. 2-OG (and 2-AG) are substrates for a family of monoacylglycerol kinases, which produce lyso-phosphatidic acids, although these products would be expected to be retained in the chloroform phase (see e.g. Simpson et al., 1991; Waggoner et al., 2004). However, other esterases may be involved. In this respect, the esterase domain of human neuropathy target esterase can also hydrolyze 2-OG (and 2-AG), and can be inhibited by the serine hydrolase inhibitor MAFP (van Tienhoven et al., 2002). Hence, we have referred to this process throughout as ‘2-OG hydrolysis' rather than as MAGL activity, except when the recombinant enzyme has been used.
RNA extraction and polymerase chain reactions
RNA extraction and polymerase chain reactions (PCR) were undertaken by standard methods, using RNAqueous-4PCR kit for RNA isolation, a first strand cDNA synthesis kit using oligo(dT)15 primers for reverse transcription (RT) into single-stranded cDNA and a PC-960G Gradient Thermal Cycler (Corbett Research, Mortlake, Australia) for PCR. The sequences of primers used were: rat MAGL sense 5′-TTGACCTGTACAACTCCGACC-3′ and antisense 5′-TAGCTCCTGCCACTGCTATCC-3′; mouse MAGL sense 5′-GGTCAATGCAGACGGACAGTACCTCTTTTG-3′ and antisense 5′-GTTTACTTCATGGAGGACGGAGTTGGTCAC-3′; mouse glyceraldehyde-3-phosphate dehydrogenase sense 5′-ACTTTGTCAAGCTCATTTCC-3′ and antisense 5′-TGCAGCGAACTTTATTGATG-3′. The expression of the target genes was compared at the logarithmic phase of the PCR reaction. No amplification was detected in samples where the RT reaction had been omitted. The PCR products for the C6 and RBL2H3 cells were purified using a QIAquick purification kit using the protocol supplied by the manufacturer, and the identities were confirmed using a Thermo-Sequenase-TM-II DYEnamic ET terminator cycle sequencing kit with sequences analysed on an ABI 377 XL DNA Sequencer (Applied Biosystems, Foster City, CA, USA).
Statistical analyses
Curve fitting and statistical comparisons were undertaken using the statistical package in the GraphPad Prism computer programme (GraphPad Software Inc., San Diego, CA, USA). IC50 values were determined using the sigmoidal dose–response curve (variable slope) analysis provided by the programme.
Materials
2-OG [glycerol – 1,2,3-3H] (0.74 TBq mmol−1) and AEA [ethanolamine-1-3H] (2.22 TBq mmol−1) were obtained from American Radiolabeled Chemicals, Inc. (St Louis, MO, USA). MAFP was obtained from Tocris Bioscience (Ellisville, MO, USA). Nonradioactive AEA, URB597, URB602 and URB754 were obtained from Cayman Chemical Co. (Ann Arbor, MI, USA). Fatty acid-free bovine serum albumin and unlabelled 2-OG were obtained from Sigma Aldrich Inc. (St Louis, MO, USA). RNAqueous-4PCR kit for RNA isolation was obtained from Ambion Inc. (Austin, TX, USA). First strand cDNA synthesis Kit and PCR Core Kit were obtained from Roche (Mannheim, Germany). Oligonucleotide primers were obtained from Invitrogen (Stockholm, Sweden). QIAquick purification kit was obtained from Qiagen Ltd (Crawley, UK). Thermo-Sequenase-TMII DYEnamic ET terminator cycle sequencing kit was obtained from Amersham Biosciences (Uppsala, Sweden).
Results
Selectivity of URB602 and URB754 for MAGL vs FAAH
In Figure 1a, the effects of URB602 on the hydrolysis of 2-OG by cytosolic and membrane-bound preparations from rat brain are shown and compared with its effects on AEA hydrolysis. URB602 showed no obvious selectivity: the pI50 values for cytosolic 2-OG hydrolysis and membrane fraction AEA hydrolysis were 4.60±0.05 and 4.76±0.05, respectively (corresponding to IC50 values of 25 and 17 μM, respectively). The data shown in the main figure was from experiments conducted in Belgium, but a similar lack of selectivity was seen when corresponding experiments were conducted in Sweden (inset to Figure 1a). Effects on human recombinant MAGL were also investigated; 2-OG hydrolysis rates of 95±0.5, 95±0.6, 73±8, 48±9 and 54±3% of vehicle controls were found at URB602 concentrations of 0.3, 3, 10, 30 and 300 μM, respectively (means±s.e.m., n=3).
URB754 was a weak inhibitor of the hydrolysis of 2-OG by the brain preparations (Figure 1b). The cytosolic activity was the most affected, with a pI50 value for URB754 of 4.31±0.07, corresponding to an IC50 value of 48 μM. Membrane-bound 2-OG metabolism (in the presence of 3 μM URB597 to prevent FAAH-catalysed hydrolysis of this substrate) and AEA hydrolysis were only inhibited by about 30% at the highest concentrations of the compound tested. At concentrations of URB754 of 0.3, 3, 10, 30 and 300 μM, the 2-OG hydrolysis rates produced by human recombinant MAGL were 97±4, 81±7, 93±2, 101±0.9 and 91±4% of vehicle controls, respectively (means±s.e.m., n=4). Mass spectrometry analyses of the stock solutions of URB754 and URB602 used in these experiments confirmed the levels of purity of the compounds stated by the manufacturers (Waters QToF2 mass spectrometer, source ESI, exact mass [M+H+]=267.1134 found for URB754; exact mass [M+Na+]=318.1458 found for URB602, analyses undertaken by Professor R Flammang, Dr P Gerbaux and Ing. M Boulvin at the Mass Spectrometry Laboratory of the University of Mons-Hainaut, Belgium).
Effects of URB602 and URB754 on the hydrolysis of 2-OG by intact cells
C6 glioma and RBL2H3 basophilic leukaemia cells expressed mRNA for MAGL (Figure 1c), and metabolized 2-OG in a time-dependent manner (data not shown). In one series of experiments, URB602 was preincubated with RBL2H3 cells for 10 min at 37°C before addition of a substrate (either 0.25 μM [3H]2-OG or [3H]AEA) and incubation for a further 5 min at 37°C. The rates of 2-OG hydrolysis in the presence of 2, 10, 50 and 100 μM URB602 were 100±3, 93±3, 83±3 and 63±2% of the corresponding controls, respectively (means±s.e.m., n=4). The corresponding values for AEA hydrolysis were 98±3, 82±5, 45±6 and 32±7%, respectively. URB754 showed little effect, concentrations of 50 and 100 μM producing rates of 101±2 and 100±3 (2-OG) and 87±3 and 82±5% (AEA) of control, respectively. In contrast, the selective FAAH inhibitor URB597 almost completely blocked the hydrolysis of AEA by the cells with little or no effect upon the metabolism of 2-OG (data not shown). The effect of 100 μM URB602 was, however, somewhat variable in these cells, as in a subsequent experiment using slightly different conditions, no inhibition of 2-OG hydrolysis was seen at the 100 μM concentration (data not shown).
In C6 cells, URB597 and URB602 had modest effects, and URB754 no effect, upon the metabolism of 2-OG, whereas URB597 potently inhibited the hydrolysis of AEA (Figure 1d). MAFP, on the other hand greatly inhibited 2-OG hydrolysis (Figure 1d), consistent with its effects upon recombinant rat MAGL (Dinh et al., 2002) and upon C6 cells when assayed at room temperature using a phenylsepharose extraction method (Brengdahl and Fowler, 2006). In an additional experiment, a longer (60 min) preincubation time was used in the presence of 1 μM URB597, to minimize any potential contribution by FAAH to the hydrolysis of 2-OG. Under these conditions, the 2-OG metabolism (as % of control) was 73±2, 97±7, 38±5 and 32±6 for 30 μM URB602, 10 μM URB754, 1 and 10 μM MAFP, respectively (means±s.e.m., n=3). In contrast, in the mouse MC3T3-E1 osteoblastic cells, which did not express mRNA for MAGL (Figure 1e), but which were able to metabolize 2-OG in a time-dependent manner that was only slightly inhibited by URB597 (Figure 1f), MAFP was a poor inhibitor (Figure 1g). Although the assay conditions for the MC3T3-E1 and C6 cells were different in terms of incubation times and vehicle concentrations, the data are consistent with the suggestion that the MAFP sensitivity of the 2-OG metabolic activity in the C6 cells correlates with the expression of MAGL. A similar result was seen for the MC3T3-E1 cells when the MAFP was preincubated with the cells for 60, rather than 10 min (data not shown).
Discussion
In the present study, we have investigated the abilities of URB602 and URB754 to inhibit MAGL in view of the diverse potencies of these compounds presented in the literature (Hohmann et al., 2005; Makara et al., 2005; Saario et al., 2006). The main findings of the study concern the selectivity of URB602 and potency of URB754. We found that URB602 inhibited the hydrolysis of 2-OG by brain cytosolic fractions with an IC50 value of 25 μM, entirely in line with the study of Hohmann et al. (2005). A concentration of 30 μM of URB602 also halved the rate of hydrolysis produced by human recombinant MAGL, a result consistent with the data of Makara et al. (2005) for rat recombinant MAGL. However, URB602 had only minor effects, compared to the effects of MAFP, on the hydrolysis of 2-OG by C6 cells that express MAGL. This would suggest that for this cell line, the compound has a limited ability to reach the 2-OG hydrolytic enzymes within the cell. Furthermore, and in contrast to the study of Hohmann et al. (2005), we did not see any selectivity of the compound for FAAH. The finding that URB602 can increase 2-AG but not AEA levels in cultures of rat brain slices (Hohmann et al., 2005) may indicate that the level of inhibition of hydrolysis required to elevate 2-AG levels is lower than for AEA. An alternative explanation is that the modes of inhibition of MAGL and FAAH by the compound are different, and that under certain conditions selectivity can be achieved. This has not been explored, simply because an at best limited degree of selectivity in vitro is unlikely to be seen in vivo.
The data obtained with URB754, showing little or no effect of this compound upon 2-OG hydrolysis by either cytosolic or membrane fractions, recombinant human MAGL, or intact C6 cells, are entirely in line with the study of Saario et al. (2006). These authors demonstrated that the compound, at a concentration of 100 μM, did not affect the hydrolysis of 2-AG by brain homogenates, cerebellar homogenates and cerebellar membranes, and at a concentration of 10 μM did not potentiate the ability of 2-AG to stimulate [35S]GTPγS binding to rat brain slices (Saario et al., 2006). The present data thus underline their suggestion that ‘the effects attributed to URB754 in fact represent activity related to another chemical entity' (Saario et al., 2006). In conclusion, our data indicate that URB602 and URB754 do not behave as selective and/or potent inhibitors of MAGL in vitro, and this places a severe limitation on their usefulness in the elucidation of the role played by this enzyme in the body.
Acknowledgments
We are grateful to Ingrid Persson and Britt Jacobsson for expert technical assistance. The research was supported by grants from the Swedish Research Council (Grant no. 12158, medicine), Konung Gustaf V's and Drottning Victorias Foundation, Gun and Bertil Stohne's Foundation and Stiftelsen för Gamla Tjänarinnor and the Research Funds of the Medical Faculty, Umeå University. SV is grateful to the foundation Wenner-Grenska Samfundet for the post doctoral fellowship, and to the Belgian National Fund for Scientific Research (FNRS) for the current grant of scientific research worker.
Abbreviations
- AEA
arachidonoylethanolamide (anandamide)
- 2-AG
2-arachidonoylgycerol
- BSA
bovine serum albumin
- FAAH
fatty acid amide hydrolase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- MAFP
methyl arachidonoyl fluorophosphonate
- MAGL
monoacylglycerol lipase
- 2-OG
2-oleoylglycerol
- URB597
3′-carbamoyl-biphenyl-3-yl-cyclohexylcarbamate
- URB602
[1,1′-biphenyl]-3-yl-carbamic acid, cyclohexyl ester
- URB754
6-methyl-2-[(4-methylphenyl)amino]-4H-3,1-benzoxazin-4-one
Conflict of interest
The authors state no conflict of interest.
References
- Boldrup L, Wilson SJ, Barbier AJ, Fowler CJ. A simple stopped assay for fatty acid amide hydrolase avoiding the use of a chloroform extraction phase. J Biochem Biophys Meth. 2004;60:171–177. doi: 10.1016/j.jbbm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Brengdahl J, Fowler CJ. A novel assay for monoacylglycerol hydrolysis suitable for high-throughput screening. Anal Biochem. 2006;359:40–44. doi: 10.1016/j.ab.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, et al. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh TP, Kathuria S, Piomelli D. RNA interference suggests a primary role for monoacylglycerol lipase in the degradation of the endocannabinoid 2-arachidonoylglycerol. Mol Pharmacol. 2004;66:1260–1264. doi: 10.1124/mol.104.002071. [DOI] [PubMed] [Google Scholar]
- Goparaju SK, Ueda N, Yamaguchi H, Yamamoto S. Anandamide amidohydrolase reacting with 2-arachidonoylglycerol, another cannabinoid receptor ligand. FEBS Lett. 1998;422:69–73. doi: 10.1016/s0014-5793(97)01603-7. [DOI] [PubMed] [Google Scholar]
- Harrington CR. Lowry protein assay containing sodium dodecyl sulfate in microtiter plates for protein determination on fractions from brain tissue. Anal Biochem. 1990;186:285–287. doi: 10.1016/0003-2697(90)90081-j. [DOI] [PubMed] [Google Scholar]
- Hohmann AG, Suplita RL, Bolton NM, Neely MH, Fegley D, Mangieri R, et al. An endocannabinoid mechanism for stress-induced analgesia. Nature. 2005;435:1108–1112. doi: 10.1038/nature03658. [DOI] [PubMed] [Google Scholar]
- Holt S, Comelli F, Costa B, Fowler CJ. Inhibitors of fatty acid amide hydrolase reduce carrageenan-induced hind paw inflammation in pentobarbital-treated mice: comparison with indomethacin and possible involvement of cannabinoid receptors. Br J Pharmacol. 2005;146:467–476. doi: 10.1038/sj.bjp.0706348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayamanne A, Greenwood R, Mitchell VA, Aslan S, Piomelli D, Vaughan CW. Actions of the FAAH inhibitor URB597 in neuropathic and inflammatory chronic pain models. Br J Pharmacol. 2006;147:281–288. doi: 10.1038/sj.bjp.0706510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson K-O, Vandevoorde S, Lambert DM, Tiger G, Fowler CJ. Effects of homologues and analogues of palmitoylethanolamide upon the inactivation of the endocannabinoid anandamide. Br J Pharmacol. 2001;133:1263–1275. doi: 10.1038/sj.bjp.0704199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria S, Gaetani S, Fegley D, Valiño F, Duranti A, Tontini A, et al. Modulation of anxiety through blockade of anandamide hydrolysis. Nat Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- Makara JK, Mor M, Fegley D, Szabó SI, Kathuria S, Astarita G, et al. Selective inhibition of 2-AG enhances endocannabinoid signaling in hippocampus. Nat Neurosci. 2005;8:1139–1141. doi: 10.1038/nn1521. [DOI] [PubMed] [Google Scholar]
- Maione S, Bisogno T, de Novellis V, Palazzo E, Cristino L, Valenti M, et al. Elevation of endocannabinoid levels in the ventrolateral periaqueductal grey through inhibition of fatty acid amide hydrolase affects descending nociceptive pathways via both cannabinoid receptor type 1 and transient receptor potential vanilloid type-1 receptors. J Pharmacol Exp Ther. 2006;316:969–982. doi: 10.1124/jpet.105.093286. [DOI] [PubMed] [Google Scholar]
- Saario SM, Palomäki V, Lehtonen M, Nevalainen T, Järvinen T, Laitinen JT. URB754 has no effect on the hydrolysis or signalling capacity of 2-AG in the rat brain. Chem Biol. 2006;13:811–814. doi: 10.1016/j.chembiol.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Simpson CMF, Itabe H, Reynolds CN, King WC, Glomset JA. Swiss 3T3 cells preferentially incorporate sn-2-arachidonoyl monoacylglycerol into sn-1-stearoyl-2-arachidonoyl phosphatidylinositol. J Biol Chem. 1991;266:15902–15909. [PubMed] [Google Scholar]
- Tornqvist H, Belfrage P. Purification and some properties of a monoacylglycerol-hydrolysing enzyme of rat adipose tissue. J Biol Chem. 1976;251:813–819. [PubMed] [Google Scholar]
- Vandevoorde S, Saha B, Mahadevan A, Razdan RK, Pertwee RG, Martin BR, et al. Influence of the degree of unsaturation of the acyl side chain upon the interaction of analogues of 1-arachidonoylglycerol with monoacylglycerol lipase and fatty acid amide hydrolase. Biochem Biophys Res Commun. 2005;337:104–109. doi: 10.1016/j.bbrc.2005.09.015. [DOI] [PubMed] [Google Scholar]
- van Tienhoven M, Atkins J, Li Y, Glynn P. Human neuropathy target esterase catalyzes hydrolysis of membrane lipids. J Biol Chem. 2002;277:20942–20948. doi: 10.1074/jbc.M200330200. [DOI] [PubMed] [Google Scholar]
- Waggoner DW, Johnson LB, Mann PC, Morris V, Guastella J, Bajjalieh SM. MuLK, a eukaryotic multi-substrate lipid kinase. J Biol Chem. 2004;279:38228–38235. doi: 10.1074/jbc.M405932200. [DOI] [PubMed] [Google Scholar]