Abstract
A clinical trial of uncomplicated skin and skin structure infections (39 locations in 19 states) observed that community-associated or community-onset methicillin-resistant Staphylococcus aureus (CO-MRSA) represented 23% of all pathogens at baseline culture and 53% of 190 S. aureus isolates. CO-MRSA strains typically were Panton-Valentine leukocidin (PVL) positive (95%), contained staphylococcal cassette chromosome mec type IVa (99%), were USA300 or USA400 clones (92%), and exhibited minimal coresistances (macrolides and/or fluoroquinolones). Clinical results remained identical (89% cures) regardless of the antimicrobial used or CO-MRSA molecular patterns, PVL production, or antimicrobial susceptibility profiles.
The treatment of uncomplicated skin and skin structure infections (uSSSI) consumes a significant proportion of national health care resources, as recently quantitated by McCaig et al. (10) using statistics from national ambulatory medical care and national hospital ambulatory care surveys. When data derived from physician offices and emergency departments from 2001 to 2003 and 1992 to 1994 were compared, the number of ambulatory care visits was 11.6 million in 2001 to 2003. From 1992 to 1994 to 2001 to 2003, rates increased 59% and 31% in the outpatient and emergency departments, respectively (10). The increase was attributed to the emergence of community-associated or community-onset methicillin-resistant Staphylococcus aureus (CO-MRSA) infections. Since the recognition of CO-MRSA in the 1990s (2, 20), the understanding of the dominant strains/clones has evolved through reports of dramatic epidemic clusters, some with fatal consequences (1), as well as by thoughtfully performed epidemiologic or molecular investigations (5, 8, 11, 12, 19). CO-MRSA has numerous characteristics that differentiate it from hospital-acquired MRSA, including (i) younger affected patient population (5, 12), (ii) methicillin resistance produced via staphylococcal cassette chromosome mec (SCCmec) type IVa (2, 11-13, 18, 20), (iii) high presence of Panton-Valentine leukocidin (PVL) (2, 9, 11, 20), and (iv) the presence of dominant epidemic clones, classified as USA300 or USA400 (18). The serious consequence of these uSSSI cases has been progression to complicated SSSI requiring hospitalization (4 to 23%) (5, 10), including potentially fatal necrotizing fasciitis or pneumonia (1). These features and concerns regarding limited treatment options among orally administered antimicrobial agents have led to adjusted treatment guidelines and suggested therapeutic paradigm shifts (7, 16).
To update our knowledge of CO-MRSA and facilitate an understanding of clinical outcomes using contemporary treatment regimens, S. aureus isolates derived from a recent phase IV, prospective, investigator-blinded, randomized uSSSI clinical trial (6) were examined to determine the impact of antibiogram patterns and pathogen (S. aureus) molecular characteristics. These organisms were obtained from patients presenting in outpatient clinical practices or emergency departments with community-onset infections (39 locations in 19 states).
(The data summarized in this paper were presented at the 46th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, 27 to 30 September 2006 [7a].)
A total of 190 S. aureus isolates were available from the multicenter uSSSI study (6). The isolates comprised 171 baseline isolates, 151 of which were from clinically and bacteriologically evaluable patients and 19 of which were posttreatment isolates; 2 baseline isolates were not available for testing. These pathogens were studied by molecular methods to characterize SCCmec type, the presence of PVL, agr type, and pulsed-field gel electrophoresis (PFGE) patterns. CO-MRSA isolates (101 strains [53%]) were detected by the reference broth microdilution method (3) using an oxacillin breakpoint of ≥4 μg/ml as resistant (4). Other comparison agents (erythromycin, ciprofloxacin, penicillin, ampicillin, piperacillin-tazobactam, cephalothin [surrogate for cephalexin], cefdinir, clindamycin, quinupristin-dalfopristin, tetracyclines, trimethoprim-sulfamethoxazole, gentamicin, rifampin, imipenem, and vancomycin) were also tested to identify profiles of cross- or coresistance with results interpreted by Clinical and Laboratory Standards Institute (CLSI) criteria (4).
PCR amplification of PVL genes (lukF-PV and lukS-PV) was performed on 101 CO-MRSA strains and 89 methicillin-susceptible S. aureus (MSSA) strains. The following PCR primers and the procedures used were those described previously by Lina et al.: luk-PV-F (ATC ATT AGG TAA AAT GTC TGG ACA TGA TCC A) and luk-PV-R (GCA TCA AST GTA TTG GAT AGC AAA AGC) (9).
All isolates were characterized for the type of SCCmecA gene cassette using a multiplex PCR strategy (13). The primers amplified various DNA segments within SCCmec characteristic to each of types I, II, III, and IV. mecA was amplified as part of the multiplex PCR to serve as an internal control. PCR products were separated on 2% agarose gels in Tris-acetate-EDTA buffer on the Criterion Sub-cell GT system (Bio-Rad, Hercules, CA) and stained with ethidium bromide. SCCmec types were assigned based on the numbers and sizes of the amplicons obtained. The agr types were determined by use of a subset of PVL-producing and SCCmec type IVa-positive strains.
Epidemiologic typing of CO-MRSA and MSSA strains that had PVL-positive PCR tests was performed by PFGE using procedures described previously (18). Briefly, bacterial cells grown overnight were embedded in agarose, lysed, and deproteinated to isolate near-intact genomic DNA. The DNA was digested with SmaI restriction enzyme (New England Biolabs, Ipswich, MA). The restriction fragments were separated by electrophoresis on a CHEF DR II (Bio-Rad) apparatus under the following conditions: 1% agarose, 0.5× Tris-borate-EDTA, and 200 V with a switch interval of 5 to 40 s over a 21-h period. Ethidium bromide-stained gels were examined visually. PFGE patterns were compared to CO-MRSA clones that are prevalent in the United States (18). Strains were assigned the same PFGE pattern only when all bands matched. When there were one or two band differences, the strains were assigned as a subtype or variant of the major type (designated by a capital letter, e.g., A, B, C, etc.), which was assigned the same capital letter followed by an Arabic numeral (for example, A1, A2, and A3).
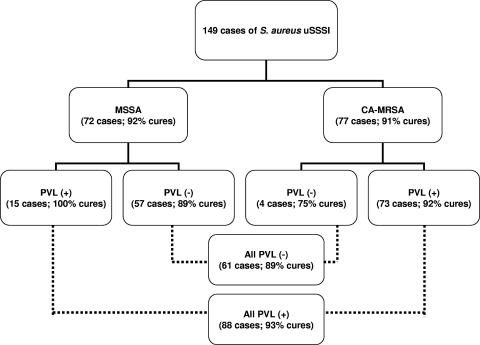
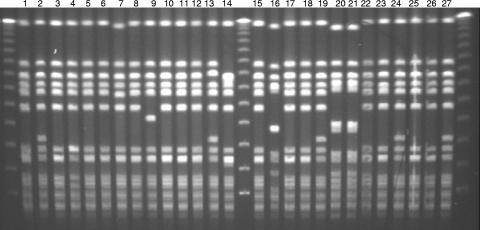
Among the 190 S. aureus isolates available for reference susceptibility testing, 101 (53%) were MRSA isolates, and among the 149 evaluable cases (2 were not available for study), 77 (52%) were caused by CO-MRSA (Fig. 1). These CO-MRSA isolates were distributed across all participating geographic locations (6). CO-MRSA isolates (Table 1) were more likely to be PVL positive (95%), have SCCmec type IVa (99%) and agr type I, be resistant to macrolides (erythromycin) and/or fluoroquinolones, and be clonally consistent with USA300 or USA400 (92%). All PVL-positive isolates contained SCCmec type IVa but varied slightly in their antibiograms. Over one-half of these CO-MRSA isolates were resistant only to erythromycin among the alternative non-β-lactam agents tested, and 37 were resistant to erythromycin and ciprofloxacin. Additional resistance rates for these tested agents were as follows (Table 1): erythromycin, 92.1%; ciprofloxacin or levofloxacin, 38.6%; tetracycline, 11.9%; clindamycin, 3.0%; rifampin, 3.0%, trimethoprim-sulfamethoxazole, 1.0%; and quinupristin-dalfopristin, 1.0%. The USA300 clonal type was highly represented (94% were usually USA300-0114, and 2% were USA400) in the PVL-positive CO-MRSA cases. In contrast, PVL-negative CO-MRSA isolates were more diverse in SCCmec types (II and IV), antibiograms, and occurrences of USA300 or USA400 clones (only 40%). An example of the PFGE patterns for 27 PVL-positive CO-MRSA isolates is shown in Fig. 2. The distribution of abscess or furuncle cases was found predominantly in the PVL-positive CO-MRSA isolates (69%) compared to only superficial wounds among the PVL-negative cases (small sample of four patients).
FIG. 1.
Clinical response results for 149 evaluable cases of S. aureus uSSSI treated with either cefdinir or cephalexin that were characterized by susceptibility to oxacillin (methicillin) and PVL production. CA-MRSA, community-acquired MRSA (6).
TABLE 1.
Results of testing of 101 strains of CO-MRSA by molecular methods
| PVL result (no. of strains, %) | SCCmecA type | agr typeb | Antibiogram resistance(s)a | No. of strains (% USA300/USA400 PFGE patterns) |
|---|---|---|---|---|
| Positive (96, 95.0) | IV | I | ER | 49 (93.3) |
| IV | NT | ER, CIP | 23 (100.0) | |
| IV | NT | ER, CIP, TC | 8 (87.5) | |
| IV | NT | CIP or none | 4 (100.0) | |
| IV | I | ER, TC | 3 (100.0) | |
| IV | I | ER, RIF | 2 (100.0) | |
| IV | I | ER, CL, RIF | 1 (0.0) | |
| IV | NT | ER, CIP, CL | 1 (100.0) | |
| IV | NT | ER, CIP, CL, T/S | 1 (100.0) | |
| IV | III | ER | 1 (100.0) | |
| IV | I | Variable | 3 (100.0) | |
| Negative (5, 5.0) | IV | NT | ER, CIP | 2 (100.0) |
| IV | NT | None | 1 (0.0) | |
| II | NT | ER, CIP | 1 (0.0) | |
| —c | NT | QD, TC | 1 (0.0) |
ER, erythromycin; CIP, ciprofloxacin; TC, tetracycline; RIF, rifampin; CL, clindamycin; QD, quinupristin-dalfopristin; T/S, trimethoprim-sulfamethoxazole.
NT, not tested against all strains.
—, unable to type.
FIG. 2.
Typical PFGE patterns of CO-MRSA uSSSI clinical trial strains showing dominant USA300 (lanes 1 to 13, 15, 17 to 19, and 22 to 27) and USA400 (lanes 16, 20, and 21) clonal patterns (18).
Results from the 89 MSSA strains indicated that only 15 (17%) were PVL positive and were quite diverse, with three different antibiograms and six unique PFGE patterns. The PVL-negative MSSA isolates displayed nine different antibiogram profiles, and the PVL-positive isolates were from abscesses or furuncles in 67% of cases compared to only 21% for the evaluable PVL-negative MSSA cases (P < 0.05).
The case outcomes of S. aureus uSSSI from clinically and bacteriologically evaluable patients treated with the two orally administered agents were analyzed by the oxacillin (methicillin) susceptibility patterns and the PVL molecular results (Fig. 1) of the baseline isolate. The clinical cure rates were not significantly different between these compounds (6), so all cases were combined for this analysis. The patient infection cure rates were essentially identical when MSSA- or MRSA-caused cases were compared (92% versus 91%, respectively). Similarly, PVL did not adversely influence the outcomes, with documented clinical cure rates of 89 to 93% (highest in PVL-positive cases).
These high levels of CO-MRSA isolates among all S. aureus isolates from year 2005 uSSSI cases confirms the elevated occurrence of MRSA (59 to 63%), high associated PVL production (98%), and clonality via USA300 strains (90 to 97%) reported previously by King et al. (8) and Moran et al. (11). The resistance patterns of these strains (erythromycin and/or ciprofloxacin) also conform to the antibiograms reported elsewhere previously (2, 5, 8, 11, 12, 14, 20) and illustrate the continued susceptibility to some older antimicrobials (clindamycin and trimethoprim-sulfamethoxazole) (2, 5, 8, 11, 12, 18). The use of clindamycin for infections caused by erythromycin-resistant S. aureus has been a concern because of inducible clindamycin resistance; however, the rates appear to be low (≤33%; none detected here) compared to those for hospital-associated MRSA (14). These agents have been suggested for suspected CO-MRSA therapy (7), but national prescription audits for uSSSI therapy indicate persisting β-lactam use (usually oral cephalosporins) (3,558 prescriptions/10,000 visits/year, or one-half of all therapies) and treatment declines in the use of lincosamides-macrolides and sulfonamide or related compounds in the last decade (10). This pattern of prescription practice was also noted previously by Naimi and colleagues (12), where 61% of CO-MRSA uSSSI cases received a β-lactam agent, which is regarded as having limited therapeutic value (3, 4).
The results from a large uSSSI clinical trial presented in this report clearly demonstrate a level of successful outcomes against CO-MRSA cases that was not significantly different than those for MSSA cases (6) or divergent from a nearly identical trial of orally administered β-lactams reported in 1997 (17). These observations confirm those of others, where measured resistances to the antimicrobials used for treatment (active or inactive) did not correlate with compromised patient outcomes (5, 11). Similarly, the presence of PVL in CO-MRSA isolates was not associated with poor cure rates or persisting infections, questioning the role of PVL in virulence. Voyich et al. (19) concluded from studies of PVL-negative (lukS/F-PV knockout) strains of USA300 or USA400 in a sepsis model that PVL was not a major virulence determinant; however, in our CO-MRSA case series, the type of infection (abscess and furuncle) was correlated with the presence of the PVL gene as reported previously (2, 9, 19).
Obviously, MRSA emergence in the community environment remains a high-priority clinical concern requiring well-constructed treatment guidelines and the promotion of continued searches for novel orally applied agents (16, 20). These CO-MRSA strains (USA300 clones) have been encountered among isolates from hospital-based bloodstream infections (34% of cases), expanding the range of public health concerns (15). These antimicrobial treatment regimens supplement the complete management of uSSSI that must consider local/topical wound care and surgical drainage (7, 16), with the consideration of expanded use of cultures to foster a better understanding of pathogen (CO-MRSA) frequency and local antibiogram patterns.
Acknowledgments
We express gratitude to the following individuals for their technical support and assistance in preparing the manuscript: N. D. O'Mara-Morrissey, T. R. Fritsche, H. S. Sader, D. J. Biedenbach, T. A. Busman, and M. G. Stilwell.
The molecular studies presented were sponsored by Abbott Laboratories.
Footnotes
Published ahead of print on 18 June 2007.
REFERENCES
- 1.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 2.Chambers, H. F. 2005. Community-associated MRSA—resistance and virulence converge. N. Engl. J. Med. 352:1485-1487. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. CLSI, Wayne, PA.
- 4.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. 16th informational supplement, M100-S16. CLSI, Wayne, PA.
- 5.Fridkin, S. K., J. C. Hageman, M. Morrison, L. T. Sanza, K. Como-Sabetti, J. A. Jernigan, K. Harriman, L. H. Harrison, R. Lynfield, and M. M. Farley. 2005. Methicillin-resistant Staphylococcus aureus disease in three communities. N. Engl. J. Med. 352:1436-1444. [DOI] [PubMed] [Google Scholar]
- 6.Giordano, P. A., D. Elston, B. K. Akinlade, K. Weber, G. F. Notario, T. A. Busman, M. Cifaldi, and A. M. Nilius. 2006. Cefdinir vs. cephalexin for mild to moderate uncomplicated skin and skin structure infections in adolescents and adults. Curr. Med. Res. Opin. 22:2419-2428. [DOI] [PubMed] [Google Scholar]
- 7.Grayson, M. L. 2006. The treatment triangle for staphylococcal infections. N. Engl. J. Med. 355:724-727. [DOI] [PubMed] [Google Scholar]
- 7a.Jones, R. N., L. Deshpande, A. Nilius, B. Akinlade, and G. Notario. 2006. Abstr. 46th Intersci. Conf. Antimicrob. Agents Chemother., abstr. L-1202. [DOI] [PMC free article] [PubMed]
- 8.King, M. D., B. J. Humphrey, Y. F. Wang, E. V. Kourbatova, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann. Intern. Med. 144:309-317. [DOI] [PubMed] [Google Scholar]
- 9.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 10.McCaig, L. F., L. C. McDonald, S. Mandal, and D. B. Jernigan. 2006. Staphylococcus aureus-associated skin and soft tissue infections in ambulatory care. Emerg. Infect. Dis. 12:1715-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran, G. J., R. N. Amii, F. M. Abrahamian, and D. A. Talan. 2005. Methicillin-resistant Staphylococcus aureus in community-acquired skin infections. Emerg. Infect. Dis. 11:928-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naimi, T. S., K. H. LeDell, K. Como-Sabetti, S. M. Borchardt, D. J. Boxrud, J. Etienne, S. K. Johnson, F. Vandenesch, S. Fridkin, C. O'Boyle, R. N. Danila, and R. Lynfield. 2003. Comparison of community- and health care-associated methicillin-resistant Staphylococcus aureus infection. JAMA 290:2976-2984. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel, M., K. B. Waites, S. A. Moser, G. A. Cloud, and C. J. Hoesley. 2006. Prevalence of inducible clindamycin resistance among community- and hospital-associated Staphylococcus aureus isolates. J. Clin. Microbiol. 44:2481-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seybold, U., E. V. Kourbatova, J. G. Johnson, S. J. Halvosa, Y. F. Wang, M. D. King, S. M. Ray, and H. M. Blumberg. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin. Infect. Dis. 42:647-656. [DOI] [PubMed] [Google Scholar]
- 16.Stevens, D. L., A. L. Bisno, H. F. Chambers, E. D. Everett, P. Dellinger, E. J. Goldstein, S. L. Gorbach, J. V. Hirschmann, E. L. Kaplan, J. G. Montoya, and J. C. Wade. 2005. Practice guidelines for the diagnosis and management of skin and soft-tissue infections. Clin. Infect. Dis. 41:1373-1406. [DOI] [PubMed] [Google Scholar]
- 17.Tack, K. J., C. H. Keyserling, J. McCarty, J. A. Hedrick, and the Cefdinir Pediatric Skin Infection Study Group. 1997. Study of use of cefdinir versus cephalexin for treatment of skin infections in pediatric patients. Antimicrob. Agents Chemother. 41:739-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tenover, F. C., L. K. McDougal, R. V. Goering, G. Killgore, S. J. Projan, J. B. Patel, and P. M. Dunman. 2006. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voyich, J. M., M. Otto, B. Mathema, K. R. Braughton, A. R. Whitney, D. Welty, R. D. Long, D. W. Dorward, D. J. Gardner, G. Lina, B. N. Kreiswirth, and F. R. Deleo. 2006. Is Panton-Valentine leukocidin the major virulence determinant in community-associated methicillin-resistant Staphylococcus aureus disease? J. Infect. Dis. 194:1761-1770. [DOI] [PubMed] [Google Scholar]
- 20.Zetola, N., J. S. Francis, E. L. Nuermberger, and W. R. Bishai. 2005. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275-286. [DOI] [PubMed] [Google Scholar]