Abstract
Previously, we had identified FOX1 and FTR1 as iron deficiency-inducible components of a high-affinity copper-dependent iron uptake pathway in Chlamydomonas. In this work, we survey the version 3.0 draft genome to identify a ferrireductase, FRE1, and two ZIP family proteins, IRT1 and IRT2, as candidate ferrous transporters based on their increased expression in iron-deficient versus iron-replete cells. In a parallel proteomic approach, we identified FEA1 and FEA2 as the major proteins secreted by iron-deficient Chlamydomonas reinhardtii. The recovery of FEA1 and FEA2 from the medium of Chlamydomonas strain CC425 cultures is strictly correlated with iron nutrition status, and the accumulation of the corresponding mRNAs parallels that of the Chlamydomonas FOX1 and FTR1 mRNAs, although the magnitude of regulation is more dramatic for the FEA genes. Like the FOX1 and FTR1 genes, the FEA genes do not respond to copper, zinc, or manganese deficiency. The 5′ flanking untranscribed sequences from the FEA1, FTR1, and FOX1 genes confer iron deficiency-dependent expression of ARS2, suggesting that the iron assimilation pathway is under transcriptional control by iron nutrition. Genetic analysis suggests that the secreted proteins FEA1 and FEA2 facilitate high-affinity iron uptake, perhaps by concentrating iron in the vicinity of the cell. Homologues of FEA1 and FRE1 were identified previously as high-CO2-responsive genes, HCR1 and HCR2, in Chlorococcum littorale, suggesting that components of the iron assimilation pathway are responsive to carbon nutrition. These iron response components are placed in a proposed iron assimilation pathway for Chlamydomonas.
Although iron is abundant in soil, its bioavailability in the aerobic world is poor because of the relative insolubility of Fe(III) and the tight binding of iron to organic chelators (reviewed in references 33, 44, and 56). Iron is therefore one of the essential nutrients that limits virtually all forms of life, and its assimilation involves mechanisms for iron capture from otherwise inaccessible forms plus mechanisms for transport. Many organisms, especially microbes, use multiple pathways for iron uptake because of the varied forms and amounts of iron supply in nature. At the same time, because of its propensity for redox chemistry, especially in an aerobic environment, iron can be toxic to cells. Therefore, the metabolism of iron is very tightly regulated (reviewed in references 9, 13, 22, 41, and 75).
Fungi.
Because of the importance of iron nutrition to life, the mechanisms of iron assimilation and distribution have been studied in many eukaryotic organisms. One well-studied pathway for iron assimilation was discovered initially through studies of iron metabolism in Saccharomyces cerevisiae. In this pathway, iron is first solubilized by reduction of ferric to ferrous iron and is then available for uptake by high-affinity transporters such as the Fet3p/Ftr1p complex (3, 14, 27, 49, 52, 72, 81, 92). There are several transcriptionally regulated FRE genes with different substrate selectivity in S. cerevisiae. Fre1p and Fre2p are the major cell surface iron reductases that reduce ferric citrate and siderophore-bound iron, Fre3p shows selectivity for iron in complex with hydroxamate-type siderophores, and Fre4p has low-affinity activity on rhodotorulic acid. Fet3p is a plasma membrane-associated multicopper oxidase that reconverts Fe2+ to Fe3+, which is a substrate for the associated trivalent cation-specific permease Ftr1p. This type of pathway is broadly distributed in the fungi, and an analogous mechanism for iron distribution occurs also in mammals; accordingly, iron metabolism in these organisms shows a requirement for copper nutrition (see, e.g., references 4, 23, 46, 48, and 60). More recently, analysis of the iron regulon in S. cerevisiae revealed the importance of a ferrichrome transporter and also three mannoproteins located in the cell wall for utilization of iron bound to a subset of siderophores such as ferrichrome (45, 66). These pathways allow the organism to utilize a range of iron sources.
Animals.
Iron metabolism in multicellular organisms is much more complex. A diheme ferrireductase, Dcytb, whose expression is increased at the RNA level during iron deficiency, is involved in iron uptake in the duodenal brush borders of animals (54). A divalent cation transporter, DCT1 or DMT1, related to the Nramp family of transporters, then takes up ferrous iron (29, 35). Dietary iron needs to be distributed systemically. Iron movement involves sequential redox chemistry (i.e., oxidation of ferrous to ferric iron by multicopper oxidases ceruloplasmin and hephaestin and reduction by ferrireductases) and membrane transport by ferroportin/Ireg1/MTP1 (1, 19, 39, 53, 86). Extracellular iron is circulated in a nontoxic form, tightly bound to transferrin, which is taken up by the transferrin receptor, and intracellular iron is stored in oxidized form in ferritin (reviewed in references 41 and 82).
Plants.
Two iron assimilation mechanisms in plants, called strategy I and strategy II, have been studied (reviewed in references 13 and 40). Both are copper independent. In strategy I, soil acidification followed by iron reduction by a root plasma membrane reductase releases Fe2+, which can be taken up by one of the iron deficiency-induced plasma membrane IRT transporters (10, 21, 71, 84). The IRT transporters belong to the so-called ZIP family of transporters found in fungi, plants, and animals. Members of the family function to transport divalent cations (reviewed in reference 34). In strategy II, discovered in grasses, the plants secrete phytosiderophores that solubilize Fe3+ by chelation (56, 85). The iron-chelate complex is then taken up by specific high-affinity YSL transporters (12). Functional homologues of the Nramp transporters are found in strategy I and strategy II plants, but the roles of individual gene products in iron assimilation versus distribution have not been detailed yet (83). As for fungi and animals, inducible iron reductases are also key enzymes in iron utilization by plants. In Arabidopsis, a ferrireductase encoded by FRO2 is involved in root iron uptake, and an NADH-dependent ferrireductase encoded by NFR in maize is implicated in ferrireductase activity (5, 7, 71, 90).
Algae.
Iron metabolism has been studied also in Chlamydomonas, a unicellular green alga that serves as an excellent model for classical, molecular, and reverse genetic analysis of chloroplast function and of eukaryotic cilia. The involvement of inducible cell surface reductases, which appears to be a general requirement for iron metabolism in the Eukarya, is well documented in Chlamydomonas as well as in many other algae (20, 51, 77, 87), although the identities of the enzymes are not yet known. A major component in the iron assimilation pathway is a ferroxidase/ferric transporter system, analogous to the molecules found in fungi and animals (42, 47). The multicopper oxidase, encoded by FOX1, although related to the multicopper oxidases of fungi and animals, is clearly also distinct with respect to the arrangement of copper binding sites and is likely to have resulted from domain rearrangement during evolution. The 959-residue protein is associated with the plasma membrane and is coordinately expressed with the FTR1 gene, encoding a putative ferric transporter related to those in fungi. However, iron utilization by wild-type Chlamydomonas does not show a requirement for copper, suggesting that an alternative pathway for iron uptake may be induced in copper-deficient cells (47). The components of the copper-independent iron uptake pathway of Chlamydomonas are not yet known. The availability of a draft genome for Chlamydomonas opened the door to the possibility of identifying additional components in iron homeostasis by testing candidate genes, homologous to those identified in other eukaryotes, for their pattern of expression relative to iron and copper nutrition.
Diverse iron assimilation pathways operate in various algae, including siderophore-mediated mechanisms in a Scenedesmus species and a transferrin-dependent pathway in Dunaliella salina (6, 28, 88). The D. salina extracellular, plasma membrane-associated transferrin is proposed to bind ferric ion generated by a surface-exposed plasma ferroxidase (64). This suggests that secreted molecules might be important in iron acquisition pathways in green algae. Therefore, we compared the secretomes of iron-deficient versus iron-replete Chlamydomonas cells to identify two related proteins, FEA1 and FEA2.
MATERIALS AND METHODS
Strains and culture.
The strains used in this work were obtained from the Chlamydomonas culture collection (Duke University) and grown in the light (60 to 80 μmol m−2 s−1) in Tris-acetate-phosphate (TAP) medium. For experiments involving iron nutrition, wild-type strains and transformants of strain CC425 were maintained in standard TAP medium and transferred to Fe-free TAP medium supplemented with Fe-EDTA from a stock solution at the indicated concentrations (58). Previously, we had identified three stages of iron nutrition based on appearance of “chlorosis,” expression of marker genes and proteins (FOX1, ferroxidase), and rate of cell growth (55, 58). Iron-deficient cells (1 to 3 μM supplementation) are not chlorotic and exhibit growth rates and growth densities (at stationary phase) comparable to those iron-replete cells (20 μM), but they show increased expression of iron assimilation components. Iron-limited cells (<0.5 μM) are chlorotic (having 50% of the chlorophyll content of replete cultures), are inhibited in the rate and extent of cell division, and show maximal up-regulation of iron assimilation genes. The iron concentrations used in specific experiments are indicated in each figure legend. In experiments involving iron limitation where it was necessary to avoid carryover of iron from the starter culture, log-phase cells (5 × 106 to 7 × 106 cells/ml) were collected by centrifugation (1,700 × g, 5 min), washed once in iron-free TAP medium, and inoculated into the desired iron-limited medium to a density of 5 × 104 cells/ml. Growth in copper-free medium has been described previously (68). For analysis of gene expression in response to manganese or zinc deficiency, strains were grown in standard TAP medium and transferred (1:1,000 dilution of a log-phase culture [4 × 106 to 8 × 106 cells/ml] at each transfer) to TAP medium lacking the test nutrient. Cells were collected at densities indicated in the figure legends.
Genetic analysis.
Strain CC124 or CC125 (which can grow on medium containing low iron) were crossed to strain CC424 or CC425 (which require higher iron for growth). Opposite mating types were mated and dissected as described by Harris (38). Resulting progeny were scored for 2:2 segregation of arginine auxotrophy and analyzed for retention of FEA proteins in washed cells and for growth (assessed visually) in medium containing limiting (0.1 μM) iron (provided as ferrous sulfate). For testing FEA retention, cells grown in medium containing 1 μM iron were collected by centrifugation at 1,000 × g, washed twice in 1 volume of 10 mM sodium phosphate [pH 7.0], and resuspended in a minimum volume.
Isolation of proteins secreted by Chlamydomonas.
Cells of Chlamydomonas strain CC400 (cw15 mt+) were grown to mid-log phase (∼ 4 × 106 cells/ml) in TAP medium and collected under sterile conditions by centrifugation (3,800 × g, 5 min). Cells were washed twice in sterile TAP medium without Fe to remove traces of iron and inoculated into fresh TAP medium containing 0.1, 1, or 18 μM Fe-EDTA at 2.5 × 105 cells/ml. Cultures were grown for 4 days under constant light (75 μmol m−2 s−1, 150 rpm). Cells were counted before collection, and the cultures were adjusted to 0.1 M NaCl and 1 mM ɛ-amino-n-caproic acid and agitated (150 rpm) for 10 min before the cells were removed from the growth medium by centrifugation (4,000 × g, 5 min. 4°C). The supernatant was collected and centrifuged again (8,000 × g, 10 min, 4°C) to remove any remaining cells. Ammonium sulfate was added to 80% saturation, and the solution was stirred (1 h, 4°C). Precipitated proteins were collected by centrifugation (30,000 × g, 90 min, 4°C); resuspended in 5 ml of a solution containing 50 mM Tris-Cl, 10 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1 mM ɛ-amino-n-caproic acid, pH 8.0; and reprecipitated by adding 100% ice-cold trichloroacetic acid (10% [wt/vol] final concentration) and mixing by gentle rocking (1 h, 4°C). Precipitated proteins were collected by centrifugation (19,000 × g, 30 min, 4°C) and resuspended in a solution containing 0.1 M Na2CO3 and 0.1 M dithiothreitol, with volumes adjusted to reflect an equal cell number per ml in each sample. Proteins in each sample were resolved on a sodium dodecyl sulfate (SDS)-containing polyacrylamide gel (12% monomer) and visualized by staining with Coomassie blue.
Immunoblot analysis.
For immunoblot analysis, proteins were separated on an SDS-containing polyacrylamide gel (12% monomer) and transferred onto polyvinylidene difluoride (0.45 μm; Millipore) for 1 h at 4°C under constant voltage (100 V) in 25 mM Tris, 192 mM glycine, 0.01% SDS, 20% methanol. Membranes were blocked with 5% dry milk in TBS (10 mM Tris-HCl, 150 mM NaCl, pH 7.5) plus Tween 20 (0.05%, wt/vol). A 1:50,000 dilution of Chlamydomonas anti-FEA1 was used as the primary antibody, and a 1:5,000 dilution of goat anti-rabbit horseradish peroxidase was used as the secondary antibody. Signals were detected using an Amersham ECL kit.
Identification of polypeptides.
The 43- and 38-kDa bands were excised by a spot excision robot (Proteome Works; Bio-Rad) and deposited into 96-well plates. Gel spots were washed and digested with sequencing-grade trypsin (Promega, Madison, WI), and the resulting tryptic peptides were extracted using standard protocols (79). The trypsin digestion and extraction and peptide spotting onto a matrix-assisted laser desorption ionization (MALDI) target were accomplished by a robotic liquid handling workstation (MassPrep; Micromass-Waters, Beverly, MA). MALDI peptide fingerprint mass spectra were acquired with a MALDI time-of-flight (TOF) instrument (M@LDI-R; Micromass-Waters, Beverly, MA), using α-cyano-4-hydroxycinnamic acid (Sigma) as the matrix. The mass spectrometry data from MALDI mass spectrometry peptide fingerprint mass spectra were searched against the nonredundant protein sequence database at NCBI and also the gene models from version 2 of the draft Chlamydomonas genome using the Mascot (Matrix Science, London, United Kingdom) search programs. Positive protein identification was based on standard Mascot criteria for statistical analysis of the MALDI peptide fingerprint mass spectra.
Nucleic acid analysis.
Total Chlamydomonas RNA was prepared and analyzed by hybridization as described previously (68). The insert in plasmid MXL014c06 from Kazusa DNA Research Institute, corresponding to expressed sequence tag (EST) BP093819, was sequenced on one strand by Agencourt Biosciences. There were no differences compared to the sequence available on the JGI browser.
5′ end identification.
The 5′ end of FRE1 mRNA was identified using the 5′ rapid amplification of DNA ends system (Invitrogen) as described by the manufacturer, using the protocol for GC-rich sequences (5% dimethyl sulfoxide). The primers were 5′ GTT GTC CAA GAC ACG CAT GG, 5′ ACC ATG TCC AGG ACG CTC AG, 5′ TTA ATT GCG CGT CCC AGG, and 5′ TAT TGG GAG TGG GGG AAG AG. The product from the final nested PCR was cloned into pCR2.1 Topo (Invitrogen) and sequenced (Agencourt Biosciences). The sequence was assembled with the sequence of the insert in plasmid MXL014c06.
For FEA2, the automated gene model was truncated at the 5′ end relative to the FEA1 model. To verify the 5′ end of the FEA2 gene model, a primer (5′ GCG CTG CGC CCC CAA GCGT) designed upstream of the predicted 5′ end (and corresponding to the 5′ end of the FEA1 model) was used with an internal primer (5′ AGG TTG GTC AGG GTG TTG TCC) to amplify a 511-bp fragment from cDNA, which was sequenced and used to generate an improved gene model.
RNA blot hybridization.
Five micrograms of RNA was loaded per lane. The probe used to detect FOX1 transcripts was generated by EcoRI-XhoI digestion of clone HCL036d03 (Kazusa DNA Research Institute; GenBank accession no. AV641563). For the detection of FEA1 transcripts, plasmid (9-18)1 (74) was digested with EcoRI and XhoI. The 1.0-kb fragment was isolated and used as a probe. For FEA2, an amplification product generated by using the primers listed in Table 1 was used as the probe. For CBLP, a 915-bp EcoRI fragment from the cDNA insert in plasmid pcf8-13 was used (78). The CBLP hybridization signal is used for normalization between samples because it does not vary as a function of metal nutrition. Specific activities of probes ranged from 5.7 to 8.4 × 108 cpm μg−1 for the experiments shown here. The blots were exposed at −80°C to film (XRP-1; Eastman-Kodak, Rochester, NY) with two intensifying screens and were developed after a few hours of exposure.
TABLE 1.
Primers used for real-time PCR
| Gene name | Protein IDa | % Efficiencyb | Primer pairc |
|---|---|---|---|
| ATX1 | 163414 | 87 | TCCTGGGAAAGCTGGATGGA |
| CCACTCCTGGCAAAAGCACA | |||
| CBR1 | 163751 | 100 | GCGCCGTGGACTTTGTCATC |
| CTTCAGGATGGCGTTCAGCA | |||
| CTP4 | 195998 | 97 | ACGGATGACGAACAGCAGCA |
| CTCACACAGCCCCTTCCACA | |||
| FEA2 | 173281 | 102 | GCTGCCACCATCACCTTT |
| AGCACCAGCTCGCGCATCAGGTC | |||
| FOX1 | 184156 | 92 | GCTGGTGGCGGGGCCGGACGGCAG |
| CTCACATCGAAGTCGGCGCTCAG | |||
| FOX2 | 129541 | 97 | GTGCAAATGGGCGAACACAA |
| CCGGAGCCAGTAACGAACCA | |||
| FRE1 | 205609 | 96 | CGCCAACGTAACCGAAATGG |
| CGTGCGTGAGGATGGTCTTG | |||
| FTR1 | 137142 | 100 | TCATCATCTACTGCGGCTTCTGGAG |
| TAGCCTCACCCGCCTCAAGCGCCGC | |||
| IRT1 | 206299 | 97 | CGACCCCAAGGACAAGGTCA |
| TCACTCCACCGGCAACTTCA | |||
| IRT2 | 196635 | 92 | GAACCTGACCACCGCCCTAA |
| CCGAACAGGATGCCGAACAC | |||
| RBOL1 | 145439 | 112 | GGCCAACCCAAAATGGTCAA |
| TCGTCGTAGCCCAGCACACT | |||
| RBOL2 | 189216 | 87 | CACAAAGGCCAACATCAGG |
| AACACCTCTGCGTGTACGTG | |||
| UBQ2 | 196845 | 96 | GCGATTTCTCGTTGGGCAGT |
| TGGCCCATCCACTTGTCCTT | |||
| ZIP2 | 196644 | 104 | GGGCTCTTCGCACGTCTTCA |
| AGGGGACCAGGAAGCCAAAA | |||
| ZIP4 | 174212 | 97 | GGGGCTACTGATGGCGGTGA |
| GTAGATGGGTGCGGCGATGA |
Corresponding to the version 3.0 draft genome.
The percent efficiency for each primer pair is calculated based on the theoretical doubling of product at each cycle. For 100% efficient primers, the amount of product doubles at every cycle.
The primer pairs for each gene model are shown. The upper row of each set corresponds to the forward direction for the gene model and the lower row to the reverse direction. All primer sequences are written 5′ to 3′.
Quantitative real-time PCR.
Genomic DNA was removed from the total RNA preparation by treatment with RQ1 DNase (Promega) according to the manufacturer's instructions. cDNA, primed with oligo(dT), was generated with reverse transcriptase (Gibco/BRL), also according to the manufacturer's instructions, and used in the amplification reaction directly after dilution. The amplification reaction was carried out with reagents from the iQ SYBR green Supermix qPCR kit (Bio-Rad Laboratories). Each reaction mixture contained the vendor's master mix, 0.3 μM of each primer, and cDNA corresponding to 5 ng/μl input RNA in the reverse transcriptase reaction. Primers that were not described previously (2) are listed in Table 1. The primers were tested for efficiency, and the identity of the product verified by sequencing. The reaction conditions for the Opticon 2 from MJ Research were as follows: 95°C for 15 min, followed by cycles of 95°C for 10 s, 65°C for 30 s, and 72°C for 30 s up to a total of 40 cycles. The fluorescence was measured at each cycle at 72°C and 83°C. The 2−ΔΔCT method was used to analyze the data based on the fluorescence at 83°C (50). Melting curves were performed after the PCR to assess the presence of a unique final product, and the product was sequenced from one reaction to verify that it represented the gene of interest. The data are presented as the fold change in mRNA abundance, normalized to the endogenous reference gene (CBLP), relative to the RNA sample from cells grown in 18 μM Fe (considered Fe replete). A second reference gene (UBQ2) was also used in some experiments. Neither UBQ2 nor CBLP had changed abundance under the conditions tested in this work.
Reporter construct and its analysis.
Upstream regions of the FEA1 (formerly known as H43), FOX1, and FTR1 genes were identified initially in the first draft of the Chlamydomonas genome (http://genome.jgi-psf.org/chlre1/chlre1.home.html). Primers (for FEA1, forward 5′FEA1-fwd1 [5′-CGG GGT ACC AGG ACA GAG TGC GTG TGGC-3′] and reverse 5′FEA1-rev1 [5′-CGG GGT ACC TGG TTA ACT GTG CGA CGGG-3′], corresponding to positions −1157 to +43 relative to the 5′ end of the transcript; for FOX1, forward 5′FOX1-fwd1 [5′-CGG GGT ACC CAC GCG ACA TAT AGC CGG GAT-3′] and reverse 5′FOX1-rev1 [5′-CCG GGT ACC CTT TTC GGC TTT TGC CTG TT-3′], corresponding to positions −1130 to +10 relative to the 5′ end of the transcript; for FTR1, forward 5′FTR1-fwd1 [5′-CGG GGT ACC TGC TCT GGA ATC TTG TAC TCC-3′] and reverse 5′FTR1-rev1 [5′-CGG GGT ACC CGG CTT GCA AGT TAG AGT GCT-3′], corresponding to positions −1280 to −1 relative to the 5′ end of the transcript) were designed to amplify the regions from Chlamydomonas genomic DNA (strain CC125). The resulting fragments were verified by restriction digest, cloned into the unique KpnI site of plasmid pJD100 containing the minimal promoter from the β-tubulin-encoding gene fused to a promoterless ARS2 (designated ptubB2Δ3,2,1/ars) (16). The resulting plasmids were linearized with PvuI (p5′FEA1-ARS2 and p5′FTR1-ARS2) or BsaI (p5′FOX1-ARS2) and introduced into strain CC425 (arg2) by cotransformation with EcoRI-linearized pArg7.8 carrying the ARG7 gene encoding argininosuccinyl lyase (17) as described previously (67). Transformants were selected for their ability to grow on TAP medium without arginine supplementation. To test reporter expression in the transformants, the plates were sprayed with a freshly diluted (10 mM frozen stock) solution of 2 mM 5-bromo-4-chloro-3-indolyl sulfate (Sigma) as described previously (15). Arylsulfatase activity was assayed quantitatively as described previously (18). The presence of the test construct in the various strains was ascertained by amplification using construct-specific primers 5′FEA1-fwd2 (5′-CGG GGT ACC CAC ATT GAA AAA CGA GCGC-3′), 5′FOX1-fwd2 (5′-CGG GGT ACC GTA TCG GCG AAG GTC AGA CG-3′), and 5′FTR1-fwd2 (5′-CGG GGT ACC GGC AGG CGC GGG CGC CTG CT-3′), with reporter-specific primer Rev-ARS2 (5′-ATG GCA GGG GAG GAA CCG GTT-3′), which produces a fragment of approximately 9 × 102 bp corresponding to approximately 5 × 102 bp of the upstream region for each gene plus 1 × 102 bp of the TUB2 minimal promoter plus 3 × 102 bp of the genomic ARS2 sequence.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in GenBank under accession numbers EF042874 and DQ223113.
RESULTS
Test of candidate transporters and reductases for function in iron assimilation.
We queried the draft genome of Chlamydomonas (initially version 2 and subsequently version 3) by BLAST to identify gene models corresponding to homologues of metal transporters and ferrireductases of other organisms, including fungi, plants, and animals (55). Since the known components of iron assimilation, including FOX1, FTR1, ATX1, and FER1, were regulated through changes in RNA abundance (47), we sought to distinguish the relevance of candidates by screening their pattern of expression as a function of iron nutrition.
(i) FRE1.
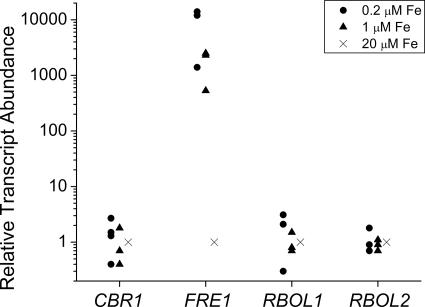
For the reductases, four good candidates were identified, corresponding to protein IDs 205609, 189216, 145439, and 163751 in the version 3.0 draft genome (Fig. 1). The expression of one gene (representing model 205609), now named FRE1, is dramatically regulated by iron deficiency (up to 103-fold in medium containing 1 μM iron in different experiments with different wild-type strains) and shows the same pattern of regulation as do the FOX1 and FTR1 genes (see Fig. 5A). Specifically, the FRE1 mRNA is more highly induced in cells maintained in iron-deficient (1 to 3 μΜ) compared to iron-limited (<0.5 μΜ) conditions (see Materials and Methods). A full-length sequence for the mRNA was derived by assembling the sequence of the product from 5′ rapid amplification of DNA ends with the sequence of the insert in plasmid MXL014c06. The FRE1 gene product is similar to the HCR2 gene product from Chlorococcum along its entire length, but its central portion, which includes the transmembrane ferric reductase motif (Pfam accession PF01794), is most related to the corresponding domain of plant FRO genes. Chlorococcum HCR2 was named for its increased expression in response to high CO2 levels but was shown also to be responsive to iron nutrition (76, 77). The version 3.0 draft genome shows three other related genes; the expression of two linked genes (corresponding to 189216 and 145439) is not affected by either iron or copper. These genes were named RBOL1 and RBOL2 because of their relationship to plant respiratory burst oxidase proteins. We found also a good gene model (163751) for a cytochrome b5 reductase related to the maize NFR gene product (5). However, this gene is also unresponsive to iron and copper nutrition on the basis of RNA abundance.
FIG. 1.
Transcripts for one of four candidate ferrireductases are increased in iron-deficient medium. Candidate genes were analyzed by quantitative real-time reverse transcription-PCR for expression during iron-deficient growth. Strains 17D−, CC1021, and CC125 were tested on 0.2, 1, or 20 μM iron. Each 20-μl reaction mixture contained cDNA equivalent to 100 ng of input total mRNA. Fold induction was calculated according to the 2−ΔΔCT method (50). Each data point is the average of a technical triplicate and represents an individual experiment.
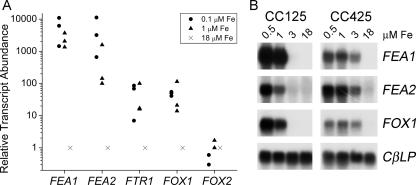
FIG. 5.
FEA1 and FEA2 are expressed coordinately with FOX1 and FTR1 genes under iron-deficient conditions. (A) Real-time PCR quantitation of gene expression in CC125. See the legend to Fig. 1. Each data point is the average of a technical triplicate and represents an individual experiment. (B) Blot hybridization. Wild-type (CC125) or wall-less (CC425) cells were grown in TAP medium with iron supplementation as indicated. RNA was isolated after 5 days of growth, corresponding to cell densities in the range of 5 × 106 to 7 × 106 cell/ml. Five micrograms of total RNA was loaded in each lane and analyzed by blot hybridization. The expression of CBLP was monitored as a loading control.
(ii) IRTs.
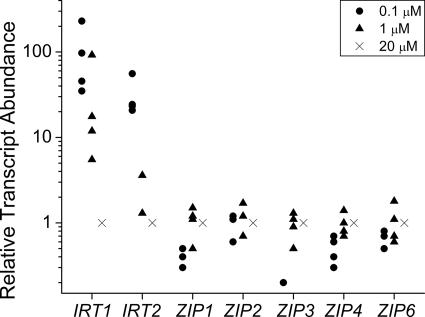
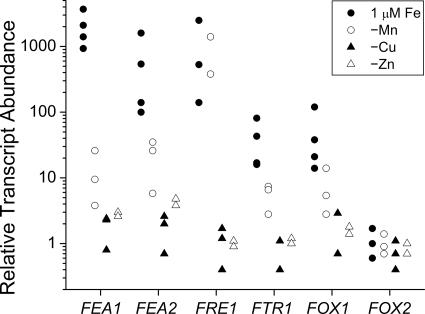
Besides the reductases, we identified homologues of metal transporters belonging to various distinct families (37, 55). These include the CTR/COPT transporters, which function in copper assimilation; the NRAMPs (also called DMT1 or DCT1), which are proton-coupled divalent cation transporters; the heavy-metal-transporting P-type ATPases (referred to as HMAs in plants); the cation diffusion facilitators (also called MTPs in plants); and the ZIP transporters (30, 34, 63, 65, 80). We tested the abundance of mRNAs for Chlamydomonas homologues of these proteins as a function of iron nutrition to determine whether the pattern of RNA accumulation of any might indicate a significant role in iron assimilation. Both copper-supplemented and copper-deficient cells were analyzed, since a copper-independent pathway for iron uptake might be expressed only in a copper-deficient situation (25, 47). None of the candidate transporters showed regulation of the magnitude noted for FRE1, FTR1, and FOX1 expression (data not shown). Because of the potential toxicity of iron, high-affinity iron uptake is very tightly regulated in most organisms. Therefore, it seemed unlikely that any of these molecules might contribute substantially to iron uptake in a situation of deficiency or limitation. On the other hand, the mRNAs for two ZIP family transporters, which might contribute to iron assimilation in a replete cell, show about a 102-fold difference in abundance in response to iron but not zinc, copper, or manganese nutrition (Fig. 2, and data not shown). The corresponding genes were therefore named IRT1 and IRT2.
FIG. 2.
Survey of candidate metal transporter genes for expression in iron-deficient medium. Candidate genes for transporters were analyzed by real time reverse transcription-PCR for expression in strains CC125 and CC1021 grown in 0.1, 1, or 20 μM iron. Relative transcript abundance was calculated as described in the legend to Fig. 1.
FEA1 and FEA2 are secreted by iron-deficient cells.
Extracellular molecules are components of iron assimilation in other chlorophyte algae, which prompted us to search for these in Chlamydomonas. A transferrin-like molecule is known in a related chlamydomonad alga, Dunaliella (28), but we did not find homologues of the Dunaliella protein in the Chlamydomonas genome. Therefore, we used the approach of previous studies in which extracellular phosphatases, sulfatases, and carbonic anhydrases that facilitate growth on low phosphate, sulfate, or CO2, respectively, were identified (see, e.g., references 8, 18, 62, and 69). The secreted proteins are easily isolated from culture supernatants of cell wall-deficient Chlamydomonas strains because the proteins are not retained with the cells but are lost into the medium.
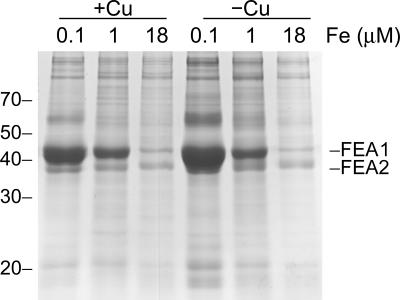
We compared proteins secreted by cells grown in 0.1 μM versus 1 μM versus 18 μM Fe (the concentration of iron in the standard TAP medium). Two proteins with mobilities of around 40 kDa appeared progressively more abundant as the iron content of the medium was reduced (Fig. 3). The bands were excised and subjected to in-gel trypsin digestion, and the resulting tryptic fragments were analyzed by MALDI-TOF mass spectrometry. The 43-kDa protein was identified as a previously known protein, H43, and the 38-kDa one was identified as a previously unknown homologue by searching against the predicted gene models from the draft genome of Chlamydomonas reinhardtii (www.jgi.doe.gov). The homologue corresponds to protein ID 173281, which is immediately adjacent to H43 (protein ID 129929 in version 3.0 of the genome). Rubinelli et al. had previously identified the H43 gene in a screen for genes displaying differential expression under cadmium treatment (74). Nevertheless, they suggested that the gene encodes an iron assimilation component based on its increased expression in iron-deficient Chlamydomonas and its ability to stimulate the growth of a fet3 fet4 strain of Saccharomyces cerevisiae. H43 was renamed FEA1 (for Fe assimilation) in anticipation of such a function, and the homologue was named FEA2. The identification of FEA1 is based on nine peptides corresponding to coverage of 34% of the predicted protein, with a root mean square (RMS) error of 66 ppm (Table 2). For FEA2, 11 peptides, representing 43% coverage, were matched, with an RMS error of 22 ppm. Occasionally peptides corresponding to FEA1 were found also in the 38-kDa band, but the identification of FEA2 in the 38-kDa band is unambiguous because of the presence of unique peptide masses. We attempted to identify the 60-kDa protein, which is apparently an iron deficiency-inducible protein by mass spectrometry, but have been unsuccessful thus far.
FIG. 3.
FEA1 and FEA2 are secreted by iron-deficient cells. Proteins secreted into the medium by a Chlamydomonas cw15 strain (CC400) were collected by ammonium sulfate/trichloroacetic acid precipitation and analyzed after electrophoretic separation on SDS-containing polyacrylamide (12% monomer) gels. Each lane contained extract corresponding to an equivalent number (7.3 × 107) of cells. The major proteins revealed by Coomassie blue staining were analyzed by MALDI-TOF mass spectrometry after in-gel trypsin digestion.
TABLE 2.
Identification of FEA proteins by mass spectrometrya
| Band (kDa) | Identity | Protein IDb | Coverage (%) | Peptides identifiedc |
|---|---|---|---|---|
| 43 | FEA1 | 129929 | 34 | 105DTSSLDAAIR114, 122ATLAAGLIQVGTLK135, 136YHLHEVDEAYNK147, 210SYVNTAMINTVNEMLAAAR228, 229LSTLNIQAYDAAR241, 271RPAA EVEDAK280, 281TMIAVHWAYLEPMLK295, 301ASAVTELHHQLTASK315, 336SSELGAPQSAIIAANWK352 |
| 38 | FEA2 | 173281 | 43 | 84SFSGFASYATSGPELLHDSLAMGR107, 110TWLDVAIR117, 118TAFAEQNRPLVEGLILVAGVK138, 139YGLHEVDEGATK150, 231DNGTLSSAVYNASRVDLMR249, 250ELVLLGLQGVLHSSYK265, 274RPDADLEEAK283, 312TALTAARPNYMDVR325, 327AVVSVADAMGR337, 339MAEIGTPIHDR349, 353GWAGCSASR361 |
Proteins were excised from an SDS gel, digested with trypsin, and analyzed by MALDI-TOF mass spectrometry as described in Materials and Methods. RMS errors for identification were 66 ppm and 22 ppm for FEA1 and FEA2, respectively.
Protein IDs correspond to the Chlamydomonas genome version 3.0.
The position of the peptide in the protein model (accession no. AB042099 for H43 and derived from the gene model for FEA2) is indicated by subscripts. The identification was repeated with independent protein preparations.
The original gene model for FEA2 was incomplete at the 5′ end because of the absence of EST coverage. Therefore, we amplified the 5′ end of the corresponding RNA isolated from iron-deficient cells. The sequence (accession no. DQ223113) was used to generate an improved model.
FEA1 and FEA2 share 43% sequence identity, and the gene arrangement is a presumed consequence of gene duplication (Fig. 4). FEA1 is predicted to encode a 362-amino-acid protein, while FEA2 is predicted to be 363 amino acids; both sequences include a predicted signal peptide. The difference in migration may result from different posttranslational processing. Given that a peptide close to the predicted C terminus of FEA1 was identified in the 38-kDa band, we attribute the presence of FEA1 in the 38-kDa band to N-terminal degradation of the 43-kDa species, but it is certainly possible that there may be differently modified forms of the protein (e.g., resulting from glycosylation or processing). FEA1 is severalfold more abundant than FEA2.
FIG. 4.
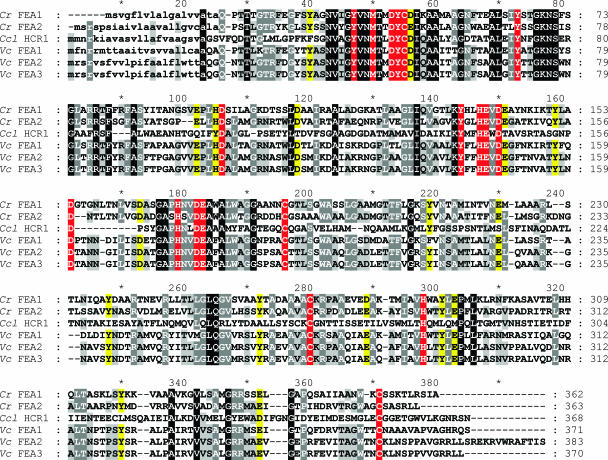
Conserved residues in the FEA proteins. FEA1 (BAA94959) and FEA2 (protein ID 173281 in the version three draft genome) of C. reinhardtii (Cr), Hcr1 (BAA22844) of Chlorococcum littorale (Ccl), and C_370140 (Fea1), C_520002 (Fea2), and C_520026 (Fea3) of V. carteri (Vc) (version 1.0 draft genome) were aligned using Multalin (http://prodes.toulouse.inra.fr/multalin/multalin.html) (11). Signal peptides, indicated by lowercase letters, and cleavage sites were predicted using TargetP (http://www.cbs.dtu.dk/services/TargetP/) (24, 59). All are predicted to be secreted proteins. Residues are indicated by white text on a black background if identical in all six sequences or on a gray background if identical in five of the six or by black text on a gray background if four of six are conserved. Potential metal binding ligands are indicated with a red background if they are identical or with a yellow background if the residues represent conservative replacements. Asterisks mark every 10th residue.
A BLAST search of genome databases and sequences at NCBI (May 2007) indicates that the FEA proteins are found in several chlorophyte algae, including Scenedesmus obliquus, Chlorococcum littorale, and Volvox carteri, and a dinoflagellate, Heterocapsa triquerta. V. carteri encodes three FEA-like proteins; two are orthologues of FEA1 and FEA2 based on synteny, and a third is unlinked. A multiple alignment identified several conserved residues, including some (Cys, Asp/Glu, Tyr, and His) with functional groups that are suitable ligands for either Fe(II) or Fe(III) binding, but no other motifs that speak to function.
We noted that the copper nutrition state did not modify the pattern of FEA1 and FEA2 accumulation (Fig. 3), indicating that these molecules were not part of the putative copper-independent high-affinity iron uptake pathway. Neither was the pattern of any other secreted protein modified by copper nutrition, arguing against a role for secreted proteins in copper assimilation.
FEA1 and FEA2 are expressed coordinately with FOX1 and FTR1.
A plasma membrane ferroxidase (encoded by FOX1) was the first iron assimilation component to be identified in Chlamydomonas (42, 47). As expected for an assimilatory component, FOX1 expression is increased prior to the appearance of iron deficiency symptoms in Chlamydomonas (58). We note that the same is true for FEA1 (Fig. 5). FEA1 is induced coordinately with FOX1, and both genes are nearly fully activated at 1.0 μM Fe in TAP medium, at which point chlorosis is not yet evident. Given the relatively low abundance of the FEA2 polypeptide recovered (Fig. 3) and the absence of ESTs in the database, we were surprised to note that FEA2 mRNA is also increased substantially in iron deficiency and that its pattern of expression parallels that of FEA1 (Fig. 5). On the other hand, a second gene, FOX2, encoding a putative ferroxidase, although expressed, is not differentially regulated by iron nutrition status. Quantitative PCR indicates that the fold induction of FEA1 and FEA2 is much more dramatic than that of FOX1. FOX1 mRNA is about 102-fold induced in 0.1 versus 18 μM Fe, while FEA1 and FEA2 are induced about 103 to 104-fold. The exact fold induction varies, depending upon the strain analyzed and the cell density of the culture when it was sampled for RNA extraction. RNA blot hybridization experiments confirmed this dramatic up-regulation as well as the strain-specific level of gene expression (Fig. 5B). Absolute quantitation of FEA1 versus FEA2 mRNAs indicates that FEA1 is present at about 10 times the stoichiometry of FEA2 in an iron-replete cell but that this changes to a 100-fold difference in iron deficiency. This is consistent with the occurrence of over 30 FEA1 ESTs but no FEA2 ESTs.
When we analyzed RNAs prepared from cells adapted to copper, manganese, or zinc deficiency, we noted that the iron assimilation components FOX1, FTR1, FEA1, and FEA2 are induced primarily by iron deficiency (Fig. 6). For example, FEA1 expression is not significantly changed by copper or zinc deficiency. There is a small increase in the expression of the FEA genes in manganese-deficient cells, but this is only a few percent of the change noted in iron deficiency and is in fact attributable to secondary iron deficiency in manganese-deficient cells (2).
FIG. 6.
Metal-selective response of FEA gene expression. Transcript abundance was assessed by real-time PCR (see the legend to Fig. 1). For copper and manganese deficiency, cells were adapted to deficiency by three sequential transfers (1:1,000 dilution at each transfer) into medium lacking the trace element, while for zinc deficiency, cells were sampled after the second transfer into zinc-free medium. For iron deficiency, the cells were washed once and transferred from iron-replete (18 μM) to iron-deficient (1 μM) medium and sampled when they reached a density of 5 × 106 to 7 × 106 cell/ml. The various RNAs collected from strains CC125, CC425, and CC1021 were tested for marker gene expression characteristic for each deficiency (CYC6 for copper deficiency, GPX1 for manganese deficiency, and ZRT1 for zinc deficiency). For each experiment, the level of expression is normalized to the replete condition. Each data point is the average of a technical triplicate and represents an individual experiment.
FEA1 and FEA2 are differently regulated by high CO2 levels.
HCR1, a Chlorococcum littorale homologue of the Chlamydomonas FEA genes, had been identified initially as a gene induced by high CO2 levels, and a recent study showed that FEA1 was also induced by high CO2 levels (36, 76). Therefore, we tested the patterns of both FEA1 and FEA2 expression in response to CO2 in wild-type cells and in a mutant (cia5) which is defective in the carbon-concentrating mechanism and hence unresponsive to CO2 nutrition (31, 57). We found that the FEA1 gene is down-regulated in low CO2 in a pathway dependent on CIA5 but that FEA2 is not (Table 3). When we repeated the analysis with the lcr1 mutant, another carbon-concentrating mechanism mutant (91), we reached the same conclusion (data not shown).
TABLE 3.
CCM1-mediated expression of iron-responsive genesa
| Gene | Relative transcript abundanceb
|
|
|---|---|---|
| Wild type | cia5 | |
| CA1 | 4.1 × 103 | 3.0 |
| FEA1 | −2.8 × 102 | −5.0 |
| FEA2 | −1.7 | −2.0 |
| FTR1 | −2.5 | −1.8 |
| FOX1 | −5.5 | −1.1 |
| FOX2 | 2.0 | 5.9 |
| CBR1 | −1.6 | −2.2 |
| FRE1 | −2.6 | 1.4 |
| RBOL1 | 2.4 | 1.2 |
| RBOL2 | −1.2 | 1.0 |
Iron-responsive genes were assessed by real-time PCR on low-CO2-grown wild-type and cia5 mutant cells (described in reference 31).
Expression was first normalized to CBLP expression, the average threshold cycle value was calculated from technical triplicates, and expression was calculated relative to high-CO2-grown cells. Expression was calculated by the 2−ΔΔCT method (50). Negative numbers represent a suppression of gene expression compared to that in the high-CO2-grown cells.
FEA1, FOX1, and FTR1 are transcriptionally regulated by iron.
Regulation of iron assimilation occurs at the transcriptional level in many microorganisms but at the posttranscriptional level in animal cells (reviewed in references 73 and 75). To distinguish the operation of an iron nutrition-responsive pathway in Chlamydomonas, we fused the 5′ flanking DNA upstream of the putative 5′ end of the mRNA (where +1 is defined by the sequence of the most 5′ EST defined by accession no. BE237853) to a reporter gene, ARS2, driven by a minimal promoter from the TUB2 gene to generate the test constructs p5′-FOX1-ARS2, p5′-FTR1-ARS2, and p5′-FEA1-ARS2 (see Materials and Methods) (constructs available from the Chlamydomonas culture collection). The constructs were each introduced into Chlamydomonas strain CC425 by cotransformation with pArg7.8 and selection for growth in the absence of arginine. For each construct, colonies that expressed the reporter gene were analyzed to assess iron-responsive expression. In parallel, colonies resulting from cotransformation of pJD100 (containing no FEA1, FOX1, or FTR1 sequences) and pArg7.8 were analyzed. No colony appearing from the control transformation ever showed significant arylsulfatase expression (one representative is shown in Table 4). Quantitation of ARS2 expression by enzyme assay (analysis of two representative colonies for each construct is shown in Table 4) confirmed that the 5′ flanking sequences from the FEA1, FOX1, and FTR1 genes confer iron responsiveness to a reporter gene. Furthermore, it appears that the magnitude of regulation of these genes is recapitulated with the reporter genes, because the constructs carrying FEA1 sequences appeared generally to show much greater reporter gene activity.
TABLE 4.
Arylsulfatase activity in strains containing transgenic reporter genes
| Reporter construct | Arylsulfatase activity (pmol of p-nitrophenol/min/106 cells)a with:
|
|
|---|---|---|
| 1 μM Fe | 200 μM Fe | |
| 5′FOX1-ARS2 1 | 10 ± 1.2 | 3.3 ± 0.2 |
| 5′FOX1-ARS2 2 | 27 ± 1.5 | 3.0 ± 0.2 |
| 5′FTR1-ARS2 1 | 13 ± 1.1 | 3.0 ± 0.2 |
| 5′FTR1-ARS2 2 | 15 ± 1.6 | 0.3 ± 0.1 |
| 5′FEA1-ARS2 1 | 86 ± 1.9 | 0.2 ± 0.1 |
| 5′FEA1-ARS2 2 | 29 ± 5.0 | 1.6 ± 0.3 |
| pJD100 | 0.1 ± 0.03 | 0.1 ± 0.02 |
The arylsulfatase activity of each strain was measured with p-nitrophenyl sulfate as the substrate (see Materials and Methods). Data represent average values from two independent experiments ± standard deviations.
We conclude that components of the iron assimilation pathway in Chlamydomonas are transcriptionally regulated by iron availability in the medium. We suspect that regulation occurs at least in part by activation of gene expression in low Fe (as opposed to repression in high Fe), because the arylsulfatase-expressing colonies carrying the 5′FEA1-ARS2, 5′FTR1-ARS2, or 5′FOX1-ARS2 construct always showed much higher expression than colonies carrying pJD100 (where ARS2 is driven by a minimal promoter). Nevertheless, we cannot rule out the presence of repressing sequences, nor can we exclude posttranscriptional mechanisms overlaid on the primary transcriptional response because we did not test for these.
Function of FEA1 and FEA2.
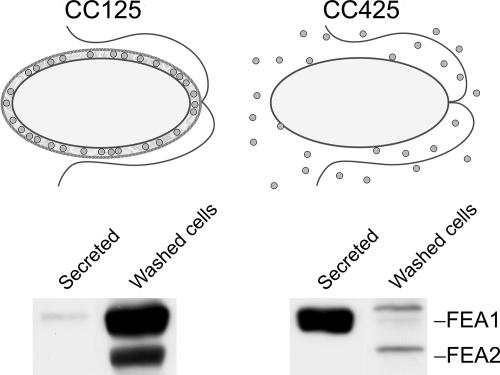
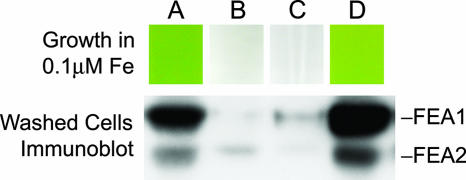
FEA1 and FEA2 are secreted to the medium in Chlamydomonas strain CC425 (which is defective with respect to cell wall structure), suggesting that the proteins are probably located in the extracellular space in a wild-type cell (Fig. 7). Therefore, we suggest that the proteins function as periplasmic substrate binding proteins to concentrate substrates in the vicinity of assimilatory transporters. Since FEA proteins were completely lost to the medium in strain CC425 (Fig. 7), we were able to assess the function of cell-associated FEA proteins in the context of iron nutrition. The rationale is as follows: a strain that loses the protein to the medium would be analogous to a loss-of-function fea mutant. Therefore, we analyzed the amount of cell-associated FEA proteins and iron nutrition-dependent growth of spores resulting from a cross of strain CC425 to strain CC124 (Fig. 8). We analyzed seven complete tetrads and noted that strains that had no cell-associated FEA1 and FEA2 were unable to grow in medium containing 0.1 to 0.2 μM iron (P < 0.01). We conclude that the FEA proteins are not essential for iron uptake but rather that they facilitate iron uptake by functioning as iron binding proteins in the extracellular space.
FIG. 7.
Functional model for FEA proteins. Top, FEA proteins (circles) associate within the periplasm in a cell wall-containing Chlamydomonas strain. In a cell wall-less mutant, FEA proteins are lost to the medium and unable to function in iron assimilation. Bottom, secreted and cell-associated fractions of CC125 (walled) and CC425 (wall-deficient) cells were isolated and separated by denaturing gel electrophoresis. FEA proteins were visualized by immunoblotting. Each lane corresponds to ∼2 × 106 cells.
FIG. 8.
Loss of FEA proteins results in poor growth on low-iron medium. Spores from a cw15 arg2 mt+ × CW15 ARG2 mt− tetrad were analyzed for growth on iron-limited (0.1 μM) TAP-Arg medium (top) and cell-associated FEA proteins by immunoblot analysis (bottom) of washed iron-deficient cells (grown on TAP-Arg with 1 μM Fe).
DISCUSSION
Studies of the impact of micronutrients on interpretation of the genome are generally facilitated in microorganisms because of the ease with which the growth medium can be manipulated, and Chlamydomonas reinhardtii has emerged as a powerful model for the study of the impact of nutrition on the photosynthetic apparatus (32, 55, 89). The photosynthetic apparatus, like the respiratory chain, is a major iron-utilizing pathway in a photosynthetic cell. Recently, we showed that the organization and abundance of chlorophyll-containing proteins are modified in a programmed pathway that responds sequentially and progressively to iron depletion (58). The mechanisms for sensing and responding to iron status are, however, unknown. In this work, we surveyed all candidate metal transporters in the genome and applied subproteomic analysis to expand our knowledge of components involved in iron assimilation in Chlamydomonas.
FEA1 and FEA2.
Two homologous proteins, FEA1 and FEA2, are identified as extracellular proteins on the basis of their recovery from the growth medium after removal of cells. The recovery of both proteins is greater in strain CC425 (which lacks a cell wall and loses proteins to the medium) than in wild-type cells, which confirms their extracellular location (Fig. 7 and data not shown). Because their accumulation increases with progressively greater iron deficiency, we hypothesized that the molecules must function in iron assimilation. Indeed, we noted that cells containing FEA proteins appeared to have a greater facility for iron utilization (Fig. 8). Cells that lose FEA proteins to the medium grow more poorly in iron-deficient medium.
FEA1 is severalfold more abundant than FEA2, especially in iron-deficient cells (Fig. 3). In wild-type cells (CC125), we note that FEA1 is already maximally increased in expression when the iron content of the medium is reduced to 1 μM, while FEA2 is maximally increased only in iron-limited cells (<0.2 μM iron). It is possible that the two proteins might be functionally different with respect to iron binding affinity. Interestingly, Chlamydomonas FEA1 was identified independently as a “high-CO2-responsive” gene (36), and this is confirmed in our study (Table 3). For Chlorococcum littorale, both HCR1 and HCR2, homologues of FEA1 and FRE1, are high-CO2-responsive genes (76). We do not understand the basis for regulation of FEA1 by CO2, but it might relate to modification of iron demand by carbon availability. Another possibility is that the response may be related to a role for (bi)carbonate in facilitating iron binding to FEA. We note that FEA2 is not regulated by CO2, and this may reflect specialization of function after gene duplication. The Volvox genome encodes three FEA proteins; perhaps this relates to its multicellularity and different metabolic demands of vegetative versus reproductive cells.
Pathways of iron assimilation in algae appear to be quite diverse. For instance, the Ostreococcus spp. genomes do not seem to encode homologues of the putative high-affinity ferric transporter (FTR1) found in Chlamydomonas, Volvox, and Physcomitrella or of the ferroxidase discovered in Chlamydomonas and Dunaliella, and it is suggested that they may synthesize siderophores (61). D. salina but not V. carteri or C. reinhardtii (all within the Chlamydomonadaceae) uses an extracellular transferrin-like molecule for high-affinity iron uptake (28). Our analysis of FEA function in Chlamydomonas suggests that periplasmic iron binding may be important for acquiring iron in a situation of limitation, but each organism may have devised/acquired different means for accomplishing this. An FEA homologue was among the ESTs from a dinoflagellate, indicating that the proteins are not restricted to the chlorophyte algae.
Regulation of iron assimilation pathway genes.
Reporter gene analysis confirmed that FEA1, FOX1, and FTR1 are regulated largely if not entirely at the transcriptional level (Table 4). The magnitude of regulation of the FEA1 and FRE1 genes makes their promoters useful targets for dissection of nutritional iron signaling in Chlamydomonas. It is likely that the signaling pathway responds primarily to iron status rather than to stress resulting from iron deficiency, because the changes in gene expression seem to be highly selective for iron deficiency (Fig. 6). Zinc-deficient cells are also growth inhibited, but the FOX1, FTR1, FRE1, and FEA genes are not up-regulated in this situation. Manganese-deficient cells do show increased expression of these genes, but this is attributed to secondary iron deficiency (2). The iron content of severely manganese-deficient cells is comparable to the iron content of cells grown in 1 μM iron. The FRE1 gene is much more responsive to manganese deficiency than are the FOX1, FTR1, and FEA genes, suggesting that it may respond also directly to manganese nutrition.
Copper-independent iron uptake.
In previous work, we had suggested that Chlamydomonas cells expressed a high-affinity, copper-independent uptake system that was expressed in copper-deficient medium (25, 47). We tested copper-deficient cells for FEA gene expression and found the proteins unchanged in copper deficiency and the mRNAs minimally changed (Fig. 3 and 6), arguing against the function of the FEA proteins as substitutes for the FOX1/FTR1 pathway of Chlamydomonas (47).
We identified a number of gene models encoding candidate transporters and reductases and tested the pattern of expression with respect to transition metal nutrition to identify other components in iron assimilation. This approach did identify a putative reductase gene, FRE1, but none of the other genes showed changes in gene expression comparable to those noted for FOX1 and FTR1 (47). We looked also for iron-responsive expression in combination with copper deficiency in the hope of identifying the transport pathway that functioned in the copper-deficient Chlamydomonas cell, but again, no candidate was revealed by this approach. This does not rule out a function of these transporters in iron homeostasis, because it is possible that there may be more than one nutritional iron signaling pathway and therefore changes in RNA abundance may play only a small role in the regulation of some assimilatory molecules. For instance, FOX2 mRNA abundance is not affected by iron deficiency in the experiments presented in this work, but the protein appears to increase in abundance when cells are maintained in iron deficiency for prolonged periods (70).
Analysis of the Chlamydomonas genome indicates that some pathways, especially those related to chloroplast function, are homologous to those in plants, while other pathways are more similar to those in animals or occasionally to those in other protists (e.g., the pathways related to mitochondrial function). The components of iron assimilation reflect this “mix-and-match” approach to pathway acquisition. The reductase is more similar to plant and other algal rather than animal or fungal homologues, the ferric transporter is related to the homologues in fungi, and the multicopper oxidase (although unique in terms of domain arrangement) shows a better sequence relationship to the animal enzymes than to the fungal ones.
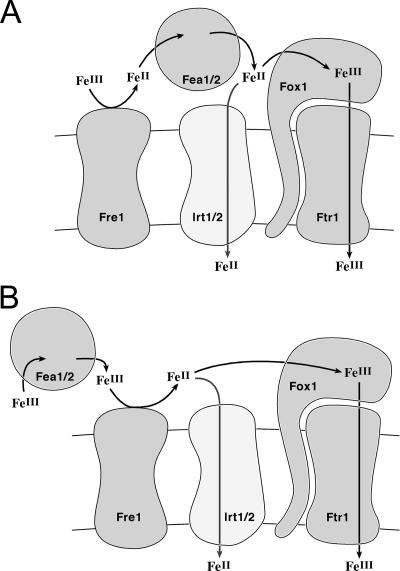
Proposed mechanism of FEA1.
In this work, we identify FRE1 and the FEA genes as the most dramatically induced genes in iron-deficient Chlamydomonas. Although the ferroxidase had been noted previously to be one of the most abundant plasma membrane proteins in iron-deficient Chlamydomonas (43), we show here that the magnitude of regulation is much greater for FRE1 and the FEA genes, and they respond most dramatically to iron deficiency rather than to other nutrient deficiencies. This is compatible with a site of action in an extracellular location as the first steps in a multistep iron assimilation pathway.
How might FEA1 facilitate iron delivery given its periplasmic location in Chlamydomonas? In one model (Fig. 9), FEA1 functions merely to concentrate iron in the periplasmic space in the vicinity of low-affinity transporters, which makes them more effective. This model is analogous to the situation for inorganic carbon uptake where a secreted carbonic anhydrase provides the substrate for a specific transporter (26), and it is compatible with the finding that FEA1 can facilitate iron uptake in a heterologous system (74).
FIG. 9.
Model for iron assimilation by Chlamydomonas. The components of an iron assimilation pathway are shown. Ferric iron may be released from chelated forms by reduction to the ferrous form. The FEA proteins may bind the ferrous iron to keep it soluble and available as a substrate for the ferroxidase/ferric transporter complex. Alternatively, FEA proteins might bind ferric ion, in which case they would function upstream of FRE1. FRE1 was found in a plasma membrane subproteome (70), which is consistent with the model proposed here.
Many organisms secrete extracellular iron binding molecules such as transferrins or siderophores when they are iron deprived. Although a transferrin-like molecule has been identified in Dunaliella spp. (28), the Chlamydomonas genome does not appear to encode proteins related to the transferrins. FEA1 may be a functionally analogous equivalent (74). S. cerevisiae induces the cell wall mannoproteins to facilitate use of iron bound to siderophores synthesized by other organisms (66). The FEA proteins are not similar in sequence to the Fit proteins, but they may have an analogous function.
A multiple alignment of the FEA proteins indicates a number of conserved residues that could provide iron binding ligands. Among these are thiols, imidazoles, carboxylates, and hydroxyl groups (Fig. 4). As soft ligands, the thiols, which are important for function, would be more compatible with Fe(II) binding, or of course they may be important for disulfide bond formation and protein stability in the extracellular space. In terms of a pathway for iron assimilation, it is attractive to place the FEA proteins either as Fe(II) carriers between the reductase and the ferroxidase (Fig. 9) or as Fe(III) carriers that make substrate available to the reductase.
Acknowledgments
We thank the laboratory of Joseph Loo and the UCLA Mass Spectrometry and Proteomics Technology Center, especially Sheng Yin, Charisa Cottonham, and James Kerwin, for their advice, suggestions, and help in the identification of FEA1 and FEA2. We are grateful to the Joint Genome Institute for the draft sequences of the Chlamydomonas reinhardtii genome and the Volvox carteri genome, to Arthur Grossman's laboratory and the Kazusa DNA Research Institute for providing cDNA clones, to Yoshihiro Shiraiwa for antibodies against Chlamydomonas FEA1, to Hideya Fukuzawa for providing the RNAs for the experiment shown in Table 3, and to Joanne Long for some of the RNAs. We thank Laurens Mets for help with crosses, Jeffrey Moseley for advice on tetrad analysis, and an anonymous reviewer for many suggestions on improving the work.
This work is supported by grants from the Department of Energy (DE-FG02-04ER15529, to S.S.M.) and the National Institutes of Health (GM42143, to S.S.M.). M.D.A. was supported in part by Institutional and Individual Ruth L. Kirschstein National Research Service Awards GM07185 and GM077066 and J.A.d.C. in part by a postdoctoral fellowship from the Spanish Ministry for Education. The Proteomics Center was established and equipped by a generous grant from the W. M. Keck Foundation with additional support from the UCLA Johnsson Comprehensive Cancer Center, which is also gratefully acknowledged.
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Abboud, S., and D. J. Haile. 2000. A novel mammalian iron-regulated protein involved in intracellular iron metabolism. J. Biol. Chem. 275:19906-19912. [DOI] [PubMed] [Google Scholar]
- 2.Allen, M. D., J. Kropat, S. Tottey, J. A. Del Campo, and S. S. Merchant. 2007. Manganese deficiency in Chlamydomonas results in loss of photosystem II and MnSOD function, sensitivity to peroxides, and secondary phosphorus and iron deficiency. Plant Physiol. 143:263-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Askwith, C., D. Eide, A. Van Ho, P. S. Bernard, L. Li, S. Davis-Kaplan, D. M. Sipe, and J. Kaplan. 1994. The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Askwith, C., and J. Kaplan. 1997. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J. Biol. Chem. 272:401-405. [DOI] [PubMed] [Google Scholar]
- 5.Bagnaresi, P., S. Thoiron, M. Mansion, M. Rossignol, P. Pupillo, and J. F. Briat. 1999. Cloning and characterization of a maize cytochrome-b5 reductase with Fe3+-chelate reduction capability. Biochem. J. 338:499-505. [PMC free article] [PubMed] [Google Scholar]
- 6.Benderliev, K. M., and N. I. Ivanova. 1994. High-affinity siderophore-mediated iron-transport system in the green-alga Scenedesmus incrassatulus. Planta 193:163-166. [Google Scholar]
- 7.Brüggemann, W., K. Maaskantel, and P. R. Moog. 1993. Iron uptake by leaf mesophyll-cells—the role of the plasma membrane-bound ferric-chelate reductase. Planta 190:151-155. [Google Scholar]
- 8.Coleman, J. R., J. A. Berry, R. K. Togasaki, and A. R. Grossman. 1984. Identification of extracellular carbonic anhydrase of Chlamydomonas reinhardtii. Plant Physiol. 76:472-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connolly, E. L., and M. Guerinot. 2002. Iron stress in plants. Genome Biol. 3:1024.1-1024.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connolly, E. L., N. H. Campbell, N. Grotz, C. L. Prichard, and M. L. Guerinot. 2003. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 133:1102-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curie, C., Z. Panaviene, C. Loulergue, S. L. Dellaporta, J. F. Briat, and E. L. Walker. 2001. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346-349. [DOI] [PubMed] [Google Scholar]
- 13.Curie, C., and J.-F. Briat. 2003. Iron transport and signaling in plants. Annu. Rev. Plant Biol. 54:183-206. [DOI] [PubMed] [Google Scholar]
- 14.Dancis, A., D. G. Roman, G. J. Anderson, A. G. Hinnebusch, and R. D. Klausner. 1992. Ferric reductase of Saccharomyces cerevisiae: molecular characterization, role in iron uptake, and transcriptional control by iron. Proc. Natl. Acad. Sci. USA 89:3869-3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davies, J. P., D. P. Weeks, and A. R. Grossman. 1992. Expression of the arylsulfatase gene from the β2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 20:2959-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, J. P., and A. R. Grossman. 1994. Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol. Cell. Biol. 14:5165-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Debuchy, R., S. Purton, and J. D. Rochaix. 1989. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J. 8:2803-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Hostos, E. L., R. K. Togasaki, and A. Grossman. 1988. Purification and biosynthesis of a derepressible periplasmic arylsulfatase from Chlamydomonas reinhardtii. J. Cell Biol. 106:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donovan, A., A. Brownlie, Y. Zhou, J. Shepard, S. J. Pratt, J. Moynihan, B. H. Paw, A. Drejer, B. Barut, A. Zapata, T. C. Law, C. Brugnara, S. E. Lux, G. S. Pinkus, J. L. Pinkus, P. D. Kingsley, J. Palis, M. D. Fleming, N. C. Andrews, and L. I. Zon. 2000. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403:776-781. [DOI] [PubMed] [Google Scholar]
- 20.Eckhardt, U., and T. J. Buckhout. 1998. Iron assimilation in Chlamydomonas reinhardtii involves ferric reduction and is similar to strategy I higher plants. J. Exp. Bot. 49:1219-1226. [Google Scholar]
- 21.Eide, D., M. Broderius, J. Fett, and M. L. Guerinot. 1996. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93:5624-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eide, D. J. 2000. Metal ion transport in eukaryotic microorganisms: insights from Saccharomyces cerevisiae. Adv. Microb. Physiol. 43:1-38. [DOI] [PubMed] [Google Scholar]
- 23.Eisenstein, R. S. 2000. Discovery of the ceruloplasmin homologue hephaestin: new insight into the copper/iron connection. Nutr. Rev. 58:22-26. [DOI] [PubMed] [Google Scholar]
- 24.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 25.Eriksson, M., J. L. Moseley, S. Tottey, J. A. del Campo, J. M. Quinn, Y. Kim, and S. Merchant. 2004. Genetic dissection of nutritional copper signaling in Chlamydomonas distinguishes regulatory and target genes. Genetics 168:795-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fett, J. P., and J. R. Coleman. 1994. Regulation of periplasmic carbonic anhydrase expression in Chlamydomonas reinhardtii by acetate and pH. Plant Physiol. 106:103-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finegold, A. A., K. P. Shatwell, A. W. Segal, R. D. Klausner, and A. Dancis. 1996. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J. Biol. Chem. 271:31021-31024. [DOI] [PubMed] [Google Scholar]
- 28.Fisher, M., A. Zamir, and U. Pick. 1998. Iron uptake by the halotolerant alga Dunaliella is mediated by a plasma membrane transferrin. J. Biol. Chem. 273:17553-17558. [DOI] [PubMed] [Google Scholar]
- 29.Fleming, M. D., M. A. Romano, M. A. Su, L. M. Garrick, M. D. Garrick, and N. C. Andrews. 1998. Nramp2 is mutated in the anemic Belgrade (b) rat: evidence of a role for Nramp2 in endosomal iron transport. Proc. Natl. Acad. Sci. USA 95:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forbes, J. R., and P. Gros. 2001. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 9:397-403. [DOI] [PubMed] [Google Scholar]
- 31.Fukuzawa, H., K. Miura, K. Ishizaki, K. I. Kucho, T. Saito, T. Kohinata, and K. Ohyama. 2001. Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc. Natl. Acad. Sci. USA 98:5347-5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grossman, A. 2000. Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 151:201-224. [DOI] [PubMed] [Google Scholar]
- 33.Guerinot, M. L., and Y. Yi. 1994. Iron: nutritious, noxious, and not readily available. Plant Physiol. 104:815-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerinot, M. L. 2000. The ZIP family of metal transporters. Biochim. Biophys. Acta 1465:190-198. [DOI] [PubMed] [Google Scholar]
- 35.Gunshin, H., B. Mackenzie, U. V. Berger, Y. Gunshin, M. F. Romero, W. F. Boron, S. Nussberger, J. L. Gollan, and M. A. Hediger. 1997. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388:482-488. [DOI] [PubMed] [Google Scholar]
- 36.Hanawa, Y., M. Watanabe, Y. Karatsu, H. Fukuzawa, and Y. Shiraiwa. 2007. Induction of a high-CO2-inducible, periplasmic protein, H43, and its application as a high-CO2-responsive marker for study of the high-CO2-sensing mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 48:299-309. [DOI] [PubMed] [Google Scholar]
- 37.Hanikenne, M., U. Krämer, V. Demoulin, and D. Baurain. 2005. A comparative inventory of metal transporters in the green alga Chlamydomonas reinhardtii and the red alga Cyanidioschizon merolae. Plant Physiol. 137:428-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris, E. H. 1989. The Chlamydomonas sourcebook: a comprehensive guide to biology and laboratory use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 39.Harris, Z. L., A. P. Durley, T. K. Man, and J. D. Gitlin. 1999. Targeted gene disruption reveals an essential role for ceruloplasmin in cellular iron efflux. Proc. Natl. Acad. Sci. USA 96:10812-10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hell, R., and U. W. Stephan. 2003. Iron uptake, trafficking and homeostasis in plants. Planta 216:541-551. [DOI] [PubMed] [Google Scholar]
- 41.Hentze, M. W., M. U. Muckenthaler, and N. C. Andrews. 2004. Balancing acts: molecular control of mammalian iron metabolism. Cell 117:285-297. [DOI] [PubMed] [Google Scholar]
- 42.Herbik, A., C. Bölling, and T. J. Buckhout. 2002. The involvement of a multicopper oxidase in iron uptake by the green algae Chlamydomonas reinhardtii. Plant Physiol. 130:2039-2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbik, A., S. Haebel, and T. J. Buckhout. 2002. Is a ferroxidase involved in the high-affinity iron uptake in Chlamydomonas reinhardtii? Plant Soil 241:1-9. [Google Scholar]
- 44.Kim, S. A., and M. L. Guerinot. 2007. Mining iron: iron uptake and transport in plants. FEBS Lett. 581:2273-2280. [DOI] [PubMed] [Google Scholar]
- 45.Kim, Y., C. W. Yun, and C. C. Philpott. 2002. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J 21:3632-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kosman, D. J. 2003. Molecular mechanisms of iron uptake in fungi. Mol. Microbiol. 47:1185-1197. [DOI] [PubMed] [Google Scholar]
- 47.La Fontaine, S., J. M. Quinn, S. S. Nakamoto, M. D. Page, V. Göhre, J. L. Moseley, J. Kropat, and S. Merchant. 2002. Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot. Cell 1:736-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lan, C. Y., G. Rodarte, L. A. Murillo, T. Jones, R. W. Davis, J. Dungan, G. Newport, and N. Agabian. 2004. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 53:1451-1469. [DOI] [PubMed] [Google Scholar]
- 49.Lesuisse, E., M. Casteras-Simon, and P. Labbe. 1996. Evidence for the Saccharomyces cerevisiae ferrireductase system being a multicomponent electron transport chain. J. Biol. Chem. 271:13578-13583. [DOI] [PubMed] [Google Scholar]
- 50.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 51.Lynnes, J. A., T. L. M. Derzaph, and H. G. Weger. 1998. Iron limitation results in induction of ferricyanide reductase and ferric chelate reductase activities in Chlamydomonas reinhardtii. Planta 204:360-365. [Google Scholar]
- 52.Martins, L. J., L. T. Jensen, J. R. Simon, G. L. Keller, D. R. Winge, and J. R. Simons. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716-23721. [DOI] [PubMed] [Google Scholar]
- 53.McKie, A. T., P. Marciani, A. Rolfs, K. Brennan, K. Wehr, D. Barrow, S. Miret, A. Bomford, T. J. Peters, F. Farzaneh, M. A. Hediger, M. W. Hentze, and R. J. Simpson. 2000. A novel duodenal iron-regulated transporter, IREG1, implicated in the basolateral transfer of iron to the circulation. Mol. Cell 5:299-309. [DOI] [PubMed] [Google Scholar]
- 54.McKie, A. T., D. Barrow, G. O. Latunde-Dada, A. Rolfs, G. Sager, E. Mudaly, M. Mudaly, C. Richardson, D. Barlow, A. Bomford, T. J. Peters, K. B. Raja, S. Shirali, M. A. Hediger, F. Farzaneh, and R. J. Simpson. 2001. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science 291:1755-1759. [DOI] [PubMed] [Google Scholar]
- 55.Merchant, S. S., M. D. Allen, J. Kropat, J. L. Moseley, J. C. Long, S. Tottey, and A. M. Terauchi. 2006. Between a rock and a hard place: trace element nutrition in Chlamydomonas. Biochim. Biophys. Acta 1763:578-594. [DOI] [PubMed] [Google Scholar]
- 56.Mori, S. 1999. Iron acquisition by plants. Curr. Opin. Plant Biol. 2:250-253. [DOI] [PubMed] [Google Scholar]
- 57.Moroney, J. V., and R. A. Ynalvez. 2007. Proposed carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii. Eukaryot. Cell 6:1251-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moseley, J. L., T. Allinger, S. Herzog, P. Hoerth, E. Wehinger, S. Merchant, and M. Hippler. 2002. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 21:6709-6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 60.Nittis, T., and J. D. Gitlin. 2002. The copper-iron connection: hereditary aceruloplasminemia. Semin. Hematol. 39:282-289. [DOI] [PubMed] [Google Scholar]
- 61.Palenik, B., J. Grimwood, A. Aerts, P. Rouzé, A. Salamov, N. Putnam, C. Dupont, R. Jorgensen, E. Derelle, S. Rombauts, K. Zhou, R. Otillar, S. S. Merchant, S. Podell, T. Gaasterland, C. Napoli, K. Gendler, A. Manuell, V. Tai, O. Vallon, G. Piganeau, S. Jancek, M. Heijde, K. Jabbari, C. Bowler, M. Lohr, S. Robbens, G. Werner, I. Dubchak, G. J. Pazour, Q. Ren, I. Paulsen, C. Delwiche, J. Schmutz, D. Rokhsar, Y. Van de Peer, H. Moreau, and I. V. Grigoriev. 2007. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 104:7705-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patni, N. J., S. W. Dhawale, and S. Aaronson. 1977. Extracellular phosphatases of Chlamydomonas reinhardi and their regulation. J. Bacteriol. 130:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paulsen, I. T., and M. H. Saier, Jr. 1997. A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156:99-103. [DOI] [PubMed] [Google Scholar]
- 64.Paz, Y., A. Katz, and U. Pick. 2007. A multicopper ferroxidase involved in iron binding to transferrins in Dunaliella salina plasma membranes. J. Biol. Chem. 282:8658-8666. [DOI] [PubMed] [Google Scholar]
- 65.Petris, M. J. 2004. The SLC31 (Ctr) copper transporter family. Pflugers Arch. 447:752-755. [DOI] [PubMed] [Google Scholar]
- 66.Protchenko, O., T. Ferea, J. Rashford, J. Tiedeman, P. O. Brown, D. Botstein, and C. C. Philpott. 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:49244-49250. [DOI] [PubMed] [Google Scholar]
- 67.Quinn, J. M., and S. Merchant. 1995. Two copper-responsive elements associated with the Chlamydomonas Cyc6 gene function as targets for transcriptional activators. Plant Cell 7:623-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinn, J. M., and S. Merchant. 1998. Copper-responsive gene expression during adaptation to copper deficiency. Methods Enzymol. 297:263-279. [DOI] [PubMed] [Google Scholar]
- 69.Quisel, J. D., D. D. Wykoff, and A. R. Grossman. 1996. Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol. 111:839-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reinhardt, I., S. Haebel, A. Herbik, and T. J. Buckhout. 2006. Proteomic studies under iron stress: iron deficiency-induced regulation of protein synthesis in the green alga Chlamydomonas reinhardtii, p. 371-393. In L. L. Barton and J. Abadia (ed.), Iron nutrition in plants and rhizospheric microorganisms. Springer, New York, NY.
- 71.Robinson, N. J., C. M. Procter, E. L. Connolly, and M. L. Guerinot. 1999. A ferric-chelate reductase for iron uptake from soils. Nature 397:694-697. [DOI] [PubMed] [Google Scholar]
- 72.Roman, D. G., A. Dancis, G. J. Anderson, and R. D. Klausner. 1993. The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and encodes a protein that is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol. Cell. Biol. 13:4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rouault, T. A. 2006. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2:406-414. [DOI] [PubMed] [Google Scholar]
- 74.Rubinelli, P., S. Siripornadulsil, F. Gao-Rubinelli, and R. T. Sayre. 2002. Cadmium- and iron-stress-inducible gene expression in the green alga Chlamydomonas reinhardtii: evidence for H43 protein function in iron assimilation. Planta 215:1-13. [DOI] [PubMed] [Google Scholar]
- 75.Rutherford, J. C., and A. J. Bird. 2004. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell. 3:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sasaki, T., N. Kurano, and S. Miyachi. 1998. Cloning and characterization of high-CO2-specific cDNAs from a marine microalga, Chlorococcum littorale, and effect of CO2 concentration and iron deficiency on the gene expression. Plant Cell Physiol. 39:131-138. [DOI] [PubMed] [Google Scholar]
- 77.Sasaki, T., N. Kurano, and S. Miyachi. 1998. Induction of ferric reductase activity and of iron uptake capacity in Chlorococcum littorale cells under extremely high-CO2 and iron-deficient conditions. Plant Cell Physiol. 39:405-410. [Google Scholar]
- 78.Schloss, J. A. 1990. A Chlamydomonas gene encodes a G protein beta subunit-like polypeptide. Mol. Gen. Genet. 221:443-452. [DOI] [PubMed] [Google Scholar]
- 79.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins on silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 80.Solioz, M., and C. Vulpe. 1996. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem. Sci. 21:237-241. [PubMed] [Google Scholar]
- 81.Stearman, R., D. S. Yuan, Y. Yamaguchi-Iwai, R. D. Klausner, and A. Dancis. 1996. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science 271:1552-1557. [DOI] [PubMed] [Google Scholar]
- 82.Theil, E. C. 2003. Ferritin: at the crossroads of iron and oxygen metabolism. J. Nutr. 133:1549S-1553S. [DOI] [PubMed] [Google Scholar]
- 83.Thomine, S., R. Wang, J. M. Ward, N. M. Crawford, and J. I. Schroeder. 2000. Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. USA 97:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vert, G., N. Grotz, F. Dédaldéchamp, F. Gaymard, M. L. Guerinot, J. F. Briat, and C. Curie. 2002. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14:1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.von Wiren, N., S. Mori, H. Marschner, and V. Romheld. 1994. Iron inefficiency in maize mutant ys1 (Zea mays L. cv Yellow-Stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiol. 106:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vulpe, C. D., Y. M. Kuo, T. L. Murphy, L. Cowley, C. Askwith, N. Libina, J. Gitschier, and G. J. Anderson. 1999. Hephaestin, a ceruloplasmin homologue implicated in intestinal iron transport, is defective in the sla mouse. Nat. Genet. 21:195-199. [DOI] [PubMed] [Google Scholar]
- 87.Weger, H. G., J. K. Middlemiss, and C. D. Petterson. 2002. Ferric chelate reductase activity as affected by the iron-limited growth rate in four species of unicellular green algae (Chlorophyta). J. Phycol. 38:513-519. [Google Scholar]
- 88.Weger, H. G., C. J. Matz, R. S. Magnus, C. N. Walker, M. B. Fink, and R. G. Treble. 2006. Differences between two green algae in biological availability of iron bound to strong chelators. Can. J. Bot. 84:400-411. [Google Scholar]
- 89.Wykoff, D. D., J. P. Davies, A. Melis, and A. R. Grossman. 1998. The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol. 117:129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yi, Y., and M. L. Guerinot. 1996. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 10:835-844. [DOI] [PubMed] [Google Scholar]
- 91.Yoshioka, S., F. Taniguchi, K. Miura, T. Inoue, T. Yamano, and H. Fukuzawa. 2004. The novel Myb transcription factor LCR1 regulates the CO2-responsive gene Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii. Plant Cell 16:1466-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yun, C. W., M. Bauler, R. E. Moore, P. E. Klebba, and C. C. Philpott. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:10218-10223. [DOI] [PubMed] [Google Scholar]