Abstract
Connector enhancer of KSR (CNK) is a multidomain-containing protein previously identified as a positive regulator of the RAS/MAPK pathway in Drosophila. Using transfection experiments and an RNAi-based rescue assay in Drosophila S2 cells, we demonstrate that CNK has antagonistic properties with respect to RAF activity. We show that CNK’s N-terminal region contains two domains (SAM and CRIC) that are essential for RAF function. Unexpectedly, we also report that the C-terminal region of CNK contains a short bipartite element that strongly inhibits RAF catalytic function. Interestingly, CNK’s opposite properties appear to prevent signaling leakage from RAF to MEK in the absence of upstream signals, but then transforms into a potent RAF activator upon signal activation. Together, these findings suggest that CNK not only participates in the elusive RAF activation process, but might also contribute to the switch-like behavior of the MAPK module.
Keywords: CNK/RAS-MAPK module/RNAi /signal transduction
Introduction
The extracellular-regulated kinase/mitogen-activated protein kinase (ERK/MAPK) module, herein referred to as the MAPK module, is defined as a group of three kinases that is comprised of specific isoforms of the serine/threonine kinase RAF, the dual-specificity MAPK/ERK kinase (MEK) and the proline-directed serine/threonine kinase ERK/MAPK. This module transmits signals mainly received from the small GTPase RAS to control a number of critical cellular events such as proliferation, differentiation and survival (for review, see English et al., 1999). Early characterization of this signaling pathway identified a simple relationship among the core components, whereby upon RAS activation, RAF is recruited to the plasma membrane by RAS, which in turn triggers a phosphorylation cascade from RAF to MAPK. In-depth investigations of each individual step, however, are now unraveling a surprisingly complex process that involves additional proteins whose respective role is either partially or not understood (for review, see Kolch, 2000).
RAS was recognized early on as a major player in RAF activation, principally by its ability to recruit RAF to the plasma membrane through an interaction between its effector loop region and the RAS-binding domain (RBD) on RAF (for review, see Avruch et al., 1994). However, as this event did not appear sufficient to activate mammalian Raf-1 in vitro, additional molecules were predicted to participate in RAF activation. A search for proteins that could bind and modify RAF activity identified the 14-3-3 protein family as potential RAF regulators (for review, see Morrison and Cutler, 1997). These abundant proteins bind as dimers a wide range of targets through the recognition of specific sequence motifs, some of which require threonine or serine phosphorylation for binding (for review, see Tzivion and Avruch, 2002). RAF proteins contain two evolutionarily-conserved 14-3-3 binding sites; one surrounding phospho-serine 259 (pS259) and the second at phospho-serine 621 (pS621) in Raf-1 (Muslin et al., 1996). Growing evidence now suggests that 14-3-3-binding to these sites has opposite effects on RAF. Whereas pS621 occupancy seems critical for RAF activity (Thorson et al., 1998; Yip-Schneider et al., 2000), pS259 binding correlates with inactive RAF (Dhillon et al., 2002; Light et al., 2002), possibly by forcing RAF to adopt an inactive conformation and/or by sequestering RAF in the cytoplasm. Displacement of 14-3-3 from pS259, an event apparently triggered by RAS-binding and accompanied by pS259 dephosphorylation, appears to be one of the critical events leading to RAF activation. Despite its importance, this event does not fully account for RAF activation since mutations disrupting the pS259 site modestly enhance RAF catalytic function (Dhillon et al., 2002; Light et al., 2002).
Genetic and yeast two-hybrid screens conducted over the years have identified additional putative components of the RAS/MAPK pathway (for review, see Kolch 2000). As some of these appear to modulate RAF function, their molecular characterization might unveil key aspects to solve at last the mystery surrounding RAF activation. For instance, genetic screens in Drosophila and Caenorhabditis elegans identified kinase suppressor of Ras (ksr), an evolutionarily conserved gene encoding a putative protein kinase structurally related to RAF (Kornfeld et al., 1995; Sundaram and Han, 1995; Therrien et al., 1995). Functional studies revealed that KSR facilitates signaling from RAF to MAPK essentially by its ability to bring together the three kinases of the MAPK module (for reviews, see Morrison, 2001; Raabe and Rapp, 2002; Roy and Therrien, 2002). Besides its importance for efficient MEK and MAPK activation, KSR also appears to control RAF activity since depletion of endogenous KSR by RNA interference (RNAi) impaired RAF catalytic function in Drosophila S2 cells (Anselmo et al., 2002). It is unclear, however, whether this effect depends on KSR’s scaffolding property as recently suggested (Roy et al., 2002) or is mediated by another mechanism.
Another potential RAF regulator is connector enhancer of KSR (CNK), a multidomain-containing protein conserved among metazoans, which was originally identified in a KSR-dependent genetic screen in Drosophila (Therrien et al., 1998). As for other bona fide components of the RTK/RAS/MAPK pathway in Drosophila, CNK is required for photoreceptor cell differentiation, wing vein formation as well as for imaginal disc cell proliferation and/or survival (Therrien et al., 1998). Genetic epistasis experiments positioned CNK downstream of RAS, but upstream or in parallel to RAF, thereby suggesting that CNK might be regulating RAF activity (Therrien et al., 1998). Consistent with that possibility, CNK was found to associate with the catalytic domain of RAF (Therrien et al., 1998) and depletion of endogenous CNK by RNAi in S2 cells abolished insulin-induced RAF activation (Anselmo et al., 2002). The role of CNK with respect to RAF is probably not restricted to Drosophila since a rat homolog, named Maguin, has recently been found to associate with Raf-1 in rat brain extracts (Yao et al., 2000).
Here, using a CNK-dependent MAPK activation assay in S2 cells combined to a novel RNAi-based rescue protocol, we show that CNK has both a positive and a negative impact on RAF function. We found that CNK, through two of its N-terminal domains, integrates RAS signals to control MEK phosphorylation by RAF. In contrast, we found that CNK’s ability to associate with RAF is mediated by a short bipartite element that acts as an inhibitor of RAF catalytic function. Finally, we present evidence that the opposite functions of CNK amplify signaling difference between the off and on states of a KSR/RAF/MEK complex, which might contribute to the switch-like behavior of the MAPK module. Together, these findings identify CNK as a novel type of signal regulator that specifically controls RAF function.
Results
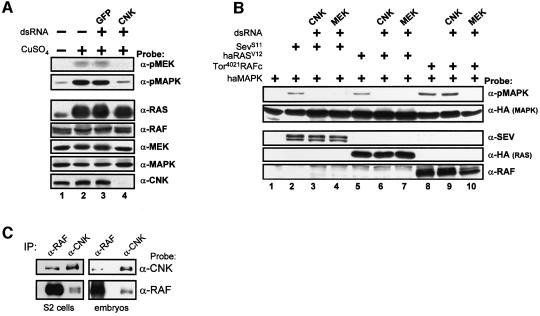
To delineate biochemically the position of CNK with respect to the components of the RAS/MAPK module, we depleted endogenous CNK by RNAi in a stable RASV12-expressing S2 cell line and assessed its effect on endogenous MEK and MAPK activation. As shown in Figure 1A, reduction of CNK by the addition of double-stranded (ds) CNK RNA specifically abrogated MEK and MAPK activation, as revealed by the decrease in phosphorylated (activated) MEK and MAPK. These results demonstrated that CNK is required downstream of RAS for activation of the MAPK module. We next examined the effect of removing CNK on activated RAF-induced MAPK activation. Compared to the activated receptor tyrosine kinase Sevenless (SevS11) or RASV12, which did not activate MAPK upon CNK or MEK depletion (Figure 1B, lanes 3, 4, 6 and 7), activated RAF (Tor4021RAFc) was still fully capable of activating MAPK upon CNK depletion, but not when MEK was eliminated (Figure 1B, lanes 9 and 10). Together, these results strongly suggest that CNK is acting between RAS and RAF.
Fig. 1. CNK activity is required downstream of RAS, but upstream of RAF. (A) Untreated (–) or CuSO4-treated (+) RASV12 cells were either incubated in the absence (–) or in the presence (+) of the indicated dsRNAs. pMEK and pMAPK levels, as well as endogenous RAS, RAF, MEK, MAPK and CNK levels were assessed by immunoblot analysis using the antibodies indicated to the right. The results shown here and thereafter are representative of at least three similar experiments. (B) S2 cells were transfected with the haMAPK reporter construct (0.3 µg) either alone (lane 1) or together (+) with the indicated combinations of SEVS11 (0.4 µg), haRASV12 (0.4 µg) or Tor4021RAFc (0.2 µg) constructs and the dsCNK or dsMEK RNAs (0.5 µg). Cells were lysed 16 h post-induction of expression and pMAPK levels were determined. Protein levels were determined as indicated. (C) Three milligrams of protein from plain S2 cells or 0–14 h Drosophila embryos were immunoprecipitated (IP) using either α-RAF or α-CNK antibodies.
Overexpression of CNK has been found previously to associate with endogenous RAF in S2 cells. Furthermore, a C-terminal fragment of CNK has also been reported to interact directly with the catalytic domain of RAF (Therrien et al., 1998). To demonstrate that a CNK/RAF complex does exist in vivo, we immunoprecipitated plain S2 cell or Drosophila embryo extracts using either anti-RAF or anti-CNK antibodies and probed immunoblots with either antibodies to look for co-immunoprecipitation. As shown in Figure 1C, the anti-RAF antibodies brought down endogenous CNK (∼10% of total NP-40-soluble CNK) and, likewise, the anti-CNK monoclonal antibody co-immunoprecipitated endogenous RAF (∼5% of total NP-40-soluble RAF) in both S2 cells and embryos. These results thus demonstrate the existence of a CNK/RAF complex in vivo. Together with the fact that CNK activity appears to be required upstream of RAF, these findings strongly suggest that CNK directly regulates RAF function.
CNK has opposite effects on RAF function
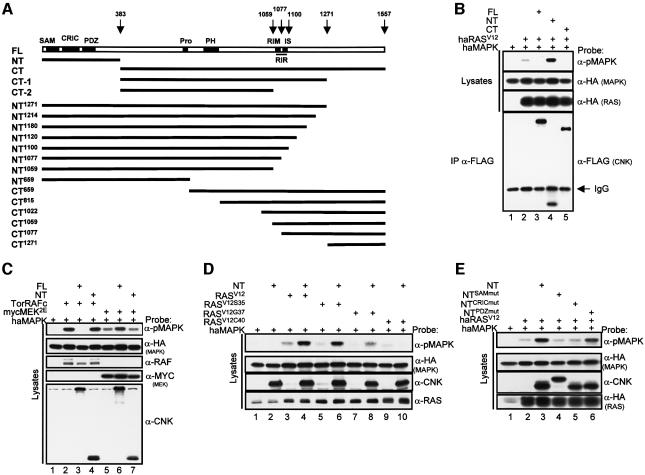
To characterize the molecular event(s) within the RAS/MAPK pathway that is/are regulated by CNK activity, we examined whether forced expression of CNK could modulate MAPK activation in S2 cells. For this purpose, we used three Flag epitope-tagged CNK constructs (Figure 2A), namely, full-length (FL), N-terminal (NT) and C-terminal (CT) that had previously been shown to modulate RASV12-mediated signaling in the developing Drosophila eye (Therrien et al., 1999).
Fig. 2. Opposite behavior of CNK in the RAS/MAPK pathway. (A) Schematic representation of Drosophila FL–CNK (top open box) with its various domains/elements (black boxes): sterile alpha motif (SAM); conserved region in CNK (CRIC); PSD-95/DLG-1/ZO-1 (PDZ); proline-rich stretch (Pro); pleckstrin homology (PH); RIR; RIM; IS. Numbers on top correspond to amino acid positions frequently referred to in the text. Solid lines, labeled to the left, denote the various CNK deletion constructs used in this study. Every CNK construct contains one copy of the Flag epitope at the N-terminus. (B) S2 cells were transfected with haMAPK alone (lane 1) or with the indicated combinations of haRASV12 (0.25 µg), FL–CNK (FL; 1.65 µg), NT–CNK (NT; 0.1 µg) and CT–CNK (CT; 1.65 µg) constructs. Cells were lysed 36 h post-induction and pMAPK levels were determined. Owing to a poor detection of Flag-tagged CNK constructs directly from cell lysates using the anti-Flag antibody, their levels were examined by immunoprecipitation (IP). (C) S2 cells were transfected with haMAPK alone (0.3 µg) or with the indicated combinations of Tor4021RAFc (0.2 µg), mycMEK2E (0.3 µg), FL (0.6 µg) or NT (0.1 µg) constructs. Cells were lysed 16 h post-induction. The flag-tagged CNK variants were examined using anti-CNK. (D) S2 cells were transfected and analyzed as in (C) using the indicated combinations of haMAPK (0.6 µg), RASV12 (0.6 µg), RASV12S35 (0.1 µg), RASV12G37 (0.25 µg), RASV12C40 (0.6 µg) and NT (0.25 µg). Various amounts of the RAS constructs were used to adjust for their apparent difference in expression levels. The RAS proteins were not tagged and thus were monitored using an anti-Drosophila RAS monoclonal antibody, which also detected endogenous RAS as seen in lanes 1 and 2. (E) S2 cells were transfected using the indicated combinations of haMAPK (0.6 µg), haRASV12 (0.25 µg), and wild-type or mutant NT constructs (0.25 µg). The mutated SAM domain (SAMmut) has a two amino acid change in conserved residues (amino acids W17S and I18S) that are critical for structural integrity (Stapleton et al., 1999). For unclear reasons, this NT–CNK mutant migrates differently from the other NT constructs (lane 4). We have generated another mutant version of the SAM domain (L71K), which changes an amino acid shown to be critical for dimer formation of the EphA4 receptor SAM domain, but does not appear to alter the structural integrity of the domain (Stapleton et al., 1999). This mutated SAM domain (NTL71K–CNK) migrates normally, and like the SAMW17S–I18S mutant, it does not cooperate with RASV12 (data not shown). NTCRICmut has a three amino acid deletion (A162-H163-R164) in the CRIC region similar to the mutation found in a Drosophila cnk loss-of-function allele (Therrien et al., 1998). Finally, NTPDZmut has a two amino acid change (G217S and F218S) in highly conserved residues of the PDZ domains (Ponting et al., 1997).
We assayed for MAPK activation by monitoring the phosphorylated levels of HA-tagged MAPK as performed above for endogenous MAPK. When expressed alone, none of the CNK constructs elevated phospho-MAPK (pMAPK) levels (data not shown and Figure 2D, lane 2 for NT–CNK). However, compared to HA-tagged RASV12 expressed alone (Figure 2B, lane 2), co-expression of FL–CNK and CT–CNK inhibited MAPK activation (Figure 2B, lanes 3 and 5, respectively), whereas NT–CNK stimulated MAPK activation (Figure 2B, lane 4). Therefore, these results indicate that forced expression of CNK affects RAS-mediated MAPK activation and also suggest that CNK comprises both positively- and negatively-acting regions.
Because CNK appears to be required between RAS and RAF (Figure 1B), we reasoned that the opposite effects of CNK could be due to a modulation of RAF function. To investigate this possibility, we examined the ability of FL–CNK and NT–CNK to alter MAPK activation induced by activated RAF or activated MEK (myc-tagged MEK2E). If FL–CNK blocked a positive step upstream of RAF, there should be no effect on MAPK activation induced by activated forms of RAF or MEK. In contrast, if CNK blocked a step downstream of RAF, it should either inhibit RAF or both RAF and MEK activities depending on the position of the inhibitory event. Strikingly, we found that FL–CNK (like CT–CNK, data not shown), completely prevented MAPK activation induced by activated RAF (Figure 2C, lane 3), but not by activated MEK (lane 6) [the apparent slight positive effect of FL–CNK on pMAPK levels induced by MEK2E (lane 6) was not reproducible]. These results therefore indicate that the negative influence of CNK occurs at a step between RAF and MEK. We applied the same logic to position the positive effect of NT–CNK and concluded that NT–CNK exerts its positive effect in a RAS-dependent manner between RAS and RAF as NT–CNK was inert on its own and did not cooperate with either activated RAF or MEK (Figure 2C, lanes 4 and 7, respectively).
Therrien et al. (1999) previously reported that NT–CNK cooperated in the Drosophila eye not only with RASV12, but also with RASV12G37, which is a RAS effector loop mutant that has a much reduced capacity to send signals through the MAPK pathway owing to its impaired association with RAF (White et al. 1995). They concluded that either NT–CNK augments RAS signaling through a RASV12G37-dependent, but MAPK-independent pathway or that, if NT–CNK functions between RAS and RAF, it could rescue or compensate to some extent the defect caused by this particular effector loop mutation thereby permitting RAF activation. To distinguish between these possibilities, we co-expressed NT–CNK with RASV12 or the three RAS effector loop mutations that had been tested. These included RASV12S35, which interacts normally with RAF, and RASV12G37 and RASV12C40, which no longer interact with RAF (data not shown). As shown in Figure 2D, NT–CNK strongly augmented pMAPK levels induced by either RASV12 or RASV12S35 (lanes 3–6) and surprisingly, it also allowed RASV12G37, which is inert on its own, to activate MAPK (compare lanes 7 and 8). These data therefore suggest that NT–CNK exerts its effect not through an alternate pathway, but largely within the RAS/MAPK pathway. The fact that NT–CNK appears to compensate to some extent the inability of RASV12G37 to activate MAPK, but not for RASV12C40 (Figure 2D, compare lanes 9 and 10), indicates that these two effector loop mutations are not equivalent with respect to their defect in activating RAF.
NT–CNK comprises three conserved regions (SAM, CRIC and PDZ domains; Figure 2A). To determine which of these is required for the positive effect of NT–CNK on the MAPK module, we tested the activity of NT–CNK mutant constructs affecting each domain individually. When co-expressed with RASV12, the mutated SAM and CRIC domain variants failed to cooperate with RAS (Figure 2E, lanes 4 and 5). In contrast, the PDZ domain mutant still retained activity (Figure 2E, lane 6). These results thus indicated that the SAM and CRIC domains are critical for the ability of NT–CNK to stimulate MAPK activation by RAS. Since the CRIC mutation used in this assay corresponds to the lesion found in a cnk loss-of-function allele (Therrien et al., 1998), it strongly suggests that our assay mimics a genuine functional property of CNK (see below).
Two short amino acid sequences in CNK define a ‘RAF-inhibitory region’ that blocks MEK phosphorylation by RAF
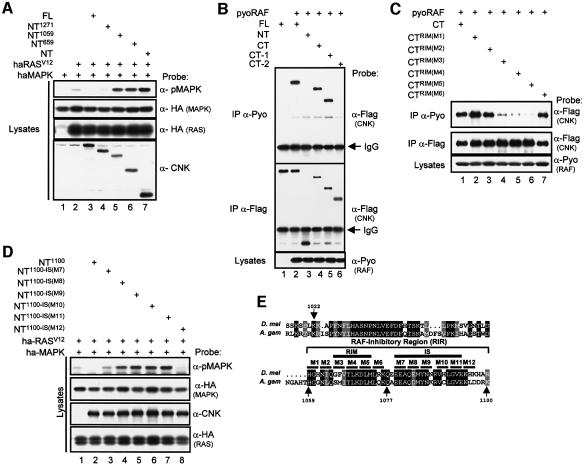
Interestingly, in addition to its positive role on the MAPK module, CNK can block RAS- or RAF-dependent MAPK activation (Figure 2). We investigated this property by first mapping the region(s) of CNK that has a negative influence on the pathway. We generated a series of C-terminal deletions of CNK (Figure 2A) and tested their ability to inhibit RAS-dependent MAPK activation. As FL–CNK, the first deletion construct (NT1271–CNK) also blocked RASV12 activity (Figure 3A, lane 4). In contrast and similar to NT–CNK, two other deletion constructs (NT1059- and NT659–CNK) no longer inhibited, but instead cooperated with RASV12 (Figure 3A, lanes 5–7). These data indicated that the C-terminal boundary of an inhibitory region, hereafter called RAF-inhibitory region (RIR), lies between amino acid position 1059 and 1271. Finer deletion constructs were then similarly tested, which positioned the RIR to a short area of ∼40 amino acids between positions 1059 and 1100 (see Supplementary figure S1, available at The EMBO Journal Online).
Fig. 3. Functional mapping of the RIR on CNK. (A) S2 cells were transfected with the indicated combinations of haMAPK (0.6 µg), haRASV12 (0.25 µg), FL and NT constructs (0.1–1 µg). Cell lysates were prepared 16 h post-induction of expression. (B) S2 cells were transfected with the indicated combinations of pyoRAF (0.7 µg), FL and CT constructs (1–1.8 µg) and NT construct (0.08 µg). Cells were lysed 36 h post-induction. For these experiments and below, pyoRAF was immunoprecipitated (IP) from cell lysates using the α-Pyo antibody and co-immunoprecipitated CNK proteins were detected using the α-Flag antibody. (C) S2 cells were transfected with the indicated combinations of pyoRAF (0.7 µg) and CT constructs (1–1.8 µg). (D) S2 cells were transfected with the indicated combinations of haMAPK (0.3 µg), haRASV12 (0.125 µg), NT constructs (0.6 µg). (E) Amino acid comparison of the Drosophila (D. mel) CNK RIR (pos. 1022–1100) to the equivalent region of A.gambiae (A. gam) CNK. Identical and conserved residues are in black and grey boxes, respectively. Positions of the ‘alanine-scanning’ mutations (M1–M12) are depicted as a solid line over the amino acid sequence. Minimal amino acid sequence for the RIM and the IS are also highlighted by a solid line over the relevant area. Although sequences within the 1022–1059 interval also appear to participate in RAF-binding (Supplementary figure S2B), these are not essential.
As CNK associates with RAF, it could be responsible for the negative effect of CNK. To address this possibility, we mapped the RAF binding site(s) on CNK to determine whether it corresponds to the RIR. CT–CNK, but not NT–CNK, was previously found to interact with RAF (Therrien et al., 1998). We first tested whether we could reproduce these findings using a transient expression assay. A polyoma (pyo) epitope-tagged RAF construct was co-expressed with either FL–, NT– or CT–CNK in S2 cells. Cell lysates were immunoprecipitated using an anti-pyo antibody and co-immunoprecipitated CNK variants were detected by probing immunoblots with an anti-flag antibody. As shown in Figure 3B (lanes 2–4), FL– and CT–CNK, but not NT–CNK, associated with RAF (∼25% of RAF is associated with FL–CNK, and ∼50% of FL–CNK is associated with RAF in these conditions). Two C-terminal deletions of CT–CNK (CT-1 and CT-2, Figure 2A) were also included in that experiment. CT-1, which ended at position 1271, still bound to RAF, whereas CT-2, which ended at position 1059, no longer interacted (Figure 3B, lanes 5 and 6). These results thus placed the C-terminal border of the RAF-binding region, hereafter called the RAF-interacting motif (RIM), in the 1059–1271 interval. The finer C- and N-terminal truncations used above to map the RIR were then used to delineate more accurately the RIM. This analysis showed that sequences in the 1059–1077 interval are critical for RAF binding (see Supplementary figure S2). Finally, we narrowed down the RIM to a nine amino acid stretch (positions 1065–1073) by testing for RAF interaction, a series of ‘alanine scanning’ mutants within the 1059–1077 interval (M1–M6, Figure 3C and E). Together, our data thus revealed that the RIM is part of the RIR, which strongly suggests that the binding of RAF by CNK is responsible for the inhibitory effect of CNK. In support of this conclusion, the three point mutations (M3–M5) that impeded RAF binding (Figure 3C) also abrogated the inhibitory effect of CNK (data not shown and see below).
Our mapping data showed that the RIR comprises additional sequences after the C-terminal end of the RIM (Figure 3E). This indicated that other sequences that are not required for RAF interaction have an inhibitory effect on RAS signaling. To define more precisely the position of these sequences, we tested a set of alanine scanning mutants within the 1077–1100 interval (M7–M12, Figure 3E) and found that mutants M7–M11 relieved the inhibitory effect of CNK (Figure 3D) but, as expected, did not prevent RAF binding (data not shown and see below). These results therefore confirmed that the RIR contains at least two distinct negative elements: the RIM that interacts with RAF and an adjacent inhibitory sequence (IS), which is required along with the RIM to inhibit signal transmission within the MAPK module.
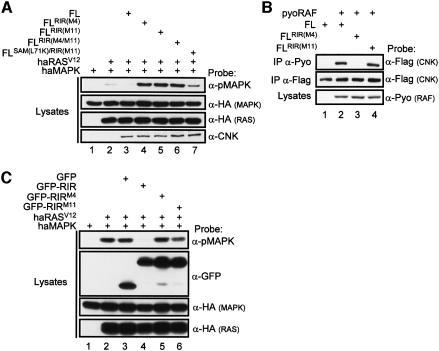
To verify whether the two negative elements (RIM and IS) also functioned accordingly in full-length CNK, we introduced the M4 mutation (affects the RIM) or the M11 mutation (affects the IS) in FL–CNK and examined their behavior with respect to RAS-induced MAPK activation and RAF-binding. Compared to FL–CNK (Figure 4A, lane 3), FLRIR(M4)–CNK and FLRIR(M11)–CNK no longer inhibited MAPK activation, but instead strongly cooperated with RASV12 (lanes 4 and 5). Furthermore, a double FLRIR(M4/M11)–CNK mutant cooperated to the same extent as either single mutants (lane 6), thus indicating that the two elements are co-required for the negative effect. As for NT–CNK (Figure 2), the ability of those mutants to cooperate with RAS appears to depend on the N-terminal domains of CNK as a double mutant version that affects both the SAM domain and the RIR barely cooperated with RASV12 (Figure 4A, lane 7). We next examined the ability of the M4 and M11 mutants to associate with RAF. As predicted, FLRIR(M4)–CNK no longer interacted with RAF (Figure 4B, lane 3), whereas FLRIR(M11)–CNK interacted normally with RAF (lane 4), thus confirming that only the RIM is essential for RAF-binding. Taken together, these results demonstrate that the RIM and IS elements, which constitute the RIR, are jointly required and sufficient to explain the inhibitory effect of CNK on the MAPK module.
Fig. 4. The negative effect of CNK is mediated by two co-required elements. (A) S2 cells were transfected with the indicated combinations of haMAPK (0.3 µg), haRASV12 (0.14 µg) and FL (0.5–0.8 µg) constructs. (B) S2 cells were transfected with the indicated combinations of pyoRAF (0.7 µg) and FL–CNK (1.5 µg) constructs. (C) S2 cells were transfected with the indicated combinations of haMAPK (0.3 µg), haRASV12 (0.2 µg) and GFP constructs (0.5 µg).
Finally, to determine whether the RIR functions autonomously, we fused it (position 1059–1100) to the C-terminal end of GFP (GFP–RIR) and examined whether this was sufficient to transpose CNK’s negative effect on GFP. As shown in Figure 4C, GFP–RIR strongly inhibited RAS-induced MAPK activation (lane 4), whereas GFP alone (lane 3) or two inactivated versions (lanes 5 and 6) of the RIR (RIRM4 or RIRM11) did not affect MAPK activation. These results therefore indicate that the RIR acts as an independent negative unit.
The RIR of CNK antagonizes RAS signaling during eye development
We wanted to determine whether the RIR of CNK also negatively influenced RAS signaling during Drosophila eye development. Intriguingly, in contrast to the data presented above, previous work showed that FL–CNK cooperated with RASV12 in the Drosophila eye (Therrien et al., 1998). We found, however, that this cooperation greatly depended on RASV12 signaling strength as well as FL–CNK expression levels, that is, FL–CNK inhibited RASV12 phenotype when a weaker RASV12 line was used (see below) or when FL–CNK levels were increased (data not shown). Nonetheless, the ability of FL–CNK to cooperate with RASV12 in the developing eye, a phenomenon not detectable in S2 cells by simply co-expressing various amounts of either protein, suggests that S2 cells might be missing a critical signal and/or factor. Although not mutually exclusive, another possibility is that there is a close relationship between the amount of activated RAS molecules and available N-terminal domains that ultimately determine RAF activation level. At low doses of RAS activity, endogenous CNK levels are probably not limiting and thus can fully mediate RAS signals for optimal RAF activation. Consequently, increasing CNK levels in that context is only providing additional RIR sequences that in turn block signal transmission. In contrast, when high levels of RAS activity are provided, endogenous CNK levels are probably limiting and thus could not fully transmit available RAS signals to RAF. Artificially increasing CNK levels in that context would provide missing CNK molecules, thereby generating greater RAF activity.
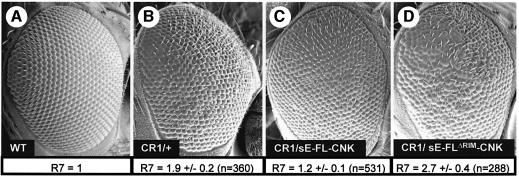
To determine whether the RIR had any inhibitory effect on RASV12 signaling in vivo, we compared the ability of FL–CNK or a CNK transgene with no RIM (FLΔRIM–CNK) to modulate RASV12 effects in the developing Drosophila eye. We crossed transgenic flies expressing the FL–CNK constructs to a strong RASV12 transgenic line (named CR2; Maixner et al., 1998). As observed previously, FL–CNK cooperated with RASV12 (data not shown). Strikingly, however, co-expression of FLΔRIM–CNK and RASV12 resulted in a complete synthetic lethality at the pupal stage with pupae presenting highly disorganized eye tissues (data not shown). This phenomenon probably resulted from a particularly strong cooperation between RASV12 and FLΔRIM–CNK. Given that both constructs are probably expressed to low levels in tissues other than the eye, the observed lethality is likely a consequence of detrimental MAPK activation in tissues required for viability. To circumvent this problem, we crossed the CNK transgenic lines to a weaker RASV12 line (CR1; Maixner et al., 1998). As expected, instead of cooperating with RASV12, FL–CNK slightly suppressed the CR1 rough eye phenotype (compare Figure 5B and C). To confirm this, we determined the average number of R7 cells per ommatidium for each genotype as a read-out for RAS/MAPK signaling (Fortini et al., 1992). The eye section results are summarized at the bottom of each SEM (Figures 5A–D). As shown in Figure 5B, CR1 had an average of 1.9 R7 cells per ommatidium. Co-expression of FL–CNK reduced this number to 1.2 (Figure 5C), which confirmed its ability to suppress RASV12 in this context. In contrast to FL–CNK, the FLΔRIM–CNK construct cooperated with RASV12 as the eyes were rougher than the CR1 parents and the average number of R7 cell per ommatidium increased to 2.7 (Figure 5D). Therefore, as in S2 cells, the RIR has a negative effect on RAS signaling during eye development. Identical results using the CR1 and CR2 lines were obtained with CNK lines that had a mutated IS element (G.Laberge and M.Therrien, unpublished results). Furthermore, and importantly, we found that the ΔRIM or IS mutant constructs were as competent as wild-type CNK at rescuing the lethality of cnk null alleles (data not shown), thus strongly suggesting that the RIR does not naturally function as a positive element (see below).
Fig. 5. CNK’s RIR antagonizes RAS signaling in vivo. (A–D) Scanning electron micrographs of adult Drosophila eyes. The genotypes are indicated on the figures, as well as the average number of R7 cells per ommatidium (± SD). n denotes the number of ommatidia analyzed and three eyes were analyzed per genotype. A t test applied on the difference in mean number of R7 cells/ommatidium between the CR1 and CR1/sE-FL–CNK genotypes or the CR1 and CR1/sE-FLΔRIM–CNK genotypes confirmed their statistical significance: P = 0.006 and 0.036, respectively.
An RNAi-based rescue assay uncovers CNK’s natural opposite effects on the MAPK module
The finding that CNK has a negative impact on the RAS/MAPK pathway is intriguing given the fact that CNK has been originally defined as a positive component for this pathway. One possibility to explain our results would be that CNK functions as a scaffold by bringing together, through independent associations, at least two signaling proteins. Therefore, as previously described for a hypothetical scaffold (Burack and Shaw, 2000), overexpression of CNK might uncouple proteins that normally require physical juxtaposition, thereby abrogating signal transmission. According to that model, the RIR might normally be required for higher order assembly of the RAS/MAPK module and thus for optimal signal transmission.
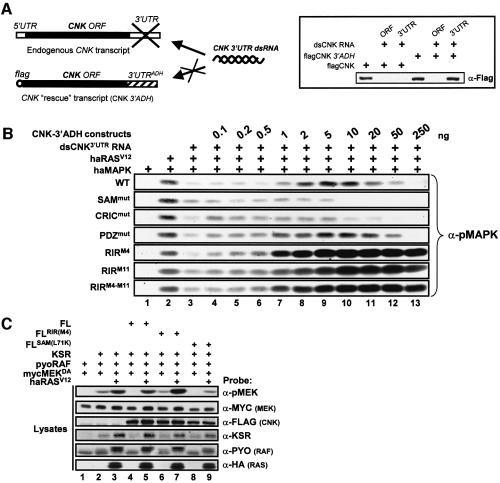
To assess unambiguously the natural effect of the RIR on the MAPK pathway, its activity needs to be monitored in non-overexpressed conditions. To that end, we devised a novel RNAi-based strategy in S2 cells that allowed us to deplete endogenous CNK levels and to restore them with exogenous, but non-targetable CNK transcripts. The ability to rescue a CNK knock-down phenotype, such as a MAPK activation defect, would indicate that relatively normal levels of exogenous CNK have been reached. This approach could then serve as a simple assay to evaluate the effect of specific CNK mutants in relatively normal stoichiometric conditions. To specifically remove endogenous CNK, we used a dsCNK RNA (dsCNK3′UTR) encompassing exclusively CNK’s 3′UTR sequences (Figure 6A), which reduced endogenous CNK levels by >90% after 4 days of culture (data not shown). Conversely, to obtain dsCNK3′UTR-resistant CNK transcripts, we replaced the natural 3′UTR sequences in the FL–CNK construct by those of the Drosophila alcohol dehydrogenase (ADH) gene. The resulting chimeric transcripts (CNK–3′ADH) were insensitive to the dsCNK3′UTR RNA (see Figure 6A; and data not shown).
Fig. 6. CNK has a natural antagonistic effect on signal transmission through the MAPK module. (A) Diagram representing the strategy used to deplete endogenous CNK by targeting its 3′UTR sequences. CNK ‘rescue’ is achieved by co-transfecting a CNK construct containing the ADH 3′UTR sequences. The specificity of the CNK 3′UTR dsRNA compared to a dsRNA targeting CNK’s open reading frame (ORF) is shown on the right: FL–CNK (0.4 µg) or FL–CNK–3′ADH (0.4 µg) constructs were transfected in S2 cells along either CNK’s 3′UTR or ORF dsRNAs (0.5 µg). Cells were lysed 36 h post-induction. (B) S2 cells were transfected with the indicated combinations of haMAPK (0.3 µg), haRAS (0.125 µg), dsCNK3′UTR RNA (20 µg) and the indicated amounts of the CNK–3′ADH variants. Cells were lysed 4 days post-induction of expression. Activated MAPK levels were detected (α-pMAPK) as well as those for haMAPK, haRASV12 and CNK-3′UTR variants (not shown). (C) S2 cells were transfected with the indicated combinations of mycMEKDA (0.3 µg), pyoRAF (0.03 µg), KSR (0.1 µg), RASV12 (0.6 µg) and FL–CNK variant (1.1 µg) constructs. Cells were lysed 36 h post-induction of expression.
Consistent with its ability to reduce endogenous CNK levels, dsCNK3′UTR RNA decreased RAS-induced MAPK activation (Figure 6B, compare lanes 2 and 3). We then determined whether introduction of a wild-type CNK–3′ADH (CNKWT–3′ADH) construct would rescue MAPK activation under those conditions. Given that CNK overexpression leads to an inhibition of RAS-induced MAPK activation (Figure 2), we conducted parallel co-transfections using a spectrum of plasmid quantities to narrow down optimal rescuing amounts. Strikingly, not only did this approach restore MAPK activation, but it also revealed a dose-dependent rescue profile identifying reproducible sub-optimal (0.1–1 ng), optimal (2–10 ng) and inhibitory (>20 ng) doses of plasmids (Figure 6B, WT panel; and data not shown). To determine whether this assay faithfully identifies CNK regions that are functionally relevant with respect to RAS-mediated MAPK activation, we tested the effect of mutations affecting the SAM, CRIC and PDZ domains, respectively. Consistent with their implication in the cooperation observed above between NT–CNK and RASV12 (Figure 2E), we found that CNKSAMmut–3′ADH and CNKCRICmut–3′ADH behave as loss-of-functions, whereas CNKPDZmut–3′ADH is as competent as wild-type CNK in restoring MAPK activation (Figure 6B). The inability of the CRIC mutation to rescue MAPK activation is consistent with its previous identification as a cnk loss-of-function allele (Therrien et al., 1998), thereby providing compelling evidence that this assay can identify bona fide functional domains/elements. In addition, these results indicate that the SAM domain is genuinely required for RAS-induced MAPK activation, whereas the PDZ domain is not involved.
Next, we tested the ability of mutations affecting the RIR to rescue MAPK activation. If the RIR normally has a positive function, its disruption should attenuate or preclude the rescue capability of a RIR mutant construct at doses optimal for WT CNK. Markedly, the three tested RIR mutants (M4, M11 or M4/M11) did not behave as loss-of-function, but rather as gain-of-function alleles (Figure 6B). Therefore, these results demonstrate that the RIR does not provide a positive input, but functions as a true inhibitory module.
CNK’s RIR prevents ‘signaling leakage’ within the KSR/RAF/MEK complex
A model to explain the two opposite roles of CNK would be that they take place at different times. For example, the negative function could be required prior to signal activation to prevent RAS-independent MEK activation by RAF. We previously reported that overexpression of KSR, RAF and MEK in S2 cells induced an association between RAF and MEK, which also resulted in a significant activation of MEK (Roy et al., 2002). Although RAS activity could increase further MEK activation, it did not appear to be critical for either KSR/RAF/MEK complex formation or MEK activation. We thus concluded that overexpression of KSR, RAF and MEK might bypass natural repressive mechanism(s) that are essential to prevent signaling in the absence of upstream signals. Given that CNK’s negative property seems to affect precisely the RAF/MEK step, we examined whether CNK could attenuate KSR-induced MEK activation by RAF.
As previously reported (Roy et al., 2002), co-expression of KSR with pyoRAF and kinase-inactivated mycMEK (mycMEKDA), resulted in MEK activation (Figure 6C, lane 2), which could be further enhanced by co-expressing RASV12 (lane 3). However, co-expression of FL–CNK along with KSR, RAF and MEK strongly blocked RAS-independent MEK activation (compare lanes 2 and 4). To determine whether the RIR was responsible for this effect, we tested the FL–CNKRIRM4 mutant in similar conditions. As shown in lane 6, this mutation obliterated the repressive effect of CNK. Similar results were obtained with either the RIRM11 or RIRM4–M11 mutations (data not shown). Interestingly, when RASV12 was introduced in the presence of FL–CNK, RAS activity counteracted much of the inhibitory effect (compare lanes 4 and 5), presumably owing to the ability of the N-terminal portion of CNK to cooperate with activated RAS. Consistent with that possibility, FL–CNKSAMmut was far less active than wild-type FL–CNK (compare lanes 5 and 9). Taken together, these results are consistent with a model whereby CNK prevents signal transmission between RAF and MEK within a KSR/RAF/MEK complex in the absence of a signal, but then facilitates signaling upon upstream activation, thus increasing significantly the signal-to-noise ratio of this signaling complex.
Discussion
CNK was originally identified as a positive component of RAS/MAPK-mediated signaling in Drosophila. In this paper, we provide evidence that the primary role of CNK in the RAS/MAPK module is to regulate RAF function. Unexpectedly, in addition to its critical role for RAF activity, we also found that CNK negatively controls RAF’s ability to phosphorylate MEK.
Three lines of evidence support the claim that CNK is essential for RAF activity. First, depletion of endogenous CNK prevented MAPK activation by RASV12, but not by activated RAF (Figure 1B). Secondly, NT–CNK cooperated with RASV12 to activate MAPK (or MEK; data not shown), but not with activated RAF (Figure 2B and C). This result not only places CNK’s positive effect between RAS and RAF, but it also suggests that this activity is RAS-dependent. Finally, NT–CNK rescued MAPK activation by RASV12G37 (Figure 2D). This finding is striking as it provides strong evidence that CNK function is intimately linked to the RAF activation mechanism. As for mammalian RAS (White et al., 1995), Drosophila RAS G37 or C40 effector mutants no longer interact with Drosophila RAF (data not shown). Since only the G37 mutant is rescued by NT–CNK co-expression (a RAF-dependent event; data not shown), it suggests that it is either a weak loss-of-function with respect to RAF binding and/or that it retained another essential function that has been lost by the C40 mutant. The G37 mutant may thus prove useful to elucidate the role of CNK in RAF activation.
We have mapped CNK’s inhibitory function to a 30 amino acid region named the RIR (see above). This region comprises at least two distinct, but co-required negative elements: the RIM and the IS elements (Figure 3E) that function together as an inhibitory unit (Figure 4A). Although its mechanism of action is unknown, the RIR appears to block signal transmission from RAF to MEK (Figures 2C and 6C) through an association between the RIM and the RAF catalytic domain. Indeed, an isolated RAF catalytic domain or the Torso-RAF catalytic domain fusion protein (Tor4021-RAFc) have been found to associate with CNK (Therrien et al., 1998; and data not shown). The association appears to be direct as it is detectable using a yeast two-hybrid interaction assay (M.Lefrançois and M.Therrien, unpublished results). The role of the IS element is unknown. It is not required for RAF binding, but it is essential for the inhibitory effect of the RIR (Figure 4). Interestingly, since only a catalytically competent RAF kinase domain associates with CNK (data not shown), it is possible that the RIR works as a RAF pseudosubstrate to control MEK phosphorylation.
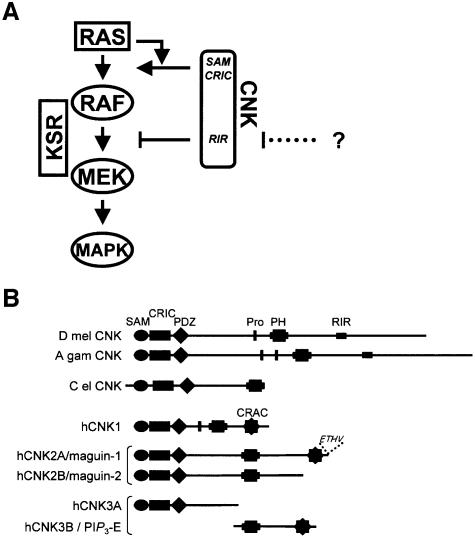
What could be the purpose of CNK’s bimodal effect? Several scenarios can be envisioned to explain our data and two of these are presented here. In quiescent cells, CNK could function together with 14-3-3 in preventing signal-independent MEK activation by RAF. This negative role might be important to ensure that no signal leaks through prior to genuine upstream activation, which otherwise might be sufficient to initiate a biological response. Upon RAS activation, CNK’s N-terminal domains would then integrate RAS signals and thus convert CNK into a positive regulator of signal transmission (Figure 7A). As CNK’s opposite action appears to augment the signal-to-noise ratio of the RAS/MAPK module (Figure 6C), it might contribute to switch-like activation of the pathway (Ferrell, 1998). Alternatively, CNK’s negative effect might have a similar role to RKIP, a recently described RAF inhibitor (Yeung et al., 1999). Namely, it might work as a rheostat to finely adjust the amount of MEK molecules activated by RAF to satisfy cell-specific requirements. For that matter, it will be interesting to determine whether the RIR is itself negatively regulated to increase signaling flow from RAF to MEK (Figure 7A). This possibility is appealing given the large difference between the ability of WT CNK and RIR-inactivated CNK constructs to rescue the MAPK activation defect (Figure 6B).
Fig. 7. CNK, a bimodal regulator of RAF function. (A) Summary of the effects of CNK on the RAS/MAPK module. The SAM and CRIC domains are two critical regions integrating RAS’s signals that then feed at a step between RAS and RAF. In contrast, the RIR inhibits MEK phosphorylation by RAF by binding to the RAF catalytic domain. The question mark and the dotted line illustrate the possibility that the RIR might itself be negatively regulated. KSR acts as a scaffold bridging RAF and MEK, thereby augmenting signal transmission (see Introduction). (B) Structural comparison of CNK homologs found in D.melanogaster (D mel; DDBJ/EMBL/GenBank accession No. AF100152), A.gambiae (A gam; DDBJ/EMBL/GenBank accession No. EAA15086), C.elegans (C el; DDBJ/EMBL/GenBank accession No. R01H10.8) and human (h). There are at least three separate genes in humans: hCNK1 (DDBJ/EMBL/GenBank accession No. AF100153), hCNK2A/B (DDBJ/EMBL/GenBank accession Nos AF418269 and AF418270, respectively) and hCNK3A/B (DDBJ/EMBL/GenBank accession Nos AK055911 and AJ310566, respectively). Although hCNK2A and hCNK2B are splicing variants, the situation is not clear for hCNK3A and hCNK3B/PIP3-E. Their respective ESTs were both mapped to the same chromosomal location (6q25.2; MapView NCBI), but are separated by ∼100 kb and have no overlapping sequences. They might thus represent two separate genes. In addition to the domain composition previously described for the CNK homologs (Therrien et al., 1998), mammalian and other CNKs found in chordates (not shown) have an additional conserved region of ∼50 amino acids of unknown function. We named this novel region of homology conserved region among chordate (CRAC) CNKs and depicted it as a black star. Similar to the rat MAGUIN homolog (Yao et al., 2000), hCNK2A contains a PDZ-binding motif (ETHV) at its C-terminus.
CNK homologs are present in other metazoans (Figure 7B). This evolutionary conservation strongly suggests that ERK/MAPK modules in other species are also regulated by a CNK activity at the level of RAF. Although the SAM and CRIC domains are relatively well conserved, the sequence corresponding to the RIR (Figure 3E) seems to be unique to Drosophila melanogaster and Anopheles gambiae. Nonetheless, rat CNK2/maguin isoforms (CNK’s closest homologs; Figure 7B) have been shown to associate with c-RAF (Yao et al., 2000), which suggests that they contain a region functionally similar to the RIR. If that were the case, it would be important to verify whether mutations disrupting its presumed negative function have any oncogenic properties. In addition, given the significance the RAS/MAPK module plays in tumor formation in humans, the identification of a short inhibitory peptide against RAF catalytic function might open new avenues for anticancer drug development.
Materials and methods
Plasmids
pMet-FL–CNK, NT–CNK and CT–CNK have been described previously (Therrien et al., 1998) and were used as starting points to generate the various CNK mutant constructs used in this study. pMet-FL–CNKΔRIM corresponds to an internal deletion from amino acids 1059 to 1077. The psE-FL–CNKΔRIM P-element construct was generated by transferring the KpnI/NotI insert from the pMet construct into the psE P-element vector (Therrien et al., 1998). Finally, the CNK-3′ADH constructs were generated by removing the natural CNK 3′UTR sequences from FL–CNK constructs, which juxtaposed downstream ADH 3′UTR sequences present in pMet.
pMet-haMAPK was generated first by inserting a PCR product corresponding to Drosophila MAPK into the EcoRI/SacI sites of pMet. A double-stranded oligonucleotide encoding three HA epitopes was then inserted in the EcoRI site of pMet-MAPK upstream of the first methionine. pMet-pyoTor4021RAFc was generated in three steps. First, an EcoRI fragment corresponding to Tor4021RAFc (Dickson et al., 1992) was inserted in the pMet vector. An AgeI site was then introduced immediately downstream from Torso’s signal peptide sequences and used to insert a PYO epitope. pMet-mycMEK2E was generated by replacing the serines 234 and 238 by glutamic acid residues.
pMet-RASV12 and effector loop mutants were described previously (Therrien et al., 1999). pMet-haRASV12, pMet-pyoRAF, pMet-KSR and pMet-mycMEKDA were described by Roy et al. (2002). pHS-SEVS11 was described previously (Therrien et al., 1998).
Cell culture, transfection and protein analysis
Cell culture, transfection, RNAi and immunoprecipitation experiments in S2 cells were performed essentially as described by Roy et al. (2002).
Cell lysates or immunoprecipitated proteins were resolved on 8 or 10% SDS–PAGE and transferred to nitrocellulose membranes. Proteins were probed using appropriate primary antibodies from the following sources: α-CNK monoclonal antibody (26A6A2) was generated by Elaine Kwan in the laboratory of Gerry Rubin. α-RAS1, α-SEV, α-PYO epitope and α-HA epitope (12CA5) mAbs were kindly provided by Gerry Rubin; α-Drosophila RAF polyclonal antibody was a kind gift from Debbie Morrison; α-MYC epitope mAb (9E10) was from Santa Cruz Biotechnology; α-MEK-1&2 and α-pMEK-1&2 polyclonal antibodies were from Cell Signaling; and α-ERK-1&2, α-dpERK-1&2 and α-FLAG mAbs were from Sigma.
Drosophila genetics and histology
Fly maintenance and crosses were conducted according to standard procedures. CR1 transgenic flies contain one copy of the sev-Ras1V12 transgene (Fortini et al., 1992) on the Cyo balancer. The sE-FL–CNK transgenic line was described previously (Therrien et al., 1998). Multiple lines expressing the sE-FL–CNKΔRIM transgene or the IS mutant version were analyzed. P-element-mediated germline transformation was performed as described by Rubin and Spradling (1982).
Adult Drosophila eye sections and scanning electron microscopy was conducted as previously described by Tomlinson and Ready (1987) and Kimmel et al. (1988), respectively.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We are grateful to David Wassarman for critical reading of the manuscript and to Debbie Morrison, Gerry Rubin and Frank Sprenger for providing reagents. We would also like to thank Alexandre Viau for help in aligning the CNK homologs and Yves Lepage for help with statistical analysis of R7 cell numbers. M.D. is the recipient of a ‘Société de Recherche sur le Cancer’ Studentship. F.R. is the recipient of a Canadian Institutes for Health Research (CIHR) Fellowship and M.T. is the recipient of a CIHR Scholarship. This work was supported by a CIHR grant to M.T.
References
- Anselmo A.N., Bumeister,R., Thomas,J.M. and White,M.A. (2002) Critical contribution of linker proteins to Raf kinase activation. J. Biol. Chem., 277, 5940–5943. [DOI] [PubMed] [Google Scholar]
- Avruch J., Zhang,X.F. and Kyriakis,J.M. (1994) Raf meets Ras: completing the framework of a signal transduction pathway. Trends Biochem. Sci., 19, 279–283. [DOI] [PubMed] [Google Scholar]
- Burack W.R. and Shaw,AS. (2000) Signal transduction: hanging on a scaffold. Curr. Opin. Cell. Biol., 12, 211–216. [DOI] [PubMed] [Google Scholar]
- Dhillon A.S., Meikle,S., Yazici,Z., Eulitz,M. and Kolch,W. (2002) Regulation of Raf-1 activation and signalling by dephosphorylation. EMBO J., 21, 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson B., Sprenger,F., Morrison,D. and Hafen,E. (1992) Raf functions downstream of Ras1 in the Sevenless signal transduction pathway. Nature, 360, 600–603. [DOI] [PubMed] [Google Scholar]
- English J., Pearson,G., Wilsbacher,J., Swantek,J., Karandikar,M., Xu,S. and Cobb,M.H. (1999) New insights into the control of MAP kinase pathways. Exp. Cell Res., 253, 255–270. [DOI] [PubMed] [Google Scholar]
- Ferrell J.E. (1998) How regulated protein translocation can produce switch-like responses. Trends Biochem. Sci., 23, 461–465. [DOI] [PubMed] [Google Scholar]
- Fortini M.E., Simon,M.A. and Rubin,G.M. (1992) Signalling by the sevenless protein tyrosine kinase is mimicked by Ras1 activation. Nature, 355, 559–561. [DOI] [PubMed] [Google Scholar]
- Kimmel B.E., Heberlein,U. and Rubin,G.M. (1990) The homeo domain protein rough is expressed in a subset of cells in the developing Drosophila eye where it can specify photoreceptor cell subtype. Genes Dev., 4, 712–727. [DOI] [PubMed] [Google Scholar]
- Kolch W. (2000) Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J., 351, 289–305. [PMC free article] [PubMed] [Google Scholar]
- Kornfeld K., Hom,D.B. and Horvitz,H.R. (1995) The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell, 83, 903–913. [DOI] [PubMed] [Google Scholar]
- Light Y., Paterson,H. and Marais,R. (2002) 14-3-3 antagonizes Ras-mediated Raf-1 recruitment to the plasma membrane to maintain signaling fidelity. Mol. Cell. Biol., 22, 4984–4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maixner A., Hecker,T.P, Phan,Q.N. and Wassarman,D.A. (1998) A screen for mutations that prevent lethality caused by expression of activated sevenless and Ras1 in the Drosophila embryo. Dev. Genet., 23, 347–361. [DOI] [PubMed] [Google Scholar]
- Morrison D.K. (2001) KSR: a MAPK scaffold of the Ras pathway? J. Cell Sci., 114, 1609–1612. [DOI] [PubMed] [Google Scholar]
- Morrison D.K. and Cutler,R.E. (1997) The complexity of Raf-1 regulation. Curr. Opin. Cell Biol., 9, 174–179. [DOI] [PubMed] [Google Scholar]
- Muslin A.J., Tanner,J.W., Allen,P.M. and Shaw,A.S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell, 84, 889–897. [DOI] [PubMed] [Google Scholar]
- Ponting C.P., Phillips,C., Davies,K.E. and Blake,D.J. (1997) PDZ domains: targeting signalling molecules to sub-membranous sites. BioEssays, 19, 469–479. [DOI] [PubMed] [Google Scholar]
- Raabe T. and Rapp,U.R. (2002) KSR–a regulator and scaffold protein of the MAPK pathway. Sci. Sig. Trans. Know. Environ., 136, PE28. [DOI] [PubMed] [Google Scholar]
- Roy F. and Therrien,M. (2002) MAP Kinase Module: The Ksr Connection. Curr. Biol., 12, R325–R327. [DOI] [PubMed] [Google Scholar]
- Roy F., Laberge,G., Douziech,M., Ferland-McCollough,D. and Therrien,M. (2002) KSR is a scaffold required for activation of the ERK/MAPK module. Genes Dev., 16, 427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G.M. and Spradling,A.C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- Stapleton D., Balan,I., Pawson,T. and Sicheri,F. (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol., 6, 44–49. [DOI] [PubMed] [Google Scholar]
- Sundaram M. and Han,M. (1995) The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell, 83, 889–901. [DOI] [PubMed] [Google Scholar]
- Therrien M., Chang,H.C., Solomon,N.M., Karim,F.D., Wassarman,D.A. and Rubin,G.M. (1995) KSR, a novel protein kinase required for RAS signal transduction. Cell, 83, 879–888. [DOI] [PubMed] [Google Scholar]
- Therrien M., Wong,A.M. and Rubin,G.M. (1998) CNK, a RAF-binding multidomain protein required for RAS signaling. Cell, 95, 343–353. [DOI] [PubMed] [Google Scholar]
- Therrien M., Wong,A.M., Kwan,E. and Rubin,G.M. (1999) Functional analysis of CNK in RAS signaling. Proc. Natl Acad. Sci. USA, 96, 13259–13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorson J.A., Yu,L.W., Hsu,A.L., Shih,N.Y., Graves,P.R., Tanner,J.W., Allen,P.M., Piwnica-Worms,H. and Shaw,A.S. (1998) 14-3-3 proteins are required for maintenance of Raf-1 phosphorylation and kinase activity. Mol. Cell. Biol., 18, 5229–5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson A. and Ready,D.F. (1987) Neuronal differentiation in the Drosophila ommatidium. Dev. Biol., 120, 366–376. [DOI] [PubMed] [Google Scholar]
- Tzivion G., and Avruch,J. (2002) 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem., 277, 3061–3064. [DOI] [PubMed] [Google Scholar]
- White M.A., Nicolette,C., Minden,A., Polverino,A., Van Aelst,L., Karin,M. and Wigler,M.H. (1995) Multiple Ras functions can contribute to mammalian cell transformation. Cell, 80, 533–541. [DOI] [PubMed] [Google Scholar]
- Yao I., Ohtsuka,T., Kawabe,H., Matsuura,Y., Takai,Y. and Hata,Y. (2000) Association of membrane-associated guanylate kinase-interacting protein-1 with Raf-1. Biochem. Biophys. Res. Commun., 270, 538–542. [DOI] [PubMed] [Google Scholar]
- Yeung K. et al. (1999) Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature, 401, 173–177. [DOI] [PubMed] [Google Scholar]
- Yip-Schneider M.T., Miao,W., Lin,A., Barnard,D.S., Tzivion,G. and Marshall,M.S. (2000) Regulation of the Raf-1 kinase domain by phosphorylation and 14-3-3 association. Biochem. J., 351, 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]