Abstract
Protein arginine methylation is a posttranslational modification that impacts cellular functions, such as RNA processing, transcription, DNA repair, and signal transduction. The majority of our knowledge regarding arginine methylation derives from studies of yeast and mammals. Here, we describe a protein arginine N-methyltransferase (PRMT), TbPRMT5, from the early-branching eukaryote Trypanosoma brucei. TbPRMT5 shares the greatest sequence similarity with PRMT5 and Skb1 type II enzymes from humans and Schizosaccharomyces pombe, respectively, although it is significantly divergent at the amino acid level from its mammalian and yeast counterparts. Recombinant TbPRMT5 displays broad substrate specificity in vitro, including methylation of a mitochondrial-gene-regulatory protein, RBP16. TbPRMT5 catalyzes the formation of ω-NG-monomethylarginine and symmetric ω-NG,NG′-dimethylarginine and does not require trypanosome cofactors for this activity. These data establish that type II PRMTs evolved early in the eukaryotic lineage. In vivo, TbPRMT5 is constitutively expressed in the bloodstream form and procyclic-form (insect host) life stages of the parasite and localizes to the cytoplasm. Genetic disruption via RNA interference in procyclic-form trypanosomes indicates that TbPRMT5 is not essential for growth in this life cycle stage. TbPRMT5-TAP ectopically expressed in procyclic-form trypanosomes is present in high-molecular-weight complexes and associates with an RG domain-containing DEAD box protein related to yeast Ded1 and two kinetoplastid-specific proteins. Thus, TbPRMT5 is likely to be involved in novel methylation-regulated functions in trypanosomes, some of which may include RNA processing and/or translation.
Protein arginine methylation is an irreversible posttranslational modification catalyzed by protein arginine methyltransferases (PRMTs). PRMTs transfer a methyl group from S-adenosyl-l-methionine (AdoMet) to the guanidino nitrogen atoms of substrate arginine residues (34). Arginine residues found within RGG, RG, or RXR motifs are common, but not exclusive, sites of methylation (reviewed in reference 60). Interestingly, a large percentage of PRMT substrates are RNA binding proteins, especially those containing RG-rich motifs (9, 57). Other common substrates include histones and transcriptional coactivators (reviewed in references 7 and 95). Arginine methylation regulates protein function by modulating subcellular localization (44, 65, 82), protein-protein interactions (6, 31), and, less frequently, protein-RNA interactions (27, 28, 86). Through these mechanisms, protein arginine methylation exerts a range of effects on several cellular processes, including transcription (18, 96), signal transduction (1, 22), DNA repair (10), and RNA processing (12, 32, 82).
Arginine methyltransferases have been identified in animals, fungi, plants, and protozoa, and in total, four classes of enzymes have been described. Type I PRMTs catalyze the formation of ω-NG-monomethylarginine (MMA) and asymmetric ω-NG,NG-dimethylarginine (aDMA). Type II enzymes catalyze the formation of MMA and symmetric ω-NG,NG′-dimethylarginine (sDMA). Type III and type IV PRMTs catalyze only MMA or δ-NG-monomethylarginine formation, respectively. Most of our knowledge about PRMTs derives from analysis of mammals and yeasts. Of the 11 putative PRMTs identified in the genome of Homo sapiens, five display clear type I activity (PRMT1, PRMT3, PRMT4/CARM1, PRMT6, and PRMT8), one displays clear type II activity (PRMT5), and work is in progress on the others (reviewed in reference 50). In contrast to humans, only three PRMTs have been described in Saccharomyces cerevisiae. They are PRMT1 and PRMT5 homologs (called HMT1p and HSL7p), as well as a novel type IV PRMT called RMT2p, which has been described only in budding yeast, although homologs are present in a variety of fungi and plants (20, 99). Genes homologous to PRMT1 and PRMT5 have also been described in the yeast Schizosaccharomyces pombe, as well as an additional type I PRMT that is a homolog of PRMT3 (4, 38, 75). Although genes encoding putative PRMTs are present in the genomes of various plants and protozoa, analysis of arginine methylation in these groups has been limited. Type II PRMT activity mediated by SKB1 specific for histone H4 is reported to control flowering time in Arabidopsis thaliana (92), whereas type I PRMT homologs have been characterized from the parasitic protozoa Trypanosoma brucei (TbPRMT1 [72]) and Toxoplasma gondii (TgCARM1 and TgPRMT1 [81]). Phylogenetic analysis suggests that PRMTs originated early in the eukaryotic lineage, since no homologs have been identified in bacteria, archaea, or the basal eukaryote Giardia lamblia (50).
T. brucei, the causative agent of African trypanosomiasis, is an early-branching eukaryote that lacks transcriptional control and displays complex mechanisms of gene expression. Transcription of protein-coding genes is polycistronic, yet the differential expression of steady-state RNA has been observed between bloodstream form (BF) and procyclic-form (PF) life stages of the parasite (37). Trypanosomes exhibit unique biology, in which gene expression is coordinated posttranscriptionally through mechanisms that include trans splicing, RNA stability, RNA editing, and translation (23, 80, 89). A vast number of RNA binding proteins are presumably required to coordinate these posttranscriptional regulatory events in T. brucei, and a few such proteins have been identified (25). Since a large percentage of PRMT substrates are RNA binding proteins, arginine methylation may be of heightened importance in trypanosomes. Both type I and type II PRMT activities have been detected in T. brucei cellular extracts (74), and an enzyme termed TbPRMT1 was shown to constitute the major type I enzyme in the organism (72). We demonstrated recently that TbPRMT1-catalyzed arginine methylation functions in mitochondrial-RNA stabilization and ribonucleoprotein formation and/or stability (41, 42). In addition to TbPRMT1, analysis of the T. brucei genome revealed the presence of four additional PRMT genes (reference 48 and this study). The enzymes encoded by these genes and their potential roles in trypanosome biology remain completely uncharacterized.
Here, we describe the identification and characterization of an evolutionarily divergent PRMT5 homolog from T. brucei, which we term TbPRMT5. Recombinant TbPRMT5 displays intrinsic type II PRMT activity, catalyzing the formation of MMA and sDMA on myelin basic protein, core histones, and an RG peptide. TbPRMT5 also methylates the mitochondrial-RNA binding protein RBP16 (45, 73) in vitro, although it catalyzes primarily MMA on this substrate. TbPRMT5 purified from trypanosome cellular extracts displays PRMT activity with properties similar to the recombinant enzyme and is present in high-molecular-weight complexes. Analysis of trypanosome proteins associated with TAP-tagged TbPRMT5 revealed the absence of proteins homologous to components of the human 20S methylosome (32, 33, 61). Instead, TbPRMT5 associates with a DEAD box-containing protein, which is related to the Ded1p and Vasa translation initiation factors described in yeast and Drosophila. This putative RNA helicase harbors an RGG-rich C terminus, suggesting it is likely an endogenous TbPRMT5 substrate. In addition, TbPRMT5 copurified with tryparedoxin peroxidase and two high-molecular-weight proteins, for which homologs are present only in the related kinetoplastid protozoa Trypanosoma cruzi and Leishmania major. Thus, TbPRMT5 apparently assembles in kinetoplastid-specific complexes, suggesting novel functions for this divergent type II PRMT.
MATERIALS AND METHODS
Plasmid constructs.
To identify type II PRMT homologs in T. brucei, a BLAST search was performed against the sequenced and partially annotated T. brucei genomic database (The Institute for Genomic Research) using the coding sequence for H. sapiens PRMT5 (GenBank accession no. O14744). A putative methyltransferase that displayed homology to human PRMT5 was identified on chromosome 10 (locus Tb10.70.7570) and was used to design oligonucleotide primers for the PCR amplification of the full-length TbPRMT5 open reading frame (ORF) from cDNA. For cDNA synthesis, total RNA was isolated from PF T. brucei brucei clone IsTaR1 stock EATRO 164, and the cDNA was reverse transcribed using oligonucleotide primer [dT]-RXS (5′-GAGAATTCTCGAGTCGACTTTTTTTTTTTTTTTTT-3′) (restriction enzyme sites of all oligonucleotides used in this study are underlined). To generate an N-terminal maltose-binding protein (MBP) fusion, the TbPRMT5 ORF was PCR amplified from cDNA using PRMT5-5′ (5′-GAGAATTCGCTAGCATGAAGCCATGTGTTTCCGCTGC-3′) forward and pMAL-PRMT5-3′ (5′-GCTCTAGACTACGTCAGCAGAGAAATGTGGCCC-3′) reverse primers and cloned into the EcoRI/XbaI restriction enzyme sites of pMAL-c2 (New England Biolabs), creating pMAL-PRMT5. The pBSC-PRMT5 plasmid construct was created for use as a template for full-length TbPRMT5 antisense riboprobe synthesis. For the generation of pBSC-PRMT5, the full-length TbPRMT5 ORF was PCR amplified from PF cDNA using PRMT5-5′ (see above) forward and PRMT5-3′ (5′-GCTCTAGACTCGAGCGTCAGCAGAGAAATGTGGCCC-3′) reverse primers, blunt ended with T4 DNA polymerase (Invitrogen), and cloned into the EcoRV site of pBluescriptII SK(−) (Stratagene). The pHD918-PRMT5 plasmid construct was created for the generation of a trypanosome stable cell line constitutively expressing a C-terminal TAP tag fusion protein. To create pHD918-PRMT5, TbPRMT5 was PCR amplified from pMAL-PRMT5 using TbPRMT5-1 (5′-CCCAAGCTTATGAAGCCATGTGTTTCCGCTGC-3′) forward and TbPRMT5-2 (5′-CTAGCTAGCGTTAACCGTCAGCAGAGAAATGTGGCCC-3′) reverse primers and cloned into the HindIII/HpaI sites of the pHD918 TAP vector (a generous gift from Christine Clayton, Heidelberg University, Heidelberg, Germany) (30, 78). To generate the pZJM-PRMT5 RNA interference (RNAi) construct, a 449-nucleotide fragment of TbPRMT5 (nucleotides 361 to 810) was PCR amplified from PF cDNA using PRMT5-5′i (5′-GCCTCGAGACTGCTTGCGGTAGCGTGTCG-3′) forward and PRMT5-3′i (5′-GCAAGCTTTTCAGCGTTGGGAAGTTCAAACGC-3′) reverse primers and cloned into the XhoI/HindIII sites of the pZJM RNAi vector (a generous gift from Paul T. England, Johns Hopkins School of Medicine) (93). All plasmid constructs were transformed into Escherichia coli strain DH5α competent cells (Invitrogen). Positive clones were selected on LB agar plates containing 100 μg/ml ampicillin and confirmed by sequencing them (Roswell Park Cancer Institute DNA Sequencing Laboratory, Buffalo, NY).
Trypanosome cell culture, transfection, and induction of RNAi.
PF T. brucei brucei clone IsTaR1 stock EATRO 164 (88) was cultured in SDM-79 as described previously (15). PF T. brucei brucei strain 29-13 (a generous gift from George A. M. Cross, Rockefeller University), which harbors integrated constructs for the expression of T7 RNA polymerase and tetracycline repressor genes, was cultured in SDM-79 supplemented with 15 μg/ml G418 and 50 μg/ml hygromycin B, as described previously (15, 94). BF T. brucei brucei strain Lister 427 (MITat 1.2) clone 221 (a generous gift from George A. M. Cross, Rockefeller University) (29, 47, 49) was cultured in HMI-9 medium as described previously (46).
Stable cell lines constitutively expressing a TbPRMT5 C-terminal TAP tag fusion protein were generated via electroporation. For transfection, log-phase PF T. brucei brucei clone IsTaR1 stock EATRO 164 was harvested by centrifugation (1,000 × g), washed in 150 ml cold EM buffer (51), and resuspended to 2 × 108 cells/ml with EM buffer. Five-hundred microliters of this cell suspension (1 × 108 cells/transfection) was transferred to chilled 2-mm-gap electroporation cuvettes containing either water (control) or 20 μg of NotI-linearized pHD918-PRMT5 plasmid DNA. The cells were pulsed twice in a Bio-Rad Gene Pulsor electroporator set at 800 V, 25 mF, and 400 Ω. After electroporation, the cells were transferred to 9.5 ml of SDM-79 medium supplemented with 15% fetal bovine serum and incubated at 27°C overnight. Antibiotic selection was applied the following day with the addition of 50 μg/ml hygromycin B. Clonal cell lines were generated by limiting dilution and confirmed by immunoblot analysis using TAP-specific antibodies. Analysis of TbPRMT5-TAP-expressing cells was performed using the clonal cell line p8.1.E8.
Stable cell lines engineered for the tetracycline-regulatable knockdown of TbPRMT5 using RNAi were generated via electroporation. For transfection, log-phase PF 29-13 cells (94) were harvested by centrifugation, washed in 150 ml cold EM buffer, and resuspended to 2.5 × 107 cells/ml. Four hundred fifty microliters of this cell suspension was aliquoted into chilled 2-mm-gap electroporation cuvettes (1.1 × 107 cells/transfection) containing either water (control) or 20 μg of Not1-linearized pZJM-PRMT5 plasmid DNA and electroporated as described above. After electroporation, the cells were transferred to 4 ml of SDM-79 supplemented with 15% fetal bovine serum, 15 μg/ml G418, and 50 μg/ml hygromycin B and incubated at 27°C overnight. Antibiotic selection was applied the following day with the addition of 2.5 μg/ml phleomycin. Clones were obtained by limiting dilution and confirmed by restriction enzyme digestion and Southern hybridization using a TbPRMT5-specific probe. TbPRMT5 RNAi (clone p1.3.E9) inductions were initiated with the addition of 1 μg/ml tetracycline.
Expression and purification of recombinant proteins.
For the expression of recombinant MBP-TbPRMT5, 4 liters of LB medium containing 100 μg/ml ampicillin was inoculated with an overnight culture of E. coli DH5α transformed with pMAL-PRMT5 and grown at 36°C and 225 rpm to an optical density (OD) of 0.4 to 0.6. The cultures were chilled on ice for 15 min to ∼20°C, and 2% (vol/vol) ethanol was added to enhance protein solubility. MBP-TbPRMT5 expression was induced with the addition of 0.3 mM isopropyl β-d-thiolgalactopyranoside for 24 h at 20°C and 225 rpm. The cells were harvested by centrifugation for 20 min at 4,000 × g and 4°C and resuspended in 50 ml lysis buffer (20 mM ethanolamine [pH 9.0], 1 M NaCl, 1 mM EDTA, 0.2% NP-40, 4 mM benzamidine, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μM leupeptin, and 300 μg/ml lysozyme [added after resuspension]) per 1-liter culture. To reduce the viscosity, the extract was incubated with 5 mM MgCl2 and 25 μg/ml DNase I (Sigma) at 4°C for 30 min with agitation. Cell lysis was completed by sonication on ice for 3 min (30-second intervals) at 50% duty cycle pulse mode. The supernatant was clarified by centrifugation at 14,000 × g for 20 min at 4°C and dialyzed overnight in 4 liters of dialysis buffer (20 mM bis-Tris [pH 7.0], 50 mM NaCl, 0.05% NP-40, 1 mM EDTA, 1 mM benzamidine, 0.5 mM PMSF, 1 μM leupeptin) at 4°C. An additional 3-h incubation in 2 liters of fresh dialysis buffer was performed the following day. The dialyzed crude extract was then incubated with 15 ml of preequilibrated amylose resin (New England Biolabs) for 2 h at 4°C with rocking. This slurry was poured through a column to pack the resin, and the supernatant was passed over the column again at a flow rate of ∼1 ml/min. The column was washed with 12 bed volumes of column buffer (dialysis buffer minus protease inhibitors) at a flow rate of ∼1 ml/min. The MBP-TbPRMT5 fusion protein was then eluted at a flow rate of ∼0.5 ml/min with 30 ml of elution buffer (20 mM bis-Tris [pH 7.0], 50 mM NaCl, 2 mM EDTA, 0.1% NP-40, 10 mM maltose, 10% [vol/vol] glycerol, 1 mM benzamidine, 1 mM PMSF, and 1 μM leupeptin) and collected in 1-ml fractions. The fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), quantified using a bovine serum albumin (BSA) standard curve, and assayed for methyltransferase activity.
Rat PRMT1 was expressed as a glutathione S-transferase (GST) fusion protein in E. coli BL-21 transformed with the pGEX-PRMT1 bacterial expression vector (a generous gift from Harvey R. Herschman, University of California at Los Angeles) (55). Cells were grown in super broth containing 0.1 mM ampicillin at 36°C and 225 rpm to an OD of 0.5 to 0.7. The expression of GST-PRMT1 was induced for 4 h with the addition of 1 mM isopropyl-β-d-thiogalactopyranoside. The cells were harvested by centrifugation at 4,000 × g for 20 min at 4°C and resuspended in 25 ml lysis buffer (50 mM Tris-HCl [pH 8.0], 50 mM NaCl, 5 mM EDTA, 0.5 mM benzamidine, 0.15 mM PMSF, and 1 μM leupeptin) per 1-liter culture. The cells were lysed by sonication on ice for 3 min (30-second intervals) at 50% duty cycle pulse mode. The crude extract was clarified by centrifugation at 14 rpm for 20 min at 4°C and loaded onto a column containing 5 ml of preequilibrated Glutathione-Sepharose 4 Fast Flow resin (Amersham Biosciences) at a flow rate of ∼0.5 ml/min. The column was washed with 10 bed volumes phosphate-buffered saline (PBS)-EDTA-PMSF buffer (PBS [pH 7.4] containing 5 mM EDTA and 0.15 mM PMSF), followed by a second wash with 10 bed volumes of PBS-EDTA buffer (PBS [pH 7.4] containing 5 mM EDTA) at a flow rate of 1.5 ml/min. GST-PRMT1 fusion protein was eluted in 2 ml fractions with 5 bed volumes glutathione buffer (50 mM Tri-HCl [pH 8.0], 10 mM reduced-form glutathione [Sigma], and 10% [vol/vol] glycerol). The peak fractions were pooled and dialyzed overnight at 4°C in two 2-liter changes of methyltransferase assay buffer (80 mM Tris-HCl [pH 8.0], 0.5 mM benzamidine, and 0.4 mM PMSF). GST-PRMT1 was then concentrated, analyzed by SDS-PAGE alongside a BSA standard curve, and assayed for methyltransferase activity. His-RBP16, His-CSD (cold shock domain), and His-RGG (RGG domain) fusion proteins were previously expressed as described previously (63). The MBP-TBRGG1 fusion protein was previously expressed as described previously (72).
In vitro methyltransferase assays.
In vitro methylation assays were performed at 36°C in the presence of 0.9 to 1 μM S-adenosyl-l-[methyl-3H]methionine ([methyl-3H]AdoMet) (Amersham; 66.0 to 80.0 Ci/mmol; 1 μCi/μl) in 80 mM Tris-HCl (pH 8.0) buffer containing 0.5 mM benzamidine and 0.4 mM PMSF protease inhibitors. Experimental details are described in the figure legends. For MBP-TbPRMT5 time course, titration, and substrate specificity, reactions were stopped by the addition of SDS-PAGE sample buffer. Samples were incubated at 95°C for 5 min and analyzed by either 12.5% or 15% polyacrylamide SDS-PAGE. Gels were stained with 0.1% (wt/vol) Coomassie brilliant blue R250 (Sigma) in 50% methanol-10% acetic acid, destained with 5% methanol-10% acetic acid, and then treated by fluorography to visualize tritiated substrates. For fluorography, gels were treated with EN3HANCE (Perkin-Elmer Life Sciences), dried at 60°C in vacuo, and exposed to Kodak X-Omat Blue XB-1 scientific imaging film at −80°C.
In vitro methylation assays performed for substrate amino acid analysis are described in the figure legends. Reactions were stopped by the addition of 10% (wt/vol) trichloroacetic acid (TCA), followed by a 30-minute incubation on ice. TCA-precipitated proteins were centrifuged at 13,000 × g for 10 min at room temperature. The supernatant was decanted, and the pellets were washed in 500 μl of acetone. Samples were centrifuged as described above, the acetone was decanted, and the pellets were allowed to dry. The TCA-precipitated pellets were then subjected to acid hydrolysis for substrate amino acid analysis.
The recombinant substrates and enzymes used in this analysis are described above. Calf thymus core histones were purchased from Roche (no. 223 565). Myelin basic protein from bovine brain was purchased from Sigma (no. M 1891). The RG peptide substrate (H-CGRGRGRGRGRGRGRG-NH2) was custom synthesized by Bachem.
Amino acid analysis.
Substrate amino acid analysis was performed essentially as described previously (32). In vitro methylation reaction mixtures were acid hydrolyzed in 200 μl of constantly boiling 6 N HCl (Sigma) for 20 h at 110°C in vacuo. Acid hydrolysates were dried at room temperature in a Savant Speed Vac, resuspended in 5 μl of water, and analyzed by thin-layer chromatography (TLC) alongside 30 nmol of MMA (acetic acid salt; Sigma), aDMA (dihydrochloride; Sigma), sDMA (dihydrochloride; Calbiochem), and AdoMet (iodide salt; MP Biochemical) standards. Samples were loaded on LK6DF silica gel 60 TLC plates (Whatman) and separated using a 2:0.5:4.5:1 ammonium hydroxide-chloroform-methanol-water solvent system. Amino acid standards were visualized with spray ninhydrin (Tokyo Chemical Industry Co., Ltd.), and methyl-3H residues were visualized by fluorography using EN3HANCE spray reagent (Perkin-Elmer Life Sciences).
For the analysis of MBP-TbPRMT5 substrates by cation-exchange chromatography of methylated amino acids obtained after acid hydrolysis, in vitro methylation reactions were performed at 36°C for 16 h in the presence of 0.35 μM MBP-TbPRMT5, 1 μM [methyl-3H]AdoMet (66.0 to 80.0 Ci/mmol), and either 20 μg calf thymus core histones or 3.5 μM His-RBP16 substrates. The reactions were stopped by precipitation with TCA and were acid hydrolyzed with 100 μl of 6 N HCl at 110°C for 20 h in vacuo in a Waters Pico-Tag Vapor Phase apparatus. The dried hydrolyzed samples were resuspended in 50 μl of water, 25 μl of which was added to 500 μl of citrate buffer (0.2 M Na+, pH 2.2) with 1.0 μmol each of unlabeled standards of MMA, aDMA, and sDMA (di-p-hydroxyazobenzene-p′-sulfonate salt; Sigma). The sample was loaded onto a cation-exchange column equilibrated with sodium citrate buffer (0.35 M Na+, pH 5.27) (the column resin was Beckman AA-15 sulfonated polystyrene beads; column length, 0.9-cm inner diameter by 7-cm column height) and eluted at 1 ml/min at 55°C.
RNA isolation and Northern blot analysis.
Total RNA was isolated from log-phase cells using the Purescript RNA Isolation Kit (Gentra Systems). A full-length [α-32P]UTP-labeled TbPRMT5 riboprobe was generated by in vitro transcription (Maxiscript T3 kit; Ambion) using NheI-linearized pBSC-PRMT5 as a template. An [α-32P]UTP-labeled tubulin riboprobe, corresponding to a 650-nucleotide stuffer sequence of the pZJM RNAi expression vector, was generated by in vitro transcription (Maxiscript T7 kit; Ambion) after digestion with XhoI and BamHI restriction endonucleases. The [γ-32P]ATP 5′-labeled tubulin oligonucleotide probe (TUB-RT; 5′-GGGGGTCGCACTTTGTC-3′; melting temperature, 46°C) was labeled by a T4 kinase forward reaction.
For the Northern blot analysis of TbPRMT5 BF versus PF expression, total RNA (20 μg) isolated from BF T. brucei strain Lister 427 (MITat 1.2) clone 221 and PF T. brucei clone IsTaR1 stock EATRO 164 was resolved on a 1.5% formaldehyde-agarose gel and transferred to Nytran (Schleicher & Schuell) by capillary action in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membrane was prehybridized for ≥1 h at 65°C in 0.15 ml hybridization solution (50% formamide, 1% SDS, 5× SSC, 1× Denhardt's solution, and 150 μg/ml denatured sheared salmon sperm DNA) per 1-cm2 blot, followed by an overnight hybridization at 65°C with ≥1 × 106 cpm full-length TbPRMT5 antisense riboprobes per ml hybridization solution. Two low-stringency washes were performed at 65°C for 15 min in 2× SSC-0.1% SDS, followed by two high-stringency washes at 65°C for 15 min in 0.1× SSC-0.1% SDS. The membrane was stripped at 80°C in Northern blot stripping solution (40 mM Tris-HCl [pH 7.5], 1% SDS, and 0.1× SSC) and reprobed for tubulin RNA as follows. Prehybridization was performed at 36°C for ≥1 h in 0.2 ml hybridization solution (10× Denhardt's solution, 5× SSC, 1% SDS, and 200 μg/ml denatured salmon sperm DNA) per 1-cm2 blot. The membrane was hybridized overnight at 36°C with a 5′-labeled tubulin oligonucleotide probe (≥1 × 106 cpm probe/ml hybridization solution). Five room temperature washes were performed in 6× SSC-1% Sarkosyl for 5 min each, followed by a final 3-min wash in 1× SSC-0.1% SDS. RNA levels were analyzed on a Bio-Rad Personal FX phosphorimager using Quantity One software.
For the analysis of TbPRMT5 RNAi, total RNA (10 μg) isolated from parental, uninduced, and induced RNAi cells on days 1, 2, 4, 6, and 8 postinduction was analyzed by Northern blotting using a full-length TbPRMT5 riboprobe as described above. To control for loading, the membrane was stripped and hybridized as described above with a tubulin-specific riboprobe. Autoradiography was performed using Kodak X-Omat AR scientific imaging film, and RNA levels were analyzed by densitometry using Bio-Rad Quantity One software.
Preparation and fractionation of trypanosome cellular extracts.
Fractionation of trypanosome cellular extracts was performed as described previously (79) with some modifications. Log-phase TbPRMT5-TAP-expressing cells were harvested by centrifugation at 10,000 × g for 10 min at 4°C. The cell pellets were washed in cold PBS, weighed, and resuspended in 3 volumes of cell mass (ml/g cell mass) low-salt hypotonic lysis buffer (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 1 mM benzamidine, 0.5 mM PMSF, 1 μM leupeptin, and 1 μg/ml pepstatin) containing 0.2% NP-40. The lysates were incubated on ice for 10 min, followed by the addition of 12 volumes low-salt hypotonic lysis buffer containing 0.5% NP-40. Cell disruption was completed by homogenization in a glass Dounce homogenizer, followed by passage through a 26-gauge needle 10 times. Crude whole-cell extracts were either quick-frozen in liquid nitrogen and stored at −80°C or further fractionated into cytosolic and nuclear extracts. To obtain a cytosolic extract, nuclei were pelleted by centrifugation at 5,000 × g for 20 min at 4°C. The supernatant (cytosolic extract) was transferred to a clean tube, quick-frozen in liquid nitrogen, and stored at −80°C. The nuclear pellet was resuspended in 5 volumes 0.25 M sucrose buffer (low-salt hypotonic lysis buffer containing 0.25 M sucrose) and centrifuged at 1,100 × g for 15 min at 4°C. The supernatant was decanted, and the pellet was resuspended in 5 volumes 0.25 M sucrose buffer. This lysate was then layered with an equal volume of 0.35 M sucrose buffer (low-salt hypotonic lysis buffer containing 0.35 M sucrose) and centrifuged at 1,100 × g for 15 min at 4°C. The supernatant was decanted, and the nuclei were resuspended in 5 volumes of 0.35 M sucrose buffer. To disrupt the nuclear membranes, the extract was sonicated on ice for 3 min (at 30-second intervals) at 50% duty cycle pulse mode and micropipet limit setting 3. The nuclear extract was then layered with 5 volumes of 0.88 M sucrose buffer (low-salt hypotonic lysis buffer containing 0.88 M sucrose) and centrifuged at 1,100 × g for 20 min at 4°C. The supernatant (nuclear extract) was transferred to a clean tube, quick-frozen in liquid nitrogen, and stored at −80°C.
The protein concentrations in subcellular fractions were quantified by Bradford assay. Cytosolic and nuclear fractionations were analyzed by Western blotting using rabbit anti-Hsp70.4 (a generous gift from J. D. Bangs, University of Wisconsin—Madison) and rabbit anti-CTD (RNAPII; a generous gift from Vivian Bellofatto, University of Medicine and Dentistry of New Jersey) antibodies, respectively. TbPRMT5-TAP localization was analyzed by immunoblotting using peroxidase-anti-peroxidase (PAP) soluble-complex antibody produced in rabbits (Sigma), which recognizes the protein A moiety of the TAP tag.
Glycerol gradient sedimentation.
For the analysis of in vivo TbPRMT5 protein complexes, 500 μl of clarified whole-cell extract prepared from 1 × 109 log-phase TbPRMT5-TAP cells was fractionated in triplicate over a 5 to 20% linear glycerol gradient. Glycerol gradients were prepared in gradient buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 1 mM benzamidine, 0.5 mM PMSF), and samples were fractionated by centrifugation at 35,000 rpm for 20 h at 4°C in a Beckman SW-41 rotor. Twenty-five 500-μl fractions were collected from the top of each glycerol gradient, resolved by denaturing (12.5 μl) and nondenaturing (10 μl) gel electrophoresis alongside 20 μg of TbPRMT5-TAP whole-cell extract, and analyzed by immunoblotting using the TAP-specific PAP reagent. The sedimentation of TbPRMT5-TAP-containing protein complexes was compared to the sedimentation of cytochrome c (1.9S), BSA (4.3S), yeast alcohol dehydrogenase (7.4S), catalase (11.3S), and thyroglobulin (19S) protein standards fractionated in a parallel glycerol gradient (also performed in triplicate).
Native gel electrophoresis.
Nondenaturing (native) electrophoresis was performed using Novex 4 to 12% polyacrylamide gradient Tris-glycine gels (Invitrogen) as described by the manufacturer. For the analysis of glycerol gradient fractions, the addition of a buoyancy reagent was omitted from the native sample buffer. A High Molecular Weight Calibration Kit for Native Electrophoresis (Amersham Biosciences) was used for molecular weight standards.
Immunoblot analysis.
For the immunoblot analysis of TbPRMT5-TAP subcellular localization, 15 μg of whole-cell, cytoplasmic, or nuclear extract was resolved by 12.5% SDS-PAGE and transferred to nitrocellulose (Bio-Rad) at 50 V for 75 min in 10 mM 3-[cyclohexylamino]-1-propanesulfonic acid buffer (pH 11.0) containing 10% methanol. The membranes were blocked for 1 h in Tris-buffered saline containing 5% (wt/vol) dry milk. For the detection of TbPRMT5-TAP, membranes were incubated with 1:2,000 PAP soluble-complex antibody (Sigma) for 1 h, and PAP was detected by chemiluminescence using the Supersignal West Pico Chemiluminescent Substrate Solution (Pierce). The membrane was then stripped and incubated overnight with either anti-TbHsp70.4 (cytosolic marker) or anti-CTD (RNAPII nuclear marker) primary antibody in Tris-buffered saline containing 2% (wt/vol) dry milk and 0.05% Tween 20. Primary antibodies were detected using horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (Pierce) secondary antibody and detected by chemiluminescence.
For the immunoblot analysis of native TbPRMT5-TAP-containing complexes, samples were resolved on Novex 4 to 12% Tris-glycine gels (Invitrogen) by nondenaturing electrophoresis and transferred to nitrocellulose for 2 h at 25 V, as described by the manufacturer. TAP-containing complexes were detected using PAP soluble-complex antibodies, as described above.
TbPRMT5-TAP tandem-affinity purification.
Ectopically expressed TbPRMT5-TAP protein was purified by tandem-affinity purification as described previously (78) with some modifications. For the analysis of native TbPRMT5-TAP-associated complexes, 1.25 × 1010 cells were harvested from either wild-type or TbPRMT5-TAP log-phase cultures. The cell pellets were washed with 100 ml of PBS-G (1× PBS containing 6 mM glucose), centrifuged at 6,090 × g for 10 min, and resuspended in 9 ml of IPP150 buffer (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40) containing 1% (wt/vol) BSA and 1 Complete Mini EDTA-free protease inhibitor tablet (Roche Diagnostics). The cells were lysed on ice for 20 min after the addition of 1% (vol/vol) Triton X-100, and the crude extract was clarified by centrifugation at 10,000 × g for 15 min at 4°C. TbPRMT5-TAP-associated complexes were purified from whole-cell extract (10 ml) over tandem IgG-Sepharose 6 Fast Flow (Amersham Biosciences) and calmodulin resin (Stratagene) columns as described previously (78). Native TbPRMT5-TAP-associated complexes were eluted in 1-ml volumes. Ten percent of the final eluate was TCA precipitated (10% final concentration) and analyzed by 12.5% SDS-PAGE, followed by silver staining. The remaining TbPRMT5-calmodulin-binding protein (CBP) final eluate was digested with trypsin and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS).
For the analysis of TbPRMT5-CBP enzymatic activity, cytosolic extract prepared from 1 liter of TbPRMT5-TAP log-phase cells was dialyzed overnight at 4°C in two 1-liter changes of IPP150 buffer containing 1 mM benzamidine, 0.5 mM PMSF, 1 μM leupeptin, and 1 μg/ml pepstatin protease inhibitors. The dialyzed extract was divided into three equal volumes and separately purified over tandem IgG-Sepharose 6 Fast Flow (Amersham Biosciences) and calmodulin resin (Stratagene) columns as described previously (78). Before final elution, the three different calmodulin columns were washed with buffer containing either 0.15 M, 0.5 M, or 1 M NaCl. The resultant eluates were dialyzed in two 1-liter changes of methyltransferase assay buffer and assayed for activity. Protein composition was analyzed by 12.5% SDS-PAGE and silver staining. The protein concentration was estimated by comparison to a BSA standard curve run on the same gel.
RESULTS
T. brucei harbors genes encoding at least two putative type II PRMTs.
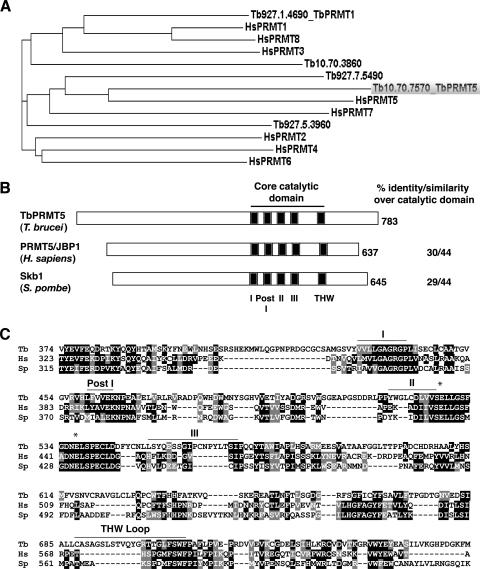
We previously reported biochemical evidence for both type I and type II PRMT activities in T. brucei cellular extracts (74), and the gene encoding the major type I PRMT (TbPRMT1) has been identified (72). To elucidate the entire complement of PRMTs in this evolutionarily ancient organism and to identify the gene(s) responsible for type II activity, we performed a protein BLAST search against the T. brucei genomic database using the H. sapiens PRMT1 (type I) and PRMT5 (type II) sequences as queries. A comparison of these BLAST results with all predicted methyltransferases in The Institute for Genomic Research's T. brucei Annotation Database identified five putative PRMTs in T. brucei. Figure 1A presents the phylogenic analysis of the putative trypanosome PRMTs compared to the known PRMTs in H. sapiens. Two trypanosome proteins (Tb10.70.7570 and Tb927.7.5490) cluster with the best-characterized human type II enzyme, PRMT5, in which the methyltransferase identified on locus Tb10.70.7570 is most similar. The other putative T. brucei type II PRMT (Tb927.7.5490) exhibits homology to mammalian PRMT7, although the trypanosome protein lacks the second putative AdoMet binding domain found in human PRMT7. Two of the five trypanosome PRMTs (Tb927.1.4690 and Tb927.5.3960), including the previously characterized TbPRMT1 (72), share clear homology with type I enzymes. Interestingly, the remaining PRMT identified at locus Tb10.70.3860 exhibits characteristics of both type I and type II enzymes. This enzyme is most similar to PRMT3, which is a type I enzyme. However, Tb10.70.3860 lacks the conserved THW loop that is found in all previously characterized type I PRMTs but which is frequently less conserved in type II enzymes. Tb10.70.3860 also harbors a substitution at the second, normally invariant glutamate residue of the double-E loop (E225D). Homologous genes that exhibit the same conserved changes are also present in the genomes of the related organisms T. cruzi and L. major, suggesting a conserved role for this enzyme in trypanosomatid biology. Taken together, our analysis indicates that the T. brucei genome encodes five putative PRMTs, including the previously characterized major type I enzyme, TbPRMT1 (72).
FIG. 1.
TbPRMT5 is a putative type II PRMT with a conserved methyltransferase domain. (A) Phylogenetic analysis of putative PRMTs identified in T. brucei with PRMT homologs from H. sapiens. Phylogenetic analysis was performed using the ClustalW multiple-sequence alignment tool. The GenBank accession numbers are as follows: HsPRMT1, NP_001527; HsPRMT2, NP_001526; HsPRMT3, NP_005779; HsPRMT4, NP_954592; HsPRMT5, O14744; HsPRMT6, Q96LA8; HsPRMT7, NP_061896; and HsPRMT8, Q9NR22. (B) Schematic representation of PRMT5 homologs from T. brucei (TbPRMT5), H. sapiens (PRMT5/JBP1; GenBank accession no. O14744), and S. pombe (Skb1; GenBank accession no. P78963) illustrating the presence of a conserved methyltransferase domain (motifs I, Post I, II, and III) and variable N termini. The percent amino acid identity/similarity over the core catalytic domain (278 amino acids in TbPRMT5) is shown. (C) A multiple-sequence alignment of core catalytic domains and flanking regions of the PRMTs shown in panel B was generated using ClustalW. Similar amino acids are shaded gray, while identical amino acids are shaded black. The conserved AdoMet binding domain (motifs I, Post I, II, and III) is indicated above the sequences. The double-E loop is denoted with asterisks. The THW domain is denoted with a bar and does not appear to be conserved in type II PRMTs. Tb, T. brucei; HS, H. sapiens; Sp, S. pombe.
To further characterize the putative PRMT5 homolog in T. brucei, the ORF of Tb10.70.7570 was amplified by reverse transcription-PCR from PF RNA, cloned into the pMAL-c2 bacterial expression vector, and sequenced. The 2,349-nucleotide ORF, which we term TbPRMT5, shares 99% nucleotide sequence identity with the T. brucei TRUE927 genomic strain and encodes a 783-amino-acid protein with the expected molecular mass and isoelectric point of 86.7 kDa and 5.52, respectively. Sequence analysis of TbPRMT5 using BLAST and CLUSTALW proteomic tools indicated that TbPRMT5 contains a conserved methyltransferase domain (amino acids 432 to 710) that displays 30/44% and 29/44% identity/similarity with PRMT5 homologs from H. sapiens (PRMT5/JBP1) and S. pombe (Skb1), respectively (Fig. 1B and C). The core catalytic domains of human (amino acids 361 to 581) and yeast (amino acids 348 to 577) PRMT5 homologs share 47/62% amino acid identity/similarity, consistent with the closer evolutionary relationship of the kingdoms Animalia and Fungi to each other than to Protista. Similar to other PRMT5 homologs, TbPRMT5 has an N-terminal addition outside the catalytic core (432 amino acids), in which no conserved domains or binding motifs have been detected. The N-terminal domain of TbPRMT5 is extended compared to that in other PRMT5 homologs by approximately 75 amino acids. The signature methyltransferase domain common to all PRMTs, including TbPRMT5 (Fig. 1C), includes an AdoMet binding domain (motifs I, Post I, II, and III), as well as a double-E loop, whose glutamate residues make up part of the arginine-binding pocket (97, 98). The catalytic core of type I PRMTs also contains a THW loop, in which the histidine residue is thought to participate in an active-site His-Asp proton relay system in the elimination of a proton from substrate arginine residues (98). As stated above, this THW loop is not conserved in type II PRMTs (50, 98), and this holds true for TbPRMT5, as well. Collectively, phylogenetic and sequence analyses indicate that TbPRMT5 represents the T. brucei homolog of mammalian PRMT5 and yeast Skb1.
Recombinant TbPRMT5 exhibits methyltransferase activity in a concentration- and time-dependent manner.
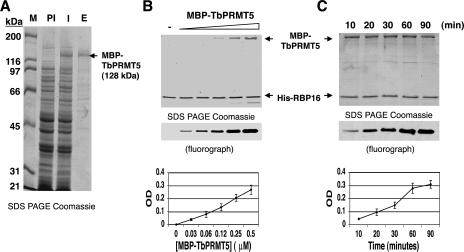
To confirm that TbPRMT5 possesses methyltransferase activity, we expressed TbPRMT5 in E. coli as an MBP fusion protein (Fig. 2). An SDS-PAGE gel showing the partial purification of MBP-TbPRMT5 by affinity chromatography on amylose resin (Fig. 2A) demonstrated that the protein was expressed as a 128-kDa fusion protein (compare lanes PI and I) and that it constituted the major protein in the column eluate (lane E). Minor contaminating proteins are not expected to exhibit PRMT activity, since prokaryotes lack this class of enzyme (50). The PRMT activity of MBP-TbPRMT5 was assayed in the presence of [methyl-3H]AdoMet and recombinant RBP16, a trypanosome RNA binding protein with an RG-rich C terminus that is methylated in vivo (74). Methylation assays were performed in the presence of either increasing (Fig. 2B) or fixed (Fig. 2C) concentrations of MBP-TbPRMT5 to determine the concentration and time dependence of the enzyme activity, respectively. These assays clearly demonstrated that RBP16 is methylated in vitro by MBP-TbPRMT5. Analysis of triplicate experiments showed that MBP-TbPRMT5 exhibited methyltransferase activity in both a concentration- and time-dependent manner. Thus, we conclude that, as predicted by its sequence, TbPRMT5 is a PRMT.
FIG. 2.
Recombinant MBP-TbPRMT5 exhibits time- and concentration-dependent in vitro methyltransferase activity. (A) Coomassie-stained 10% SDS-PAGE gel showing the partial purification of recombinant TbPRMT5. TbPRMT5 was cloned and expressed as an N-terminal MBP fusion protein and partially purified by affinity chromatography over amylose resin. M, markers; PI, preinduced; I, induced; E, eluate from amylose column. (B) Recombinant MBP-TbPRMT5 exhibits concentration-dependent in vitro methyltransferase activity. In vitro methylation assays were performed in triplicate for 1 h at 36°C in the presence of 1 μM [methyl-3H]AdoMet, 5 μM His-RBP16, and increasing concentrations of MBP-TbPRMT5. The reactions were stopped with the addition of SDS sample buffer, resolved by 15% polyacrylamide SDS-PAGE, and analyzed by fluorography. Bands were quantified using Bio-Rad Quantity One imaging software. The background OD was subtracted and set to 0. The graph shows the mean ± standard deviation of triplicate experiments. A representative fluorograph and corresponding Coomassie-stained gel are shown above the graph. The positions of MBP-TbPRMT5 and His-RBP16 in the Coomassie-stained gel are indicated with arrows. (C) Time course of MBP-TbPRMT5 in vitro methyltransferase activity. In vitro methyltransferase assays were performed in triplicate at 36°C in the presence of 1 μM [methyl-3H]AdoMet, 3 μM His-RBP16, and 0.3 μM MBP-TbPRMT5. Aliquots were taken at the indicated time points, and the reactions were stopped with the addition of SDS sample buffer. The reactions were analyzed as described for panel B, except baseline OD values were normalized to the minimal value of triplicate experiments. Labels as in panel B.
Recombinant TbPRMT5 exhibits broad substrate specificity.
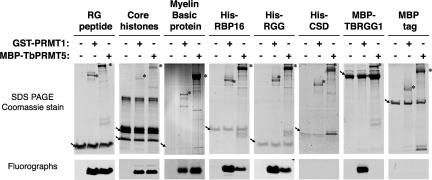
We next wanted to examine the substrate specificity of TbPRMT5. Since TbPRMT5 was predicted to be a type II PRMT, we asked whether its substrate specificity differs from that of a well-characterized type I PRMT, mammalian PRMT1 (55). To assess the substrate specificity of TbPRMT5, methylation assays were performed in the presence of [methyl-3H]AdoMet, various PRMT substrates indicated in Fig. 3, and equal molar MBP-TbPRMT5 or rat GST-PRMT1. Arginine residues found within the context of RGG, RG, or RXR motifs are common sites of methylation (reviewed in reference 60). Thus, we first determined whether TbPRMT5 methylates arginine residues in the RG context by assaying its activity toward a peptide consisting of seven RG repeats (22). TbPRMT5 robustly methylated the RG peptide in vitro to an extent similar to that of PRMT1. We next compared the substrate specificities of TbPRMT5 and PRMT1 toward well-characterized PRMT substrates that lack extensive RGG-rich regions: calf thymus core histones and myelin basic protein. While PRMT1 is reported to methylate only histone H4 (85, 87), PRMT5 has been reported to methylate histones H2A (13, 52, 76), H3 (69, 70), and H4 (52, 76). Myelin basic protein harbors MMA or sDMA modifications in vivo (5, 14) and is an in vitro substrate for human PRMT5 (13, 76). Methylation of both histone H4 and myelin basic protein reportedly occurs in the GRG context (14, 87). In our in vitro methylation assays, we found that TbPRMT5 and PRMT1 displayed very similar activities toward the core histones, methylating both histones H4 and, to a lesser extent, H2A. Both TbPRMT5 and PRMT1 exhibited methylation activities toward myelin basic protein, with TbPRMT5 showing greater activity, as expected for a type II enzyme. These data demonstrate that TbPRMT5 is highly active toward the RG peptide and suggest that proteins harboring arginine residues found within this context may also be good substrates for this enzyme.
FIG. 3.
Recombinant MBP-TbPRMT5 exhibits broad substrate specificity. MBP-TbPRMT5 exhibits in vitro methyltransferase activity toward a synthetic RG peptide, core histones H4 and H2A (arrows), myelin basic protein, and the trypanosome RNA binding protein, His-RBP16. For the core histone substrates, in vitro methylation assays were performed at 36°C for 1 h in the presence of 1 μM [methyl-3H]AdoMet, 10 μg calf thymus core histones, and 0.1 μM of either rat GST-PRMT1 or MBP-TbPRMT5. For the remaining substrates, in vitro methylation assays were performed at 36°C for 1 h in the presence of 1 μM [methyl-3H]AdoMet, 30 μM RG peptide or 1.2 μM of the indicated substrates, and 0.3 μM of either rat GST-PRMT1 or MBP-TbPRMT5. The reaction mixtures were resolved on 10 or 15% SDS-PAGE gels, and tritiated substrates were visualized by fluorography. Representative fluorographs and corresponding Coomassie-stained SDS-PAGE gels of triplicate experiments are shown. The positions of enzymes and substrates are indicated with asterisks and arrows, respectively. Note that a breakdown product of MBP-TbPRMT5 comigrates with myelin basic protein. The fluorographs correspond to the regions of the gels indicated above with arrows.
Since RNA binding proteins harboring RGG-rich motifs are preferred substrates for type I enzymes (reviewed in reference 60), we next assessed the specificity of TbPRMT5 toward similar proteins of trypanosome origin. We demonstrated (Fig. 2) that TbPRMT5 could utilize the trypanosome RBP16 protein as a substrate. RBP16 is methylated in both RGG and non-RGG contexts in vivo (74). When we compared the activity of TbPRMT5 toward this substrate with that of PRMT1, we found that TbPRMT5 exhibited relatively weak activity toward RBP16 compared to PRMT1. Analysis of the RBP16 cold shock domain (His-CSD) and RGG domain (His-RGG) (63) confirmed that both enzymes exclusively methylated residues within the RGG-containing C terminus of the protein. We next examined the activity of TbPRMT5 toward the RGG-rich T. brucei mitochondrial-RNA binding protein, TBRGG1 (Fig. 3, MBP-TBRGG1) (91). We previously demonstrated that the N-terminal RGG domain of TBRGG1 is methylated in vitro by the trypanosome type I enzyme, TbPRMT1 (72). Interestingly, TbPRMT5 is completely unable to methylate MBP-TBRGG1, although the same protein was robustly methylated by PRMT1 in a parallel assay. A control assay with the MBP tag alone confirmed that methylation of MBP-TBRGG1 by PRMT1 occurred within the TBRGG1 portion of the protein. Taken together, these results indicate that TbPRMT5 exhibits unexpectedly broad substrate specificity. Based on the robust methylation of the RG peptide and the reported in vivo methylation of histone H4 and myelin basic protein occurring on arginines in the context of GRG (14, 87), these data suggest that TbPRMT5 has a preference for methylation of arginines within the context of GRG, as opposed to those within extensive RGG motifs.
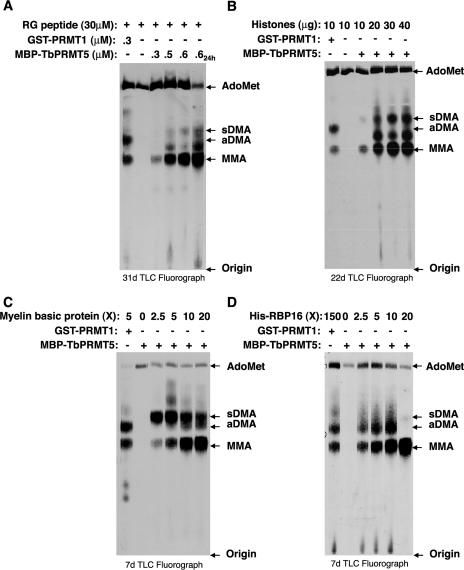
Recombinant TbPRMT5 catalyzes the formation of monomethylarginine and symmetric dimethylarginine in proteins.
TbPRMT5 is predicted by its sequence to be a type II enzyme and therefore is predicted to synthesize MMA and sDMA modifications. Since TbPRMT5 displays substrate specificity with many similarities to rat PRMT1, it was important to directly analyze the modifications catalyzed by TbPRMT5 in vitro. To this end, we determined the identity of [methyl-3H]arginine modifications in various TbPRMT5 substrates after acid hydrolysis and fractionation of substrate amino acids by TLC alongside MMA, aDMA, sDMA, and AdoMet standards (Fig. 4). Methylation assays were performed in the presence of [methyl-3H]AdoMet and either increasing concentrations of MBP-TbPRMT5 (Fig. 4A) or the indicated substrates (Fig. 4B, C, and D). Control reactions for each substrate were performed in the presence of GST-PRMT1 (Fig. 4A to D) and in the absence of either enzyme (Fig. 4A and B) or substrate (Fig. 4C and D). With all substrates, we observed that PRMT1 catalyzed the formation of MMA and aDMA, as previously demonstrated (55). The methylation profile for TbPRMT5-catalyzed reactions differed from that of PRMT1. Amino acid analysis of all four substrates clearly demonstrated that TbPRMT5 catalyzes the formation of MMA (Fig. 4A to D). In the presence of RG peptide (Fig. 4A), core histones (Fig. 4B), or myelin basic protein (Fig. 4C), TbPRMT5 also catalyzed the formation of a substantial amount of sDMA. A small amount of sDMA synthesis was observed with the RBP16 substrate at the highest substrate concentration, although MMA was clearly the most dominant modification on this substrate and aDMA also appeared to be present (Fig. 4D).
FIG. 4.
Recombinant MBP-TbPRMT5 catalyzes the formation of monomethylarginine and symmetric dimethylarginine in a substrate- and concentration-dependent manner. (A) Amino acid analysis of an RG peptide methylated in vitro by recombinant MBP-TbPRMT5. In vitro methylation assays were performed at 36°C for 1 or 24 h (far-right lane) in the presence of 1 μM [methyl-3H]AdoMet, 30 μM RG peptide substrate, and either 0.3 μM GST-PRMT1 or the indicated concentrations of MBP-TbPRMT5. Samples were acid hydrolyzed to free amino acids and resolved by TLC alongside 30 nmol of the indicated amino acid standards shown to the right, as described in Materials and Methods. (B) Amino acid analysis of methylated calf thymus core histones. Assays were performed for 16 h at 36°C in the presence of 1 μM [methyl-3H]AdoMet, 10 to 40 μg core histone substrate, and either 0.07 μM GST-PRMT1 or 0.13 μM MBP-TbPRMT5. The reactions were stopped and analyzed as described for panel A. (C) Amino acid analysis of methylated myelin basic protein. Assays were performed for 16 h at 36°C in the presence of 1 μM [methyl-3H]AdoMet, the indicated molar excess (X) of myelin basic protein substrate, and either 2.2 μM GST-PRMT1 or 0.5 μM MBP-TbPRMT5. The reactions were stopped and analyzed as described for panel A. (D) Amino acid analysis of methylated His-RBP16. Assays were performed for 16 h at 36°C in the presence of 1 μM [methyl-3H]AdoMet, the indicated molar excess (X) of His-RBP16 substrate, and either 0.05 μM GST-PRMT1 or 0.7 μM MBP-TbPRMT5. The reactions were stopped and analyzed as described for panel A.
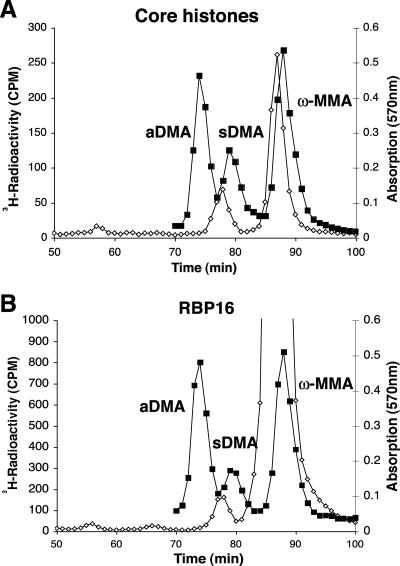
In our TLC analyses, we frequently observed an unknown [3H]methyl species that comigrated between the MMA and aDMA standards in the presence of RG peptide and core histone substrates (Fig. 4A and B). In addition, we suspected that the RBP16 [3H]methyl species observed comigrating with or near aDMA by TLC (Fig. 4D) might be the same or a similar unknown [3H]methyl species. Because of the possibility that [methyl-3H]AdoMet breakdown products might complicate the TLC analyses, especially in the presence of substrate proteins, we decided to use high-resolution cation ion-exchange amino acid analysis, in which larger amounts of hydrolyzed proteins can be fractionated, to confirm that TbPRMT5 catalyzes the production of solely MMA and sDMA and no other novel products (Fig. 5). Amino acid analysis of core histones (Fig. 5A) and His-RBP16 (Fig. 5B) substrates revealed only MMA and sDMA and did not reveal the presence of a novel [3H]methyl species that exhibited a difference in comigration compared to MMA, aDMA, or sDMA standards. These results confirmed that TbPRMT5 catalyzes the formation of MMA and sDMA in core histones. Cation-exchange chromatography also indicated that TbPRMT5 catalyzes primarily MMA, and a relatively small amount of sDMA, in the trypanosome RNA binding protein His-RBP16 (Fig. 5B). The identities of the peak radioactive fractions 76 to 79 for the His-RBP16 sample were further confirmed to be sDMA, after the samples were pooled, desalted, and analyzed by TLC alongside MMA, aDMA, and sDMA amino acid standards (data not shown). In conclusion, combined TLC and cation-exchange analyses indicate that TbPRMT5 catalyzes the formation of MMA and sDMA in proteins and is therefore a type II enzyme.
FIG. 5.
Amino acid analysis detects monomethylarginine and symmetric dimethylarginine in core histones and RBP16. Purified MBP-TbPRMT5 was incubated with the methyl-accepting substrate core histones (A) or His-RBP16 (B) and analyzed by amino acid analysis as described in Materials and Methods. TCA-precipitated and acid-hydrolyzed samples were applied to a cation-exchange column with unlabeled MMA, aDMA, and sDMA standards. To detect 3H radioactivity (open diamonds), 200 μl of each fraction was mixed with 400 μl water and 5 ml fluor. Radioactivity was determined using a Beckman LS6500 counter as an average of three 3-min counting cycles. Unlabeled standards (closed squares) were detected with a ninhydrin assay using 100 μl of each fraction (66). As expected, the 3H radiolabel for sDMA and MMA migrated slightly ahead of the nonisotopically labeled standards due to a tritium isotope effect in the ion-exchange chromatography, where three tritium atoms replace three hydrogen atoms on each methyl group (35, 40).
TbPRMT5 is constitutively expressed in BF and PF life stages of the parasite, and localizes to the cytoplasm.
Having identified TbPRMT5 as a type II PRMT, we next wanted to determine the properties of the enzyme in vivo. To assess whether TbPRMT5 is differentially expressed in BF and PF life stages of the parasite, we analyzed RNA isolated from BF and PF trypanosomes by Northern blotting using a TbPRMT5-specific probe (Fig. 6A). We detected one transcript of approximately 3,200 nucleotides, indicating the presence of combined 5′ and 3′ untranslated regions of approximately 850 nucleotides. The abundances of this transcript were similar in BF and PF life stages, indicating that TbPRMT5 is constitutively expressed throughout the T. brucei life cycle.
FIG. 6.
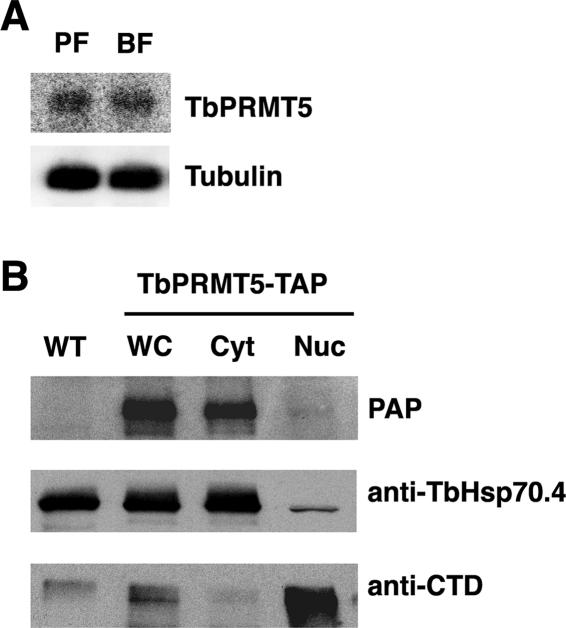
TbPRMT5 is constitutively expressed in PF and BF life stages and localizes to the cytoplasm. (A) Northern blot analysis of TbPRMT5 expression in PF and BF life stages. Twenty micrograms of total RNA isolated from log-phase PF and BF parasites was resolved on a 1.5% formaldehyde-agarose gel, transferred to a nylon membrane, and hybridized with a full-length TbPRMT5 antisense riboprobe. To normalize for loading, the membrane was stripped and hybridized with a 5′ end-labeled oligonucleotide probe complementary to tubulin mRNA. (B) TbPRMT5 localizes to the cytoplasm. To determine the subcellular localization of TbPRMT5, a PF cell line expressing TbPRMT5-TAP was subjected to subcellular fractionation, followed by immunoblot analysis. Fifteen micrograms of wild-type (WT) or TbPRMT5-TAP (WC) whole-cell extract was resolved by SDS-PAGE alongside equivalent microgram amounts of TbPRMT5-TAP cytoplasmic (Cyt) and nuclear (Nuc) extracts. The proteins were electroblotted to nitrocellulose, and the fractionation of TbPRMT5-TAP was analyzed using the TAP-specific PAP soluble-complex reagent. The efficiency of subcellular fractionation was assessed by Western blotting using antibodies against cytoplasmic (TbHsp70.4) and nuclear (RNAP CTD) proteins.
To determine the subcellular localization of TbPRMT5, we engineered a PF cell line constitutively expressing a C-terminal TAP-tagged fusion protein. Topology prediction programs did not detect a signal peptide or localization signal for TbPRMT5, and the protein was therefore predicted to localize to the cytoplasm. Whole-cell extracts were prepared from log-phase TbPRMT5-TAP-expressing cells and fractionated into cytoplasmic and nuclear subcellular fractions. The subcellular fractionation of TbPRMT5-TAP was analyzed by immunoblotting using the PAP reagent, which detects the protein A moiety of the TAP tag (Fig. 6B). As a control to assess the efficiency of cytoplasmic and nuclear fractionations, the membrane was stripped and analyzed by Western blotting using anti-TbHsp70.4 and anti-CTD antibodies, respectively. As shown in Fig. 6B, the expected 107-kDa fusion protein was expressed only in TbPRMT5-TAP-expressing cells and was absent from wild-type cells. Analysis of subcellular fractions indicated that TbPRMT5-TAP was present almost exclusively in the cytoplasmic fraction and was essentially absent from the nucleus. Therefore, similar to the localization reported for human PRMT5 (77), TbPRMT5 is predominantly localized to the cytoplasm.
TbPRMT5 associates with a homolog of the DEAD box RNA helicase, Ded1p, and novel trypanosome proteins.
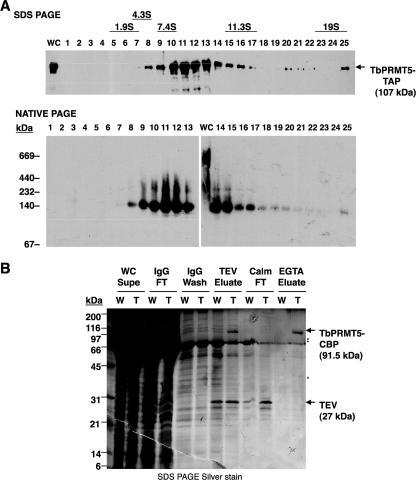
To investigate the possible biological processes in which TbPRMT5 may play a role, we utilized our PF TbPRMT5-TAP-expressing cell line to analyze the compositions of native TbPRMT5 protein complexes (Fig. 7). To address whether TbPRMT5 is present in macromolecular complexes, we subjected whole-cell extract prepared from TbPRMT5-TAP-expressing cells to glycerol gradient fractionation (Fig. 7A). Fractions from a 5 to 20% glycerol gradient were first resolved by SDS-PAGE and analyzed by immunoblotting using the TAP-specific PAP reagent (Fig. 7A, top). TbPRMT5-TAP-containing complexes sedimented predominantly at 8 to 10S, with minor complexes exhibiting sedimentation values up to 19S. Monomeric TbPRMT5-TAP was expected to exhibit sedimentation values of less than 7.4S. Therefore, the sedimentation pattern of TbPRMT5-TAP suggested the presence of both monomers and higher-order complexes. To further characterize higher-order TbPRMT5-TAP-containing complexes, we then fractionated both whole-cell extract and glycerol gradient fractions by nondenaturing PAGE (Fig. 7A, bottom). These analyses demonstrated that in whole-cell extract TbPRMT5-TAP is a component of protein complexes approximately 250 kDa and 700 kDa in mass (Fig. 7A, bottom, lane WC). The larger of these complexes is less stable, as it does not withstand glycerol gradient fractionation (Fig. 7A, bottom, fractions 1 to 25). Upon native gel analysis of 5 to 20% glycerol gradient fractionation, we observed primarily two bands with apparent molecular masses of approximately 140 kDa and 250 kDa, suggesting that monomeric and dimeric forms of the enzyme may be in equilibrium during the gradient sedimentation and native PAGE. Taken together, these results suggest that TbPRMT5 is present in relatively unstable macromolecular complexes, which may be either homo- or heteromultimers.
FIG. 7.
Analysis of in vivo TbPRMT5 protein complexes. (A) Glycerol gradient fractionation of TbPRMT5-TAP-containing complexes. Whole-cell extract prepared from PF trypanosomes ectopically expressing a TbPRMT5-TAP fusion protein was fractionated by centrifugation on a 5 to 20% glycerol gradient. The fractions were resolved by either denaturing polyacrylamide gel electrophoesis (SDS PAGE; top) or nondenaturing PAGE (native PAGE; bottom), and TbPRMT5-TAP-containing complexes were analyzed by immunoblotting using the TAP-specific PAP reagent. Twenty micrograms of whole-cell extract (WC) was analyzed by both methods for comparison. The sedimentation values and molecular masses of protein standards are indicated. (B) Tandem-affinity purification of native TbPRMT5 protein complexes. Wild type (W) or TbPRMT5-TAP (T) whole-cell extracts were fractionated by tandem-affinity purification over consecutive IgG-Sepharose and calmodulin resin columns. A silver-stained SDS-PAGE gel of TbPRMT5-CBP purification and complex composition is shown. TbPRMT5-CBP (arrow; 91.5 kDa) was eluted from IgG-Sepharose resin after TEV protease cleavage (TEV eluate), purified over calmodulin resin (Calm), and eluted under native conditions with EGTA (EGTA eluate). Major copurifying proteins present only in eluates of TAP-expressing cells (EGTA eluate; cf. lanes W and T) are indicated (asterisks). WC Supe, whole-cell supernatant; IgG FT, IgG-Sepharose resin flowthrough; TEV, TEV protease; Calm FT, calmodulin resin flowthrough.
To identify native TbPRMT5-associated proteins, we purified TbPRMT5-TAP by tandem-affinity chromatography over consecutive IgG-Sepharose and calmodulin resin columns (Fig. 7B). Tobacco etch virus (TEV) protease cleavage of the TbPRMT5-TAP chimera exposed a C-terminal CBP moiety that could then be purified over calmodulin resin and eluted under native conditions with EGTA. The composition of TbPRMT5-TAP protein complexes were analyzed by SDS-PAGE and silver staining (Fig. 7B), or the complexes were digested with trypsin and analyzed by LC-MS/MS (Table 1). The SDS-PAGE gel in Fig. 7B shows the composition of IgG-Sepharose and calmodulin resin eluates from wild-type or TbPRMT5-TAP whole-cell extracts. As expected, TEV protease (27-kDa protein) cleavage resulted in the elution of a prominent 91.5-kDa protein that was present in TAP eluates and not in wild-type eluates. This protein was not present in the calmodulin resin flowthrough but was eluted with EGTA from calmodulin resin, indicating that it was TbPRMT5-CBP. Three TbPRMT5-copurifying proteins with apparent molecular weights of 40, 75, and 85 were visible only in the TAP EGTA eluates, where they were present in approximately equivalent ratios. Mass spectrometry analysis (Table 1) did not detect TbPRMT5-interacting proteins with the predicted molecular masses of 40 and 85 kDa. Since PRMT5 homologs are reported to interact with themselves (32) and to homo-oligomerize (77), we suspect that the copurifying 85-kDa band was endogenous TbPRMT5 (the theoretical mass is 86.7 kDa). Mass spectrometry analysis of final TAP eluates did reveal the presence of a predicted 71.3-kDa protein, which may represent the 75-kDa protein observed by silver staining. This protein (Tb10.61.2130) contains an RGG-rich C terminus and a DEAD box motif, and the latter defines it as a putative RNA helicase. BLAST search analyses performed against human, yeast, and fruitfly genomes indicated that this putative RNA helicase is related to the translation initiation factors Ded1p from S. pombe (43) and S. cerevisiae (21, 26) and Drosophila melanogaster Vasa (16, 36). This protein, which we refer to as TbDED1, displays 49/64% and 41/61% amino acid identity/similarity to Ded1p and Vasa, respectively. The presence of an RGG-rich C terminus suggests that TbDED1 is a substrate for arginine methylation and further substantiates the interaction observed between this protein and TbPRMT5.
TABLE 1.
TbPRMT5-TAP-associated proteins identified by mass spectrometry
| Locus tag | Name/description | Molecular mass (kDa) | Molecular functiona | Biological processa |
|---|---|---|---|---|
| Tb09.160.4250 | TRYP1; tryparedoxin peroxidase | 22.4 | Trypanothione-disulfide reductase activity (ISS) | Electron transport (ISS) |
| Tb10.61.2130 | ATP-dependent DEAD/H RNA helicase; putative | 71.3 | ATP binding (IEA, ISS); ATP-dependent helicase activity (IEA, ISS); nucleic acid binding (IEA) | Nucleobase, nucleoside, nucleotide, and nucleic acid metabolism (ISS) |
| Tb10.389.1040 | Hypothetical protein; no conserved domains | 121.2 | Nucleic acid binding (IEA) | Unknown |
| Tb09.160.1400 | Hypothetical protein; no conserved domains | 147.2 | Unknown | Unknown |
T. brucei GeneDB Gene Ontology evidence codes: IEA, inferred by electronic annotation; ISS, inferred from sequence or structural similarity.
Mass spectrometry analysis also identified two hypothetical conserved proteins (Tb10.389.1040 and Tb09.160.1400) in TbPRMT5-TAP eluates. These hypothetical T. brucei proteins have predicted molecular masses of 121.2 and 147.2 kDa, respectively, and have no homologs outside the kinetoplastid protozoa. While they do not contain RG-rich or conserved domains, the methylation site prediction program MeMo (19; http://www.bioinfo.tsinghua.edu.cn/∼tigerchen/memo.html), identified several potential sites of arginine methylation in both proteins. In addition, Tb10.389.1040 was inferred by electronic annotation to possess nucleic acid binding activity, suggesting that this protein may play a role in nucleic acid metabolism. Another novel protein that was identified in TbPRMT5-TAP eluates was the cytosolic tryparedoxin peroxidase, which is involved in the detoxification of hydrogen peroxides produced from aerobic metabolism (90). Interestingly, no obvious homologs of the human PRMT5-containing 20S methylosome (MEP50/pICln/p37) were identified in either the T. brucei genomic database or TbPRMT5-TAP eluates. Taken together, our results suggest that TbPRMT5 is present in novel kinetoplastid-specific complexes.
Endogenous cofactors do not appear to modulate the enzymatic properties of TbPRMT5.
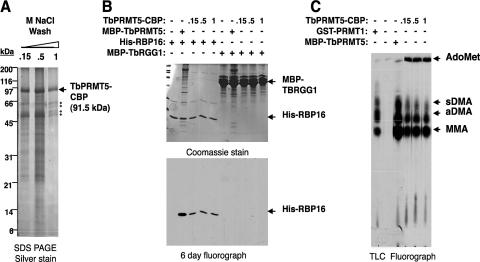
We demonstrated that bacterially expressed TbPRMT5 is competent to catalyze the formation of both MMA and sDMA in the absence of trypanosome cofactors (Fig. 2 to 5). To determine if endogenous cofactors modulate the enzymatic properties of TbPRMT5, we next examined the activity of TbPRMT5 isolated from its native context (Fig. 8). Since TbPRMT5 localizes to the cytoplasm (Fig. 6), we isolated TbPRMT5-CBP by tandem-affinity chromatography from PF cytosolic extract and assessed its substrate specificities toward RBP16 and TBRGG1 trypanosome proteins. We first assessed the affinity of TbPRMT5-associated proteins by running three separate calmodulin columns that were washed with increasing concentrations of NaCl (0.15, 0.5, or 1 M). Figure 8A shows a silver-stained SDS-PAGE gel of the final calmodulin resin eluates. A large number of proteins copurify with TbPRMT5-CBP under the two lower-salt wash conditions. However, when washes were performed with 1 M NaCl, the majority of these proteins were removed, with the exception of four major salt-resistant copurifying proteins. These salt-resistant proteins were most likely contaminating CBPs, since several were detected by mass spectrometry in native TbPRMT5-TAP eluates (data not shown). We did not observe a copurifying protein corresponding to the molecular mass of endogenous TbPRMT5 (86.7 kDa) in these eluates, although we cannot rule out the possibility that it was present below the level of detection. To determine if endogenous trypanosome cytosolic factors act to modulate the substrate specificity of TbPRMT5, we compared the activities of TbPRMT5 prepared under different salt washes to those of recombinant enzyme (Fig. 8B). Identical to the bacterially produced TbPRMT5 protein, TbPRMT5-CBP methylated RBP16 but did not display any activity toward the TBRGG1 substrate. The complete absence of TBRGG1 methylation, even with preparations washed at relatively low salt (0.15 M), indicated that the previously characterized type I enzyme, TbPRMT1, did not copurify with TbPRMT5. In the presence of the RBP16 substrate, we observed similar levels of activity with TbPRMT5-CBP prepared under all salt concentrations. Thus, TbPRMT5-associated proteins do not appear to regulate either the substrate specificity or activity of the enzyme.
FIG. 8.
The enzymatic properties of TbPRMT5-CBP purified from T. brucei are equivalent to those of recombinant enzyme. (A) Silver-stained SDS-PAGE gel showing the final eluates of TbPRMT5-CBP purified by tandem-affinity purification over IgG-Sepharose and calmodulin resins. Three separate purifications were performed. Prior to final elution, the eluates were washed with either 0.15, 0.5, or 1 M NaCl. The asterisks denote salt-resistant TbPRMT5-CBP-copurifying proteins. (B) TbPRMT5-CBP methylates His-RBP16, but not MBP-TBRGG1, in vitro. In vitro methylation assays were performed at 36°C for 1 h in the presence of 1 μM [methyl-3H]AdoMet, 1.4 μM substrate, and either 0.1 μM recombinant MBP-TbPRMT5 or 0.6 nM TbPRMT5-CBP washed with 0.15, 0.5, or 1 M NaCl prior to elution. The reaction mixtures were analyzed by SDS-PAGE (top) and fluorography (bottom). (C) Amino acid analysis of His-RBP16 methylated in vitro by TbPRMT5-CBP. In vitro methylation assays were performed at 36°C for 16 h in the presence of 0.9 μM [methyl-3H]AdoMet, 2 μM His-RBP16, and either 0.02 μM rat GST-PRMT1, 0.2 μM recombinant MBP-TbPRMT5, or 1.1 nM TbPRMT5-CBP washed prior to elution with 0.15, 0.5, or 1 M NaCl. The reactions were stopped by TCA precipitation, acid hydrolyzed to free amino acids, and analyzed by TLC, as described in Materials and Methods. All abbreviations are as in Fig. 4.
Since recombinant TbPRMT5 catalyzed predominantly MMA modifications of His-RBP16 (Fig. 4 and 5), we wanted to determine if the presence of endogenous factors would modulate the ratio of TbPRMT5-catalyzed MMA to DMA modifications (Fig. 8C). To assess this, we performed methylation assays in the presence of [3H]AdoMet, His-RBP16, and TbPRMT5-CBP washed at either 0.15, 0.5, or 1 M NaCl prior to final elution. Control reactions were performed in the absence or presence of either recombinant TbPRMT5 or rat PRMT1. The reaction mixtures were acid hydrolyzed and analyzed by TLC, as described in the legend to Fig. 4. The results presented in Fig. 8C demonstrate that recombinant TbPRMT5 and TbPRMT5-TAP catalyze similar ratios of MMA and sDMA modifications. The product of unknown identity, migrating between MMA and aDMA, that was described previously (Fig. 4) was also present in TbPRMT5 and PRMT1 reactions. Overall, these data demonstrate that TbPRMT5 ectopically expressed in PF T. brucei is present in high-molecular-weight complexes with methyltransferase activity indistinguishable from that exhibited by recombinant TbPRMT5.
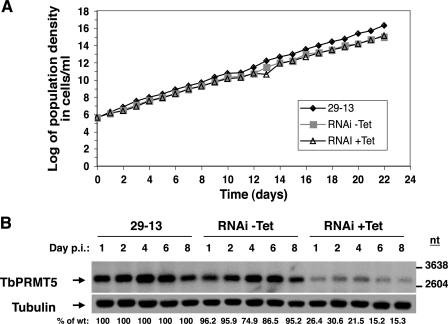
TbPRMT5 is not essential for growth in PF trypanosomes.
To determine if TbPRMT5 is essential for growth, we disrupted its expression via tetracycline-inducible RNAi and monitored cell growth daily for 22 days (Fig. 9A). Comparison of parental 29-13 cells with uninduced and induced RNAi cells indicated that TbPRMT5 was not essential for growth. To confirm that TbPRMT5 expression was disrupted, RNA was isolated on days 1, 2, 4, 6, and 8 postinduction and analyzed by Northern blotting using a TbPRMT5-specific riboprobe (Fig. 9B). After normalization to a tubulin loading control, we observed a maximum 85% reduction in TbPRMT5 mRNA by day 6 postinduction. These results indicate that TbPRMT5 is not essential for growth under normal log-phase conditions in PF trypanosomes or, alternatively, that an 85% reduction in TbPRMT5 mRNA is not great enough to confer a growth defect.
FIG. 9.
TbPRMT5 is not essential for growth in PF trypanosomes. (A) Growth curve of TbPRMT5-disrupted cells. TbPRMT5 expression in PF trypanosomes was disrupted via tetracycline-inducible RNAi. The cumulative cell density of wild-type (29-13), and tetracycline-induced (RNAi +Tet) and uninduced (RNAi −Tet) RNAi cells was monitored daily for 22 days. (B) Northern blot analysis of TbPRMT5 RNAi-disrupted cells. For the analysis of TbPRMT5 expression in RNAi-expressing cells, total RNA (10 μg) isolated on days 1, 2, 4, 6, and 8 postinduction (p.i.) from wild-type, uninduced, and induced RNAi cells was analyzed using a full-length TbPRMT5 riboprobe. As a control for loading, the membrane was stripped and rehybridized with a tubulin riboprobe. TbPRMT5 mRNA levels were analyzed by densitometry and are expressed as percentages of the wild type on each day after normalization to tubulin mRNA levels.
DISCUSSION
In this report, we characterized an evolutionarily divergent PRMT5 ortholog in T. brucei, termed TbPRMT5. TbPRMT5 is constitutively expressed in the BF and PF life stages of the parasite (Fig. 6A) and is not essential for growth in PF trypanosomes (Fig. 9). Primary sequence analysis revealed that TbPRMT5 contains a conserved methyltransferase domain, lacks the THW loop that is present in type I PRMTs but frequently absent from type II enzymes, and has an extended N terminus compared to PRMT5 homologs in higher eukaryotes (Fig. 1). Recombinant TbPRMT5 exhibits methyltransferase activity (Fig. 2) with substrate specificity toward a synthetic RG peptide, calf thymus core histone H4 and H2A, myelin basic protein, and RBP16 substrates and apparently prefers to methylate arginine residues found within the context of GRG- as opposed to RGG-rich regions (Fig. 3). Substrate amino acid analysis indicated that TbPRMT5 catalyzes the formation of MMA and sDMA, demonstrating that it is a type II enzyme, as predicted by its primary sequence (Fig. 4 and 5). Like human PRMT5 (77), TbPRMT5 localizes almost exclusively to the cytoplasm (Fig. 6B) and is present in high-molecular-weight complexes (Fig. 7A). In PF trypanosomes, TbPRMT5-TAP copurifies with a DEAD box containing putative RNA helicase, which is related to a family of DEAD box proteins that includes yeast Ded1p and Drosophila Vasa (reviewed in references 56 and 67). In addition, TbPRMT5 associates with novel proteins, including tryparedoxin peroxidase (90) and two hypothetical conserved proteins that are present only in kinetoplastids (Table 1).
TbPRMT5 displays intrinsic type II PRMT activity (Fig. 4 and 5) and does not appear to be dependent on additional cofactors for the modulation of this activity (Fig. 8). This report constitutes the first demonstration of sDMA synthesis by a bacterially expressed PRMT5 homolog. Moreover, the ability of recombinant TbPRMT5 to robustly methylate numerous substrates in vitro is in contrast to what has been observed for PRMT5 from higher eukaryotes. Several groups have reported that bacterially expressed mammalian PRMT5 is inactive (32, 50, 70), although low levels of activity have been demonstrated in some cases (77, 83). In vivo in higher eukaryotes, PRMT5 homo-oligomerizes (76, 77) and complexes with MEP50, pICln, and p37 to form the 20S methylosome, which is responsible for spliceosomal UsnRNP assembly and subsequent pre-mRNA splicing (8, 32, 33, 61). FLAG-tagged PRMT5 in the context of the methylosome efficiently methylates a variety of substrates (32), and gel filtration chromatography indicated that active forms of PRMT5 in metazoans are present only in multimeric complexes (54). It has been suggested that dimerization of PRMT5 and subsequent association with MEP50 is required to promote biological activity (50). Moreover, genetic experiments recently demonstrated that the Drosophila MEP50 homolog is required for activity and/or stability of the PRMT5 homolog in vivo (3, 39). While we did observe higher-order TbPRMT5-containing complexes in vivo, we did not find that the activity of TbPRMT5-TAP expressed in T. brucei differed significantly from that of the recombinant enzyme in the nature of the products synthesized (Fig. 4 and 8). Moreover, column washes of TbPRMT5-TAP with NaCl concentrations ranging from 150 mM to 1 M did not alter the activity of the enzyme (Fig. 8). We cannot rule out the possibility that TbPRMT5 activity or substrate specificity is modulated by factors that are not detected in our assays. Nevertheless, our data suggest that TbPRMT5 differs from PRMT5 in higher eukaryotes in that its activity is not significantly affected by its interaction with additional trypanosome-encoded factors. Consistent with the lack of cofactor requirements to promote TbPRMT5 stability and/or activity, we detected no obvious homologs of MEP50, pICln, or p37 in the T. brucei genome or in copurifying TbPRMT5 native complexes (Fig. 7 and Table 1).
We identified the mitochondrial RNA binding protein RBP16 as an in vitro substrate for TbPRMT5. It is not known, however if RBP16 is a TbPRMT5 substrate in vivo. RBP16 facilitates specific RNA-editing events, stabilizes a subset of mitochondrial mRNAs, and is a constituent of a major guide RNA-containing ribonucleoprotein (gRNP) particle in T. brucei mitochondria (42, 73). Methylation of RBP16 is critical for at least some of its mRNA stabilization functions, as well as for gRNP assembly and/or stability (41, 42). We recently demonstrated that two of three known methylarginines in RBP16 are exclusively methylated in vivo by TbPRMT1 (Arg78 and Arg85) whereas a yet-unidentified PRMT is responsible for the constitutive methylation of Arg93 (41). TbPRMT5 expressed recombinantly in either E. coli or T. brucei catalyzes predominantly MMA modifications toward RBP16 (Fig. 4D, 5B, and 8C). This preference may be due to a low affinity of TbPRMT5 for RBP16, resulting in highly distributive activity. Since RBP16 Arg93 is constitutively dimethylated and TbPRMT5 primarily monomethylates RBP16 in vitro, it is unclear whether TbPRMT5 is the other RBP16-methylating enzyme in vivo. Analysis of TbPRMT5-TAP activity copurified from trypanosome cellular extracts suggested that endogenous cofactors do not influence the substrate specificity or enzymatic properties of this enzyme (Fig. 8C). However, we cannot rule out the possibility that factors not isolated under our conditions increase the processivity of TbPRMT5 toward RBP16 in vivo. It is also feasible that monomethylation of RBP16 Arg93 by TbPRMT5 is a prerequisite for subsequent dimethylation by another enzyme. Determination of the methylation status of RBP16 in TbPRMT5 knockdown cells, as well as in cells depleted of the other three putative PRMTs, in T. brucei will provide insight into the roles of these enzymes in the apparently complicated modification of the protein.
In PF trypanosomes, TbPRMT5 associates with a putative RNA helicase, TbDED1, which is related to the Ded1p and Vasa translation initiation factors described in yeast and Drosophila, respectively. TbDED1 has an RGG-rich C terminus, which is lacking in yeast Ded1p, suggesting that TbDED1 is likely an endogenous substrate for methylation. Interestingly, the Drosophila Vasa protein possesses an RGG-rich N terminus and is methylated in vitro by the type I PRMT, DART1 (11), although its methylation status in vivo is unknown. Our results constitute the first report of any physical association between a PRMT5 homolog and Ded1p/Vasa family proteins (Table 1). However, there is evidence from Drosophila supporting a role for PRMT5-catalyzed methylation in the function of the Vasa helicase. First, Vasa and the D. melanogaster PRMT5 homolog, termed Capsuleen, were reported to act in the same germ line differentiation pathway (2, 3). Second, the Drosophila homolog of the MEP50 methylosome protein is required for proper localization of Vasa during fly development (17). The finding that Vasa, Capsuleen, and additional methylosome components act in the same biological pathways supports a functional significance for the physical association that we detect between TbPRMT5 and TbDED1. Experiments are currently under way to analyze the biological role of the TbPRMT5-TbDED1 association. In animals and fungi, roles for Ded1p/Vasa family members have been described in translation, cell cycle regulation, and differentiation (16, 43, 59, 68; reviewed in reference56). Thus, the related trypanosome protein may be involved in similar processes, which are potentially regulated by TbPRMT5. In this regard, it will be of interest to examine the effects of both TbDED1 and TbPRMT5 knockdown on cell cycle regulation in both PF and BF T. brucei, as well as the differentiation from BF to PF life stages. In addition, TbDED1 was recently reported to interact with RNA polymerase II in PF trypanosomes (24) and thus may also play a role in basal transcription.
Since trypanosomes are early-branching eukaryotes that exhibit several unique biological properties, the role of arginine methylation in these organisms is likely to diverge somewhat from that described in higher eukaryotes. In trypanosomes, genes are transcribed polycistronically and are processed into monomeric transcripts by a process mechanistically analogous to cis splicing, termed trans splicing (reviewed in reference 53). Regulation of RNA polymerase II transcription is essentially absent, and gene expression is regulated primarily at the level of RNA processing, stability, and translation (reviewed in reference 23). In mammals, PRMT5-catalyzed methylation of Sm proteins and SMN are critical for spliceosome assembly (8, 31, 32, 61, 62). Although homologous Sm proteins have been identified in T. brucei and participate in trans splicing (58, 71), trypanosome Sm proteins lack the conserved RG motifs that are methylated by the 20S methylosome in mammals. Preliminary evidence suggests that TbPRMT5 may not be involved in trans splicing, at least in PF trypanosomes (D. A. Pasternack and L. K. Read, unpublished data). Thus, it is intriguing that TbPRMT5 copurifies in novel and kinetoplastid-specific protein complexes. TbPRMT5 associates with cytosolic tryparedoxin peroxidase, which is involved in the detoxification of hydrogen peroxides produced from aerobic metabolism (90). The mitochondrial tryparedoxin peroxidase is known to regulate the oligomerization/activity of the universal minicircle sequence-binding protein (UMSBP) by redox, which is required for maintenance of the mitochondrial genome (64). Therefore, it is plausible that the association of cytosolic tryparedoxin peroxidase with TbPRMT5 may function similarly to modulate the redox state of proteins and the assembly of ribonucleoprotein complexes in vivo. We also detected the association of TbPRMT5 with two hypothetical conserved proteins for which homologs have been detected only in the genomes of the related kinetoplastid parasites T. cruzi and L. major. Neither protein contains RGG motifs, but multiple potential methylation sites have been identified in both proteins by computer prediction. While the larger of these proteins possesses no homology to any known protein or conserved domain, the 121.2-kDa hypothetical protein has been inferred by electronic annotation to possess nucleic acid binding activity, consistent with a model in which potentially novel RNA metabolic functions are affected by TbPRMT5.
PRMTs seem to have evolved early in the eukaryotic lineage, since no PRMTs have been detected in the basal eukaryote G. lamblia and only PRMT1, PRMT4 (CARM1), and PRMT5 homologs have been detected in the genome of the protozoan parasite Plasmodium falciparium (50, 81). P. falciparum is considerably less ancient than T. brucei (84), and it is interesting that its genome contains fewer putative PRMTs. The presence of five putative PRMTs in the genome of T. brucei (Fig. 1A), two of which have now been confirmed to possess PRMT activity (reference 72 and this report), may reflect a heightened importance for this posttranslational modification in order to overcome the limitations of life without transcriptional control.
Acknowledgments
This work was supported by NIH grants RO1 AI060260 to L.K.R. and R37 GM026020 to S.C. D.A.P. was supported in part by NIH Training Grant T32 AI007614.
We thank Jay Bangs and Vivian Bellofatto for providing antibodies. We thank Aswini Panigrahi and Yuko Ogata (Seattle Biomedical Research Institute) for their LC-MS/MS analysis of TbPRMT5-TAP-copurifying eluates.
Footnotes
Published ahead of print on 29 June 2007.
REFERENCES
- 1.Abramovich, C., B. Yakobson, J. Chebath, and M. Revel. 1997. A protein-arginine methyltransferase binds to the intracytoplasmic domain of the IFNAR1 chain in the type I interferon receptor. EMBO J. 16:260-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anne, J., and B. M. Mechler. 2005. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development 132:2167-2177. [DOI] [PubMed] [Google Scholar]
- 3.Anne, J., R. Ollo, A. Ephrussi, and B. M. Mechler. 2007. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134:137-146. [DOI] [PubMed] [Google Scholar]
- 4.Bachand, F., and P. A. Silver. 2004. PRMT3 is a ribosomal protein methyltransferase that affects the cellular levels of ribosomal subunits. EMBO J. 23:2641-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baldwin, G. S., and P. R. Carnegie. 1971. Isolation and partial characterization of methylated arginines from the encephalitogenic basic protein of myelin. Biochem. J. 123:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bedford, M. T., A. Frankel, M. B. Yaffe, S. Clarke, P. Leder, and S. Richard. 2000. Arginine methylation inhibits the binding of proline-rich ligands to Src homology 3, but not WW, domains. J. Biol. Chem. 275:16030-16036. [DOI] [PubMed] [Google Scholar]
- 7.Bedford, M. T., and S. Richard. 2005. Arginine methylation an emerging regulator of protein function. Mol. Cell 18:263-272. [DOI] [PubMed] [Google Scholar]
- 8.Boisvert, F. M., J. Côte, M. C. Boulanger, P. Cléroux, F. Bachand, C. Autexier, and S. Richard. 2002. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J. Cell Biol. 159:957-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boisvert, F. M., J. Côte, M. C. Boulanger, and S. Richard. 2003. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteomics 2:1319-1330. [DOI] [PubMed] [Google Scholar]
- 10.Boisvert, F. M., U. Déry, J. Y. Masson, and S. Richard. 2005. Arginine methylation of MRE11 by PRMT1 is required for DNA damage checkpoint control. Genes Dev. 19:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boulanger, M. C., T. B. Miranda, S. Clarke, M. Di Fruscio, B. Suter, P. Lasko, and S. Richard. 2004. Characterization of the Drosophila protein arginine methyltransferases DART1 and DART4. Biochem. J. 379:283-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brahms, H., L. Meheus, V. de Brabandere, U. Fischer, and R. Lührmann. 2001. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B′ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 7:1531-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Branscombe, T. L., A. Frankel, J. H. Lee, J. R. Cook, Z. Yang, S. Pestka, and S. Clarke. 2001. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem. 276:32971-32976. [DOI] [PubMed] [Google Scholar]
- 14.Brostoff, S., and E. H. Eylar. 1971. Localization of methylated arginine in the A1 protein from myelin. Proc. Natl. Acad. Sci. USA 68:765-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brun, R., and M. Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 16.Carrera, P., O. Johnstone, A. Nakamura, J. Casanova, H. Jackle, and P. Lasko. 2000. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell 5:181-187. [DOI] [PubMed] [Google Scholar]
- 17.Cavey, M., S. Hijal, X. Zhang, and B. Suter. 2005. Drosophila valois encodes a divergent WD protein that is required for Vasa localization and Oskar protein accumulation. Development 132:459-468. [DOI] [PubMed] [Google Scholar]
- 18.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 19.Chen, H., Y. Xue, N. Huang, X. Yao, and Z. Sun. 2006. MeMo: a web tool for prediction of protein methylation modifications. Nucleic Acids Res. 34:W249-W253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chern, M. K., K. N. Chang, L. F. Liu, T. C. Tam, Y. C. Liu, Y. L. Liang, and M. F. Tam. 2002. Yeast ribosomal protein L12 is a substrate of protein-arginine methyltransferase 2. J. Biol. Chem. 277:15345-15353. [DOI] [PubMed] [Google Scholar]
- 21.Chuang, R. Y., P. L. Weaver, Z. Liu, and T. H. Chang. 1997. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science 275:1468-1471. [DOI] [PubMed] [Google Scholar]
- 22.Cimato, T. R., J. Tang, Y. Xu, C. Guarnaccia, H. R. Herschman, S. Pongor, and J. M. Aletta. 2002. Nerve growth factor-mediated increases in protein methylation occur predominantly at type I arginine methylation sites and involve protein arginine methyltransferase 1. J. Neurosci. Res. 67:435-442. [DOI] [PubMed] [Google Scholar]
- 23.Clayton, C. E. 2002. Life without transcriptional control? From fly to man and back again. EMBO J. 21:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Das, A., H. Li, T. Liu, and V. Bellofatto. 2006. Biochemical characterization of Trypanosoma brucei RNA polymerase II. Mol. Biochem. Parasitol. 150:201-210. [DOI] [PubMed] [Google Scholar]
- 25.De Gaudenzi, J., A. C. Frasch, and C. Clayton. 2005. RNA-binding domain proteins in kinetoplastids: a comparative analysis. Eukaryot. Cell 4:2106-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de la Cruz, J., I. Iost, D. Kressler, and P. Linder. 1997. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:5201-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denman, R. B. 2002. Methylation of the arginine-glycine-rich region in the fragile X mental retardation protein FMRP differentially affects RNA binding. Cell. Mol. Biol. Lett. 7:877-883. [PubMed] [Google Scholar]
- 28.Dolzhanskaya, N., G. Merz, J. M. Aletta, and R. B. Denman. 2006. Methylation regulates the intracellular protein-protein and protein-RNA interactions of FMRP. J. Cell Sci. 119:1933-1946. [DOI] [PubMed] [Google Scholar]
- 29.Doyle, J. J., H. Hirumi, K. Hirumi, E. N. Lupton, and G. A. Cross. 1980. Antigenic variation in clones of animal-infective Trypanosoma brucei derived and maintained in vitro. Parasitology 80:359-369. [DOI] [PubMed] [Google Scholar]
- 30.Estevez, A. M., T. Kempf, and C. Clayton. 2001. The exosome of Trypanosoma brucei. EMBO J. 20:3831-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friesen, W. J., S. Massenet, S. Paushkin, A. Wyce, and G. Dreyfuss. 2001. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol. Cell 7:1111-1117. [DOI] [PubMed] [Google Scholar]
- 32.Friesen, W. J., S. Paushkin, A. Wyce, S. Massenet, G. S. Pesiridis, G. Van Duyne, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friesen, W. J., A. Wyce, S. Paushkin, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 277:8243-8247. [DOI] [PubMed] [Google Scholar]
- 34.Gary, J. D., and S. Clarke. 1998. RNA and protein interactions modulated by protein arginine methylation. Prog. Nucleic Acid Res. Mol. Biol. 61:65-131. [DOI] [PubMed] [Google Scholar]
- 35.Gary, J. D., W. J. Lin, M. C. Yang, H. R. Herschman, and S. Clarke. 1996. The predominant protein-arginine methyltransferase from Saccharomyces cerevisiae. J. Biol. Chem. 271:12585-12594. [DOI] [PubMed] [Google Scholar]
- 36.Gavis, E. R., L. Lunsford, S. E. Bergsten, and R. Lehmann. 1996. A conserved 90 nucleotide element mediates translational repression of nanos RNA. Development 122:2791-2800. [DOI] [PubMed] [Google Scholar]
- 37.Gibson, W. C., B. W. Swinkels, and P. Borst. 1988. Post-transcriptional control of the differential expression of phosphoglycerate kinase genes in Trypanosoma brucei. J. Mol. Biol. 201:315-325. [DOI] [PubMed] [Google Scholar]
- 38.Gilbreth, M., P. Yang, D. Wang, J. Frost, A. Polverino, M. H. Cobb, and S. Marcus. 1996. The highly conserved skb1 gene encodes a protein that interacts with Shk1, a fission yeast Ste20/PAK homolog. Proc. Natl. Acad. Sci. USA 93:13802-13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonsalvez, G. B., T. K. Rajendra, L. Tian, and A. G. Matera. 2006. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr. Biol. 16:1077-1089. [DOI] [PubMed] [Google Scholar]
- 40.Gottschling, H., and E. Freese. 1962. A tritium isotope effect on ion exchange chromatography. Nature 196:829-831. [DOI] [PubMed] [Google Scholar]
- 41.Goulah, C. C., M. Pelletier, and L. K. Read. 2006. Arginine methylation regulates mitochondrial gene expression in Trypanosoma brucei through multiple effector proteins. RNA 12:1545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulah, C. C., and L. K. Read. 2007. Differential effects of arginine methylation on RBP16 mRNA binding, gRNA binding, and gRNP formation. J. Biol. Chem. 282:7181-7190. [DOI] [PubMed] [Google Scholar]
- 43.Grallert, B., S. E. Kearsey, M. Lenhard, C. R. Carlson, P. Nurse, E. Boye, and K. Labib. 2000. A fission yeast general translation factor reveals links between protein synthesis and cell cycle controls. J. Cell Sci. 113:1447-1458. [DOI] [PubMed] [Google Scholar]
- 44.Green, D. M., K. A. Marfatia, E. B. Crafton, X. Zhang, X. Cheng, and A. H. Corbett. 2002. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. J. Biol. Chem. 277:7752-7760. [DOI] [PubMed] [Google Scholar]
- 45.Hayman, M. L., and L. K. Read. 1999. Trypanosoma brucei RBP16 is a mitochondrial Y-box family protein with guide RNA binding activity. J. Biol. Chem. 274:12067-12074. [DOI] [PubMed] [Google Scholar]
- 46.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 47.Hirumi, H., K. Hirumi, J. J. Doyle, and G. A. Cross. 1980. In vitro cloning of animal-infective bloodstream forms of Trypanosoma brucei. Parasitology 80:371-382. [DOI] [PubMed] [Google Scholar]
- 48.Janzen, C. J., J. P. Fernandez, H. Deng, R. Diaz, S. B. Hake, and G. A. Cross. 2006. Unusual histone modifications in Trypanosoma brucei. FEBS Lett. 580:2306-2310. [DOI] [PubMed] [Google Scholar]
- 49.Johnson, J. G., and G. A. Cross. 1979. Selective cleavage of variant surface glycoproteins from Trypanosoma brucei. Biochem. J. 178:689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krause, C. D., Z. H. Yang, Y. S. Kim, J. H. Lee, J. R. Cook, and S. Pestka. 2007. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 113:50-87. [DOI] [PubMed] [Google Scholar]
- 51.LaCount, D. J., B. Barrett, and J. E. Donelson. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277:17580-17588. [DOI] [PubMed] [Google Scholar]
- 52.Lee, J. H., J. R. Cook, B. P. Pollack, T. G. Kinzy, D. Norris, and S. Pestka. 2000. Hsl7p, the yeast homologue of human JBP1, is a protein methyltransferase. Biochem. Biophys. Res. Commun. 274:105-111. [DOI] [PubMed] [Google Scholar]
- 53.Liang, X. H., A. Haritan, S. Uliel, and S. Michaeli. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lim, Y., Y. H. Kwon, N. H. Won, B. H. Min, I. S. Park, W. K. Paik, and S. Kim. 2005. Multimerization of expressed protein-arginine methyltransferases during the growth and differentiation of rat liver. Biochim. Biophys. Acta 1723:240-247. [DOI] [PubMed] [Google Scholar]
- 55.Lin, W. J., J. D. Gary, M. C. Yang, S. Clarke, and H. R. Herschman. 1996. The mammalian immediate-early TIS21 protein and the leukemia-associated BTG1 protein interact with a protein-arginine N-methyltransferase. J. Biol. Chem. 271:15034-15044. [DOI] [PubMed] [Google Scholar]
- 56.Linder, P. 2003. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell 95:157-167. [DOI] [PubMed] [Google Scholar]
- 57.Liu, Q., and G. Dreyfuss. 1995. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol. Cell. Biol. 15:2800-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mandelboim, M., S. Barth, M. Biton, X. H. Liang, and S. Michaeli. 2003. Silencing of Sm proteins in Trypanosoma brucei by RNA interference captured a novel cytoplasmic intermediate in spliced leader RNA biogenesis. J. Biol. Chem. 278:51469-51478. [DOI] [PubMed] [Google Scholar]
- 59.Markussen, F. H., W. Breitwieser, and A. Ephrussi. 1997. Efficient translation and phosphorylation of Oskar require Oskar protein and the RNA helicase Vasa. Cold Spring Harbor Symp. Quant. Biol. 62:13-17. [PubMed] [Google Scholar]
- 60.McBride, A. E., and P. A. Silver. 2001. State of the arg: protein methylation at arginine comes of age. Cell 106:5-8. [DOI] [PubMed] [Google Scholar]
- 61.Meister, G., C. Eggert, D. Buhler, H. Brahms, C. Kambach, and U. Fischer. 2001. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11:1990-1994. [DOI] [PubMed] [Google Scholar]
- 62.Meister, G., and U. Fischer. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21:5853-5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller, M. M., and L. K. Read. 2003. Trypanosoma brucei: functions of RBP16 cold shock and RGG domains in macromolecular interactions. Exp. Parasitol. 105:140-148. [DOI] [PubMed] [Google Scholar]
- 64.Motyka, S. A., M. E. Drew, G. Yildirir, and P. T. Englund. 2006. Overexpression of a cytochrome b5 reductase-like protein causes kinetoplast DNA loss in Trypanosoma brucei. J. Biol. Chem. 281:18499-18506. [DOI] [PubMed] [Google Scholar]
- 65.Nichols, R. C., X. W. Wang, J. Tang, B. J. Hamilton, F. A. High, H. R. Herschman, and W. F. Rigby. 2000. The RGG domain in hnRNP A2 affects subcellular localization. Exp. Cell Res. 256:522-532. [DOI] [PubMed] [Google Scholar]
- 66.Niewmierzycka, A., and S. Clarke. 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274:814-824. [DOI] [PubMed] [Google Scholar]
- 67.Noce, T., S. Okamoto-Ito, and N. Tsunekawa. 2001. Vasa homolog genes in mammalian germ cell development. Cell Struct. Funct. 26:131-136. [DOI] [PubMed] [Google Scholar]
- 68.Obara-Ishihara, T., and H. Okayama. 1994. A B-type cyclin negatively regulates conjugation via interacting with cell cycle ‘start’ genes in fission yeast. EMBO J. 13:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pal, S., S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst, and S. Sif. 2004. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24:9630-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 23:7475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palfi, Z., S. Lücke, H. W. Lahm, W. S. Lane, V. Kruft, E. Bragado-Nilsson, B. Séraphin, and A. Bindereif. 2000. The spliceosomal snRNP core complex of Trypanosoma brucei: cloning and functional analysis reveals seven Sm protein constituents. Proc. Natl. Acad. Sci. USA 97:8967-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelletier, M., D. A. Pasternack, and L. K. Read. 2005. In vitro and in vivo analysis of the major type I protein arginine methyltransferase from Trypanosoma brucei. Mol. Biochem. Parasitol. 144:206-217. [DOI] [PubMed] [Google Scholar]
- 73.Pelletier, M., and L. K. Read. 2003. RBP16 is a multifunctional gene regulatory protein involved in editing and stabilization of specific mitochondrial mRNAs in Trypanosoma brucei. RNA 9:457-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pelletier, M., Y. Xu, X. Wang, S. Zahariev, S. Pongor, J. M. Aletta, and L. K. Read. 2001. Arginine methylation of a mitochondrial guide RNA binding protein from Trypanosoma brucei. Mol. Biochem. Parasitol. 118:49-59. [DOI] [PubMed] [Google Scholar]
- 75.Perreault, A., C. Lemieux, and F. Bachand. 2007. Regulation of the nuclear poly(A) binding protein by arginine methylation in fission yeast. J. Biol. Chem. 282:7552-7562. [DOI] [PubMed] [Google Scholar]
- 76.Pollack, B. P., S. V. Kotenko, W. He, L. S. Izotova, B. L. Barnoski, and S. Pestka. 1999. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 274:31531-31542. [DOI] [PubMed] [Google Scholar]
- 77.Rho, J., S. Choi, Y. R. Seong, W. K. Cho, S. H. Kim, and D. S. Im. 2001. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J. Biol. Chem. 276:11393-11401. [DOI] [PubMed] [Google Scholar]
- 78.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Séraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 79.Roberts, T. G., N. R. Sturm, B. K. Yee, M. C. Yu, T. Hartshorne, N. Agabian, and D. A. Campbell. 1998. Three small nucleolar RNAs identified from the spliced leader-associated RNA locus in kinetoplastid protozoans. Mol. Cell. Biol. 18:4409-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ryan, C. M., K. T. Militello, and L. K. Read. 2003. Polyadenylation regulates the stability of Trypanosoma brucei mitochondrial RNAs. J. Biol. Chem. 278:32753-32762. [DOI] [PubMed] [Google Scholar]
- 81.Saksouk, N., M. M. Bhatti, S. Kieffer, A. T. Smith, K. Musset, J. Garin, W. J. Sullivan, Jr., M. F. Cesbron-Delauw, and M. A. Hakimi. 2005. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 25:10301-10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shen, E. C., M. F. Henry, V. H. Weiss, S. R. Valentini, P. A. Silver, and M. S. Lee. 1998. Arginine methylation facilitates the nuclear export of hnRNP proteins. Genes Dev. 12:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shire, K., P. Kapoor, K. Jiang, M. N. Hing, N. Sivachandran, T. Nguyen, and L. Frappier. 2006. Regulation of the EBNA1 Epstein-Barr virus protein by serine phosphorylation and arginine methylation. J. Virol. 80:5261-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sogin, M. L., J. H. Gunderson, H. J. Elwood, R. A. Alonso, and D. A. Peattie. 1989. Phylogenetic meaning of the kingdom concept: an unusual ribosomal RNA from Giardia lamblia. Science 243:75-77. [DOI] [PubMed] [Google Scholar]
- 85.Stallcup, M. R., D. Chen, S. S. Koh, H. Ma, Y. H. Lee, H. Li, B. T. Schurter, and D. W. Aswad. 2000. Co-operation between protein-acetylating and protein-methylating co-activators in transcriptional activation. Biochem. Soc. Trans. 28:415-418. [PubMed] [Google Scholar]
- 86.Stetler, A., C. Winograd, J. Sayegh, A. Cheever, E. Patton, X. Zhang, S. Clarke, and S. Ceman. 2006. Identification and characterization of the methyl arginines in the fragile X mental retardation protein Fmrp. Hum. Mol. Genet. 15:87-96. [DOI] [PubMed] [Google Scholar]
- 87.Strahl, B. D., S. D. Briggs, C. J. Brame, J. A. Caldwell, S. S. Koh, H. Ma, R. G. Cook, J. Shabanowitz, D. F. Hunt, M. R. Stallcup, and C. D. Allis. 2001. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr. Biol. 11:996-1000. [DOI] [PubMed] [Google Scholar]
- 88.Stuart, K., E. Gobright, L. Jenni, M. Milhausen, L. Thomashow, and N. Agabian. 1984. The IsTaR 1 serodeme of Trypanosoma brucei: development of a new serodeme. J. Parasitol. 70:747-754. [PubMed] [Google Scholar]
- 89.Stuart, K. D., A. Schnaufer, N. L. Ernst, and A. K. Panigrahi. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30:97-105. [DOI] [PubMed] [Google Scholar]
- 90.Tetaud, E., C. Giroud, A. R. Prescott, D. W. Parkin, D. Baltz, N. Biteau, T. Baltz, and A. H. Fairlamb. 2001. Molecular characterisation of mitochondrial and cytosolic trypanothione-dependent tryparedoxin peroxidases in Trypanosoma brucei. Mol. Biochem. Parasitol. 116:171-183. [DOI] [PubMed] [Google Scholar]
- 91.Vanhamme, L., D. Perez-Morga, C. Marchal, D. Speijer, L. Lambert, M. Geuskens, S. Alexandre, N. Ismaïli, U. Göringer, R. Benne, and E. Pays. 1998. Trypanosoma brucei TBRGG1, a mitochondrial oligo(U)-binding protein that co-localizes with an in vitro RNA editing activity. J. Biol. Chem. 273:21825-21833. [DOI] [PubMed] [Google Scholar]
- 92.Wang, X., Y. Zhang, Q. Ma, Z. Zhang, Y. Xue, S. Bao, and K. Chong. 2007. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 26:1934-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 94.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 95.Wysocka, J., C. D. Allis, and S. Coonrod. 2006. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11:344-355. [DOI] [PubMed] [Google Scholar]
- 96.Xu, W., H. Chen, K. Du, H. Asahara, M. Tini, B. M. Emerson, M. Montminy, and R. M. Evans. 2001. A transcriptional switch mediated by cofactor methylation. Science 294:2507-2511. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, X., and X. Cheng. 2003. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 11:509-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang, X., L. Zhou, and X. Cheng. 2000. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 19:3509-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zobel-Thropp, P., J. D. Gary, and S. Clarke. 1998. δ-N-Methylarginine is a novel posttranslational modification of arginine residues in yeast proteins. J. Biol. Chem. 273:29283-29286. [DOI] [PubMed] [Google Scholar]