Abstract
The background selection hypothesis predicts a reduction in nucleotide site diversity and an excess of rare variants, owing to linkage associations with deleterious alleles. This effect is expected to be amplified in species that are predominantly self-fertilizing. To examine the predictions of the background selection hypothesis in self-fertilizing species, we sequenced 1,362 bp of adh1, a gene for alcohol dehydrogenase (Adh; alcohol:NAD+ oxidoreductase, EC 1.1.1.1), in a sample of 45 accessions of wild barley, Hordeum vulgare ssp. spontaneum, drawn from throughout the species range. The region sequenced included 786 bp of exon sequence (part of exon 4, all of exons 5–9, and part of exon 10) and 576 bp of intron sequence (all of introns 4–9). There were 19 sites polymorphic for nucleotide substitutions, 8 in introns, and 11 in exons. Of the 11 nucleotide substitutions in codons, 4 were synonymous and 7 were nonsynonymous, occurring uniquely in the sample. There was no evidence of recombination in the region studied, and the estimated effective population size (N̂e) based on synonymous sites was ≈1.8–4.2 × 105. Several tests reveal that the pattern of nonsynonymous substitutions departs significantly from neutral expectations. However, the data do not appear to be consistent with recovery from a population bottleneck, recent population expansion, selective sweep, or strong positive selection. Though several features of the data are consistent with background selection, the distributions of polymorphic synonymous and intron sites are not perturbed toward a significant excess of rare alleles as would be predicted by background selection.
The fundamental goal of population genetics is a quantitative assessment of the various forces of evolution in shaping patterns of genetic diversity. There are five forces to quantify: mutation, selection, genetic random drift, migration, and recombination. The forces of selection, genetic random drift, and migration have been notoriously difficult to quantify because they interact in complex ways. The estimation of gene genealogies (gene trees) from DNA sequence samples drawn from species or populations provides a powerful approach to this classical problem. The reasons that gene trees are potentially powerful are (i) they estimate patterns of identity-by-descent among alleles thus bringing the objects of observation into a much closer correspondence with population genetic theory, and (ii) the coalescence framework integrates over a period of time of the order of the effective population size thus greatly enhancing the power to detect selection (reviewed in ref. 1).
The primary statistic describing a sample of DNA sequences is θ, which is equal to 4Neμ (where Ne is the effective population size and μ is the mutation rate per generation), assuming that the sample has been generated by a strict drift-mutation process at equilibrium (neutral process). A number of statistical tests have been introduced to test departures of θ from the neutral null hypothesis. These tests are based on contrasts between pairs of different estimators for θ, θ̂. Thus for example, the test of Tajima (2) examines the difference between θ̂ based on the number of segregating sites versus the average number of pairwise differences taken over the sequence sample. Analogous tests based on various other contrasts have been introduced (3–5). Simonsen et al. (6) conducted a number of simulations to assess the power of these test statistics under well-defined alternative hypotheses that included (i) a selective sweep, (ii) a population bottleneck, and (iii) population subdivision. They found that the power to reject the null hypothesis of neutrality is limited for samples of fewer than 50 sequences.
In an important paper, Charlesworth et al. (7) considered the role of linkage and the flux of deleterious mutations on the shape of the distribution of neutral nucleotide site diversity. This simulation study was motivated by the observation that regions of reduced recombination in Drosophila tend to also be regions of low polymorphism (8). One potential explanation for this observation is that a selective sweep will influence nucleotide site diversity at distances proportional to s/r [where s is the selective intensity and r is the recombination fraction (9)]. Charlesworth et al. (7) showed that selection against a flux of deleterious mutations (background selection) could also lead to reductions in θ in regions of low recombination. In addition, Charlesworth et al. (7) considered the interaction of self-fertilization with background selection and concluded that neutral nucleotide site diversity is expected to be reduced in species with high selfing rates (>80%).
Empirical studies of gene genealogies in plant species are still in their infancy. Most of the published work has concentrated on genes coding for a single glycolytic enzyme, alcohol dehydrogenase (Adh; alcohol:NAD+ oxidoreductase, EC 1.1.1.1). Alcohol dehydrogenase is an important enzyme in anaerobic metabolism and it is usually coded by a small multigene family in flowering plants. The adh1 locus of maize has 9 introns and 10 exons and comprises about 3.4 kb of DNA sequence when upstream anaerobic regulatory signals are included (10). In grasses there are two or three Adh loci. adh1 and adh2 originated by duplication prior to the origin of the grass family (11). A third Adh locus is observed as a subsequent duplication in some grass lineages [e.g., Hordeum, where adh3 is a relatively recent duplicate of adh2 (12)].
DNA sequence diversity has been studied in a very small number of plant species to date. These studies have concentrated on the agronomic crops maize [Zea mays (13, 14)] and pearl millet [Pennisetum glaucum (15)]. Studies of Adh sequence diversity have also been published for Arabidopsis thaliana (16, 17) [where only a single Adh locus has been reported (18)], Arabis gemmifera (19), and Dioscorea tokoro (20). Sample sizes have usually been small, and some studies have sample sizes below 10, so it is not surprising that the null hypothesis of neutrality is usually accepted.
The current investigation was initiated to (i) examine the background selection hypothesis by studying a predominantly self-fertilizing grass species that could be contrasted to the two outcrossing (wind pollinated) grass species already investigated, and (ii) increase statistical power by sequencing a large sample of adh1 genes drawn from throughout the species range. The species selected for study is Hordeum vulgare ssp. spontaneum, or wild barley, a diploid species with seven chromosomes. Wild barley is the progenitor of cultivated barley, H. vulgare ssp. vulgare, and is distributed in the Fertile Crescent and more broadly through southwestern Asia. Wild barley is a predominantly self-pollinated species with an average outcrossing rate of approximately 1.6%, with individual population estimates rarely exceeding 5% (21); thus wild barley conforms to the conditions favoring background selection. Previous studies of isozyme variation show relatively high levels of polymorphism in wild barley (21), and studies of chloroplast DNA (cpDNA) variation (22–24) also reveal relatively high levels of cpDNA diversity compared with most plant species studied (reviewed in ref. 25).
We report total estimates of θ for wild barley that are very similar to those obtained for the outcrossing grass species Pennisetum glaucum. However, when the estimates are partitioned into nonsynonymous changes versus synonymous or intron site differences, a remarkable pattern emerges. The null hypothesis of neutrality is rejected for all tests based on nonsynonymous differences, owing to a large excess of unique polymorphisms in this class of sites. In contrast, the synonymous and intron site differences do not reject the null hypothesis. This pattern appears to be consistent with very weak selection against most amino acid changes, but the effect of background selection is not detectable at synonymous or intron sites.
MATERIALS AND METHODS
Plant Materials.
Seeds of wild barley, H. vulgare ssp. spontaneum, were obtained from the U.S. Department of Agriculture, Agricultural Research Service, National Small Grains Collection (Aberdeen, ID). The materials were drawn from throughout the natural geographic range of the species (Table 1). Seeds were germinated and greenhouse grown in 1 gallon (US) plastic pots filled with a nutrient supplemented sand and peat moss mixture. Leaf material from individual plants was harvested and DNA was prepared by using a standard protocol (26).
Table 1.
Samples of H. vulgare ssp. spontaneum used in this study
| PI number | Country of origin | Haplotype group† | PI number* | Country of origin | Haplotype group† |
|---|---|---|---|---|---|
| 211041 | Afghanistan | I | 401370 | Iran | III |
| 212305 | Afghanistan | II | 401371 | Iran | |
| 212306 | Afghanistan | 406275 | Israel | ||
| 219796 | Iraq | 406276 | Israel | III | |
| 220341 | Afghanistan | III | 420911 | Jordan | |
| 220523 | Afghanistan | 420912 | Jordan | ||
| 227019 | Iran | 420913 | Jordan | I | |
| 236386 | Syria | IV | 420915 | Jordan | II |
| 236388 | Syria | II | 420916 | Jordan | |
| 253933 | Iraq | III | 420917 | Jordan | |
| 254894 | Iraq | 466381 | Israel | II | |
| 268242 | Iran | 466460 | Israel | V | |
| 293402 | Turkmenistan | II | 531851 | Israel | III |
| 293405 | Turkmenistan | II | 531852 | Israel | I |
| 293408 | Turkmenistan | 531853 | Israel | ||
| 293409 | Turkmenistan | II | 531857 | Israel | V |
| 293411 | Tajikistan | III | 554425 | Turkey | I |
| 293412 | Tajikistan | III | 554426 | Turkey | I |
| 293413 | Azerbaijan | II | 554428 | Turkey | I |
| 293414 | Azerbaijan | II | 559556 | Turkey | II |
| 296926 | Israel | V | 560559 | Turkey | IV |
| 366446 | Afghanistan | IV | 560560 | Turkey | IV |
| 401367 | Iran | III |
National Plant Germplasm System Plant Introduction Number.
Denotes identical haplotypes with mroe than one representative for gene region sequenced.
PCR and Sequencing.
Templates for DNA sequencing were generated by using a two-step nested primer amplification procedure and standard PCR mixes (27). An initial amplification was conducted by using primers corresponding to sequence in exon 2 (F2, 5′-TACTTCTGGGAGGCCAAGG-3′) and 3′ noncoding region (3P4R, 5′-GCGAAACCGCAGACGAT-3′). The portion of this initial amplification product was used as the template for the second amplification by using primers nested within the first primers and corresponding to sequence in exon 4 (Adh322F, 5′-AGTGGAGAGTGTTGGAGAGGGCG-3′) and 3′ noncoding region internal to 3P4R (3P3R, 5′-GCCATCAGAAGCACTTG-3′). The PCR conditions for both amplification steps consisted of initial denaturation of 2 min at 95°C followed by 40 cycles of 1 min at 94°C, 2 min at 42°C, and 4 min at 72°C. This amplification procedure is specific for adh1.
Sequencing templates were purified by using a protocol (27) modified by the addition of extra chloroform extractions after PEG precipitation. Dideoxy cycle sequencing was done on an Applied Biosystems model 373A sequencer with dye-labeled terminators and Li-Cor (Lincoln, NE) 4000L and 4200L sequencers with end-labeled internal primers. All sequences were determined on both strands, with a minimum of 2-fold coverage and with 4- to 6-fold coverage for most regions. Sequences of H. vulgare ssp. spontaneum are identified throughout this paper by their associated U.S. Department of Agriculture accession numbers.
Sequence Data Analyses.
For comparative purposes, sequences of Adh from other species were obtained from GenBank, including Pennisetum glaucum (15), Arabidopsis thaliana (17–19), Arabis gemmifera (19), and several species of Zea (13). Analyses of these sequences were restricted to a common homologous region across all species.
Sequences were aligned with the program clustal w (28). The program sites (29) was used for polymorphism analysis and coalescent-based estimation of recombination rates where applicable. In those cases where there were no incompatible sites apparent recombination rate was zero. This finding was confirmed by parsimony analysis of polymorphic sites and observing a consistency index of zero for the best tree. Parsimony analyses were done by using paup (30). Tests of neutrality and determination of their associated significance were done by using the programs of Fu (5).
RESULTS
Nucleotide Sequence Polymorphism.
DNA sequence was determined for all 45 samples (Table 1). The region sequenced was 1,362 bp in length, including 786 bp of exon sequence (part of exon 4, all of exons 5–9, and part of exon 10) and 576 bp of intron sequence (all of introns 4–9). There were 19 sites polymorphic for nucleotide substitutions, 11 in exons and 8 in introns (Table 2). There is little apparent bias toward transitions, as there are 10 transitions and 9 transversions observed. Of the 11 nucleotide substitutions in codons, 4 were synonymous and 7 were nonsynonymous. The substitutions in introns occurred in a range of frequencies in the sample of 45 sequences: 1 polymorphism in 21/45 sequences, 1 polymorphism in 9/45 sequences, 2 polymorphisms in 3/45 sequences, 1 polymorphism in 2/45 sequences, and 3 polymorphisms were unique in the sample. The synonymous substitutions exhibit a similar range of frequencies: 1 polymorphism in 14/45 sequences, 1 polymorphism in 9/45 sequences, and 2 polymorphisms were unique in the sample. In contrast, all 7 nonsynonymous substitutions were unique in the sample.
Table 2.
Sequence of polymorphisms in sample of adh1 from H. vulgare ssp. spontaneum
| Position | Change* | Base(s) | Amino acid | Class | Base(s) | Amino acid | Class | |
|---|---|---|---|---|---|---|---|---|
| Exon Regions | ||||||||
| a | 60 | S T | C | Ile | Nonpolar | T | Ile | Nonpolar |
| c | 347 | N R | A | Lys | Bassic | C | Gln | Polar |
| k | 1,032 | N R | G | Ala | Nonpolar | T | Ser | Polar |
| l | 1,061 | S T | C | Phe | Aromatic | T | Phe | Aromatic |
| m | 1,075 | N R | C | Thr | Polar | A | Asn | Polar |
| n | 1,083 | N R | G | Gly | Nonpolar | C | Arg | Basic |
| o | 1,151 | S T | G | Lys | Basic | A | Lys | Basic |
| t | 1,325 | N T | C | Ser | Polar | T | Leu | Nonpolar |
| u | 1,343 | N T | T | Phe | Aromatic | C | Ser | Polar |
| v | 1,350 | S T | C | Leu | Nonpolar | T | Leu | Nonpolar |
| w | 1,355 | N R | C | Ala | Nonpolar | A | Glu | Acidic |
| Intron Regions | ||||||||
| b | 282 | Indel | G | Gap | ||||
| d | 412 | T | T | C | ||||
| e | 417 | R | T | A | ||||
| f | 607 | R | T | A | ||||
| g | 614 | R | G | T | ||||
| h | 615 | Indel | Gap | T | ||||
| i | 631 | T | T | C | ||||
| j | 738 | R | A | T | ||||
| p | 1,188 | Indel | Gap | A | ||||
| q | 1,204–1,214 | Indel | GGGAGCCCACAC | Gap | ||||
| r | 1,247 | T | A | G | ||||
| s | 1,279 | T | C | T | ||||
S, synonymous; N, nonsynonymous; T, transition; R, transversion; indel, insertion or deletion.
Change relative to consensus sequence.
All but one of the nonsynonymous substitutions were nonconservative with regard to simple biochemical classification of amino acids (i.e., polar, nonpolar, acidic, basic, aromatic, and cysteine). A similar pattern of bias toward nonconservative substitutions is seen for Arabis gemmifera. Comparison to ADH amino acid sequences from a number of plants (31) shows some amino acid polymorphisms in H. vulgare ssp. spontaneum are at sites that vary across a wide range of angiosperms, but several are at sites conserved across other plant taxa.
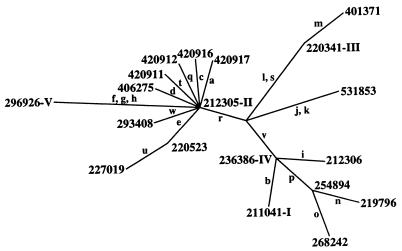
The four distinct insertion/deletion events were restricted to the introns, and these events include three involving 1 bp and one involving 11 bp. The nucleotide substitutions and insertion/deletion events defined 19 haplotypes (Fig. 1 and Table 1). No more than seven mutations separate any two haplotypes.
Figure 1.
Minimum spanning diagram showing adh1 haplotype relationships in wild barley (H. vulgare ssp. spontaneum). Superscript numerals denote haplotype group with more than one representative (Table 1). Letters adjacent branches represent specific polymorphisms (Table 2).
Intraspecific Sequence Diversity Tests of Neutrality.
A number of tests have been developed to determine significant departures from neutral evolution of sequence data. Several of the tests are based on the difference between independent estimates of nucleotide polymorphism, θ̂, which is equal to 4Neμ, where Ne is the effective population size and μ is the mutation rate per generation. The test statistic of Tajima (2), represented by T here, is the difference between θ̂ based on the number of segregating sites in a sample (Sn) and θ̂ based on the average number of pairwise differences between sequences (π). The test statistics of Fu and Li (3) are D*, the difference between θ̂ based on Sn; θ̂, based on the number of singletons (external mutations) in a sample (ηs); F*, the difference between θ̂ based on π; and θ̂ based on ηs. The expected difference between all these estimates of θ is zero for a neutral drift-mutation process at equilibrium. Another test statistic is that of Fu (4), Fs, which is based on the probability of k alleles in a sample conditioned on a given value of θ̂ based on π.
We have applied these tests to the wild barley adh1 data and determined their associated significance levels (Table 3) by using the program of Fu (5). Partitioning the data into groups (e.g., exon, intron, synonymous, nonsynonymous) allows for comparison of the evolutionary dynamics between groups. For the exon sequence data of H. vulgare ssp. spontaneum all test statistic values are significant, and none of the test statistic values are significant for the intron sequence data. Further partitioning the data into synonymous and nonsynonymous sites shows significant test statistic values for nonsynonymous sites but not for synonymous sites. Taken together, it appears that the pattern of polymorphism at nonsynonymous sites is responsible for the significant test statistic values at higher levels of site classification.
Table 3.
Estimates of nucleotide diversity, θ̂, and test statistics across a common homologous region of Adh
| Region | Length, bp | Haplotypes | θ̂/bp based on
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ηs | Sn | π | ηs | T† | D* | F* | Fs | |||
| Hordeum vulgare ssp. spontaneum (n = 45) | ||||||||||
| Overall | 1,362 | 19 | 12 | 0.00320 | 0.00182 | 0.00861 | −1.319 | −2.715‡ | −2.490‡ | −13.118§ |
| Introns | 576 | 8 | 3 | 0.00319 | 0.00229 | 0.00509 | −0.746 | −0.761 | −0.836 | −2.100 |
| Exons | 786 | 12 | 9 | 0.00320 | 0.00148 | 0.01120 | −1.510‡ | −3.473§ | −3.131§ | −7.584§ |
| Synonymous | 179.67 | 5 | 2 | 0.00509 | 0.00473 | 0.01088 | −0.144 | −1.116 | −0.897 | −0.640 |
| Nonsynonymous | 606.33 | 8 | 7 | 0.00264 | 0.00051 | 0.01129 | −2.056 | −3.950§ | −3.694§ | −9.149§ |
| Zea spp. (n = 8) | ||||||||||
| Overall | 1586 | 8 | 33 | 0.01715 | 0.01742 | 0.01821 | −0.077 | −0.104 | −0.099 | −0.488 |
| Introns | 804 | 8 | 19 | 0.02015 | 0.02092 | 0.02068 | −0.070 | −0.039 | −0.047 | −1.286 |
| Exons | 782 | 8 | 14 | 0.01381 | 0.01357 | 0.01566 | −0.084 | −0.195 | −0.171 | −2.052 |
| Synonymous | 178.33 | 8 | 9 | 0.04758 | 0.04929 | 0.04416 | 0.166 | 0.103 | 0.119 | −2.457 |
| Nonsynonymous | 601.67 | 5 | 5 | 0.00385 | 0.00302 | 0.00727 | −0.877 | −1.112 | −1.072 | −1.359 |
| Pennisetum glaucum (n = 21) | ||||||||||
| Overall | 1,359 | 13 | 7 | 0.00311 | 0.00204 | 0.00491 | −1.173 | −0.858 | −1.016 | −6.982§ |
| Introns | 573 | 9 | 5 | 0.00450 | 0.00243 | 0.00831 | −1.450‡ | −1.171 | −1.335 | −4.941§ |
| Exons | 786 | 6 | 2 | 0.00212 | 0.00176 | 0.00242 | −0.500 | −0.166 | −0.279 | −1.227 |
| Synonymous | 180.06 | 5 | 2 | 0.00772 | 0.00666 | 0.01058 | −0.381 | −0.412 | −0.428 | −0.585 |
| Nonsynonymous | 605.94 | 2 | 0 | 0.00046 | 0.00030 | 0.00000 | −0.563 | 0.603 | 0.302 | −0.137 |
| Arabidopsis thaliana (n = 17) | ||||||||||
| Overall | 1,038 | 9 | 7 | 0.00630 | 0.00685 | 0.00635 | 0.323 | −0.018 | 0.083 | 0.443 |
| Introns | 252 | 7 | 2 | 0.00958 | 0.00958 | 0.00747 | 0.002 | 0.258 | 0.196 | −1.061 |
| Exons | 786 | 8 | 5 | 0.00527 | 0.00599 | 0.00599 | 0.483 | −0.190 | 0.000 | 0.063 |
| Synonymous | 172.67 | 8 | 3 | 0.02056 | 0.02589 | 0.01635 | 0.902 | 0.279 | 0.479 | −0.091 |
| Nonsynonymous | 613.33 | 3 | 2 | 0.00097 | 0.00039 | 0.00307 | −1.366 | −1.813 | −1.775 | −1.680 |
| Arabis gemmifera (n = 8) | ||||||||||
| Overall | 1,036 | 7 | 21 | 0.00786 | 0.00509 | 0.01774 | −1.646§ | −1.812§ | −1.799§ | −1.794 |
| Introns | 250 | 4 | 5 | 0.00787 | 0.00510 | 0.01450 | −1.406‡ | −1.534‡ | −1.525‡ | −0.785 |
| Exons | 786 | 7 | 16 | 0.00785 | 0.00509 | 0.01781 | −1.617§ | −1.778§ | −1.765‡ | −2.459 |
| Synonymous | 173.58 | 3 | 2 | 0.00444 | 0.00288 | 0.01008 | −1.135 | −1.230 | −1.223 | −0.999 |
| Nonsynonymous | 612.42 | 7 | 14 | 0.00882 | 0.00572 | 0.02000 | −1.600‡ | −1.758‡ | −1.746‡ | −2.813 |
Estimates of Effective Population Size.
If a specific mutation rate, μ̂, is assumed, then estimates of θ can be used to estimate Ne, the effective population size, by using the equation N̂e = θ̂/4μ̂. We have estimated Ne by using a mean synonymous substitution rate estimate for Adh in the grass family of 6.5 × 10−9 (11). Estimated effective population size for H. vulgare ssp. spontaneum based on synonymous sites is moderately less than that estimated for Pennisetum glaucum and fairly similar to Arabidopsis thaliana and Arabis gemmifera (Table 4). The estimated effective population size for Zea spp. is an order of magnitude larger than the other species. All species display relatively large estimated effective population sizes (>105), despite a history of strong selection for domestication for a few of the species.
Table 4.
Estimates of effective population size (Ne) using synonymous sites from a common homologous region of Adh
| Taxon |
Ne using θ̂ based on
|
||
|---|---|---|---|
| Sn | π | ηs | |
| H. vulgare ssp. spontaneum | 1.958 × 105 | 1.819 × 105 | 4.185 × 105 |
| Zea spp. | 1.830 × 106 | 1.896 × 106 | 1.698 × 106 |
| Pennisetum glaucum | 2.969 × 105 | 2.562 × 105 | 4.069 × 105 |
| Arabidopsis thaliana | 7.908 × 105 | 9.958 × 105 | 6.288 × 105 |
| Arabis gemmifera | 1.708 × 105 | 1.108 × 105 | 3.877 × 105 |
Estimates of Recombination.
One motivation for examining adh1 evolution in H. vulgare ssp. spontaneum is because it is a predominantly self-fertilizing plant, in contrast to many other plants that have been examined, which are predominantly outcrossing. One implication of selfing is that the apparent recombination rate should be greatly reduced compared with outcrossing plants. Although recombination may occur, the resulting recombinant products are usually identical to the prerecombination states because of the greatly decreased heterozygosity associated with selfing. In keeping with this expectation, the estimate of 4NeC/bp (where C is recombination rate) for H. vulgare ssp. spontaneum is zero (Table 5). Both Pennisetum and Arabis also show an absence of apparent recombination in Adh. In contrast, Arabidopsis shows evidence of moderate recombination as previously noted (17), and Zea shows evidence for more recombination (ref. 13; Table 5).
Table 5.
Estimates of recombination across a common homologous region of Adh
| Taxon | Estimated γ/bp | Minimum number of intervals |
|---|---|---|
| H. vulgare ssp. spontaneum | 0.000000* | 0 |
| Zea ssp. | 0.026069 | 6 |
| Pennisetum glaucum | 0.000000* | 0 |
| Arabidopsis thaliana | 0.005037 | 2 |
| Arabis gemmifera | 0.000000* | 0 |
No incompatible sites, consistency index = 100.
DISCUSSION
The central result of this investigation is the fact that the barley data reject the null hypothesis of a pure drift mutation process based on all available statistical tests. When the data are partitioned into nonsynonymous versus synonymous and intron sites, the departures from the null hypothesis are accounted for by an excess of singleton amino acid replacements. In contrast, the distribution of synonymous and intron polymorphisms conform with neutral expectations. What processes are most likely to account for this pattern?
Recovery from a population bottleneck or a recent demographic expansion are both expected to lead to a transient excess of rare variants, but this would be true for all sites (synonymous and nonsynonymous) and consequently this explanation does not appear consistent with the observed data. Similarly, a selective sweep is not consistent with the distribution of synonymous and intron polymorphism because the recovery from a selective sweep would appear indistinguishable from recovery from a bottleneck for the region associated with a single locus. These two hypotheses are only distinguishable when estimates of θ based on genes from different regions of the genome are compared. This result follows from the fact that a bottleneck would affect all loci, whereas the impact of a selective sweep would be confined to a region associated with the locus that had been subject to positive selection.
The hypothesis that deserves serious consideration is the background selection hypothesis. It is instructive to briefly review the salient features of the background selection hypothesis. The parameters chosen for the simulations of Charlesworth et al. (7) were based on the best estimates of whole chromosome selection and mutation rates in Drosophila. The simulations reveal that the distribution of neutral sites can be skewed toward an excess of rare alleles owing to their linkage association with negatively selected mutations. This effect is dependent on suitably low values of recombination. The background selection effect was also shown to be exacerbated when the frequency of self fertilization exceeded ≈75%. How do these predictions conform to the observed barley data?
Several features of the data appear to be consistent with the background selection hypothesis. (i) Background selection is expected to be amplified in predominantly self-fertilizing species (7). (ii) There is no evidence for recombination within the barley adh1 sequences (consistent with a high frequency of self-fertilization). (iii) The amino acid replacements in the sample are all unique, as would be expected with weak negative selection. However, four additional features of the data need to be considered in concluding that selection against deleterious mutations is the primary determinant of the observed distribution of nonsynonymous changes: (i) there are nearly twice as many nonsynonymous polymorphisms as synonymous polymorphisms in the sample (seven versus four); (ii) there appears to be no restriction on the kinds of amino acid changes accepted (polar, nonpolar, acidic, basic, etc.); (iii) the frequency of deleterious genes in highly homozygous species should be very low [approximately μ/s, where s is the selection coefficient (32)]; and (iv) the estimates of effective population size are relatively large (approximately 105), so very small selective values should be effective (s ≈ 10/Ne ≈ 10−4). These observations point to very weak negative selection on the amino acid replacements at nonsynonymous sites. The mere fact that 7 nonsynonymous changes were observed in a sample of size 45 implies either very weak selection or some force favoring rare variants. (A force favoring rare variants seems to us unlikely in view of the promiscuous acceptance of amino acid replacements.)
Why is the distribution of synonymous and intron sites not perturbed toward a significant excess of rare alleles as would be predicted by the background selection hypothesis? In addressing this question, it is important to note that the observed data pertain to a very limited region of the chromosome (1,362 bp) rather than to whole chromosomes as was the case for the simulations of background selection (7). One conclusion that is consistent with the data is that there may be a background effect, because T is negative for synonymous and intron sites as predicted, but it is too weak to lead to a significant perturbation in the distribution. This result would appear to suggest rather weak selection at the whole chromosome level. A second possible explanation arises from the consideration that different chromosomal regions may be affected by different patterns of selection. Because of the very reduced levels of recombination in self-fertilizing species, the observed data represent an integration of the selective forces that affect loci over most of the chromosome. As a consequence, the factors that determine sequence diversity at adh1 in barley may be well outside the window of observation. Put differently, the dynamic at adh1 may be influenced by selection at other loci on the same chromosome, but at a considerable distance from adh1. This explanation would also help to account for the surprisingly large number of the amino acid replacements detected in the sample, because selection at the adh1 locus may be moderated by selection operating at other chromosomal loci, but in opposing directions.
A particularly intriguing aspect of Table 3 is the similarity in test statistics between barley and Arabis gemmifera. Both of these self-fertilizing species exhibit a significant departure from the neutral null hypothesis owing to an excess of singleton amino acid replacement polymorphisms, and both show no significant departure from the null hypothesis for nonsynonymous sites. In addition, Arabidopsis thaliana, with evidence of only moderate outcrossing (compared with maize), shows negative test statistics (although nonsignificant), suggesting an excess of rare variants. Taken together these data appear to provide convincing support for the background selection hypothesis.
Comparisons between barley adh1 diversity and that of other plant species reveals moderately low values of θ̂, but not dramatically below that of most other plants so far investigated. Thus the effect of background selection and self-fertilization on reducing Ne and hence θ are not dramatic (33). Taken in toto, the collection of studies of adh1 sequence diversity in plants provides little support for positive selection maintaining nucleotide sequence diversity in outcrossing or in self-fertilizing species. It will be important for future studies of plant nucleotide sequence diversity to focus on a wider set of other genetic loci to ask whether the processes affecting different loci are heterogeneous within genomes and lineages. It is only through careful empirical comparisons of diverse sets of different loci within genomes that the relative importance of selection versus drift and mutation can be assessed.
Acknowledgments
We thank A. H. D. Brown and B. S. Gaut for comments on an earlier version of the manuscript, M. Debacon and M. L. Durbin for technical assistance, and M.P.C. thanks G. A. Gilbert and M. C. Neel for encouragement. The work reported in this paper was supported in part by the Alfred P. Sloan Foundation.
ABBREVIATION
- Adh
alcohol dehydrogenase gene
Footnotes
References
- 1.Clegg M T. J Hered. 1997;88:1–7. doi: 10.1093/oxfordjournals.jhered.a023048. [DOI] [PubMed] [Google Scholar]
- 2.Tajima F. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu Y-X, Li W-H. Genetics. 1993;133:693–709. doi: 10.1093/genetics/133.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y-X. Genetics. 1996;143:557–570. doi: 10.1093/genetics/143.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fu Y-X. Genetics. 1997;147:915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simonsen K L, Churchill G A, Aquadro C F. Genetics. 1995;141:413–429. doi: 10.1093/genetics/141.1.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth B, Morgan M T, Charlesworth D. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Begun D J, Aquadro C F. Nature (London) 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 9.Hudson R R. Proc Natl Acad Sci USA. 1994;91:6815–6818. doi: 10.1073/pnas.91.15.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachs M M, Dennis E S, Gerlach W L, Peacock W J. Genetics. 1986;113:449–467. doi: 10.1093/genetics/113.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaut B S, Morton B R, McCaig B C, Clegg M T. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trick M, Dennis E S, Edwards K J R, Peacock W J. Plant Mol Biol. 1988;11:147–160. doi: 10.1007/BF00015667. [DOI] [PubMed] [Google Scholar]
- 13.Gaut B S, Clegg M T. Proc Natl Acad Sci USA. 1993;90:5095–5099. doi: 10.1073/pnas.90.11.5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanson M A, Gaut B S, Stec A O, Furstenberg S I, Goodman M M, Coe E H, Doebley J F. Genetics. 1996;143:1395–1407. doi: 10.1093/genetics/143.3.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaut B S, Clegg M T. Genetics. 1993;135:1091–1097. doi: 10.1093/genetics/135.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanfstingl U, Berry A, Kellogg E A, Costa J T, III, Rüdiger W, Ausubel F M. Genetics. 1994;138:811–828. doi: 10.1093/genetics/138.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Innan H, Tajima F, Terauchi R, Miyashita N T. Genetics. 1996;143:1761–1770. doi: 10.1093/genetics/143.4.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang C, Meyerowitz E M. Proc Natl Acad Sci USA. 1986;83:1408–1412. doi: 10.1073/pnas.83.5.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyashita N T, Innan H, Terauchi R. Mol Biol Evol. 1996;13:433–436. doi: 10.1093/oxfordjournals.molbev.a025603. [DOI] [PubMed] [Google Scholar]
- 20.Terauchi R, Terachi T, Miyashita N T. Genetics. 1997;147:1899–1914. doi: 10.1093/genetics/147.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown A H D, Zohary D, Nevo E. Heredity. 1978;41:49–62. [Google Scholar]
- 22.Clegg M T, Brown A H D, Whitfield P R. Genet Res. 1984;43:339–343. [Google Scholar]
- 23.Holwerda B C, Jana S, Crosby W L. Genetics. 1986;114:1271–1291. doi: 10.1093/genetics/114.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neale D B, Saghai-Maroof M A, Allard R W, Zang Q, Jorgensen R A. Genetics. 1988;120:1105–1110. doi: 10.1093/genetics/120.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clegg M T, Learn G H, Golenberg E M. In: Evolution at the Molecular Level. Selander R K, Clark A G, Whittam T S, editors. Sunderland, MA: Sinauer Associates; 1991. pp. 135–149. [Google Scholar]
- 26.Murray M G, Thompson W F. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley Interscience; 1997. [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hey J, Wakeley J. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swofford D L. Phylogenetic Analysis Using Parsimony, paup Portable version (UNIX) Champaign: Illinois Natural History Survey; 1992. , Version 3.0r+4 (Prerelease 0.4). [Google Scholar]
- 31.Clegg M T, Cummings M P, Durbin M L. Proc Natl Acad Sci USA. 1997;94:7791–7798. doi: 10.1073/pnas.94.15.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crow J, Kimura M. An Introduction to Population Genetics Theory. Edina, MN: Alpha Editions; 1970. [Google Scholar]
- 33.Nordborg M. Genetics. 1997;146:1501–1514. doi: 10.1093/genetics/146.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]