Abstract
The seven-transmembrane CCR5 was recently found to double as a coreceptor for a genetically diverse family of human and nonhuman primate lentiviruses. Paradoxically, the main region of the envelope protein believed to be involved in CCR5 utilization was mapped to hypervariable region 3, or V3, of the envelope glycoprotein gp120. In this study, we addressed the question of whether functional convergence in CCR5 utilization is mediated by certain V3 residues that are highly conserved among HIV type 1 (HIV-1), HIV type 2, and simian immunodeficiency virus. Site-directed mutagenesis carried out on three such V3 residues revealed that the Arg-298 of HIV-1 gp120 has an important role in CCR5 utilization. In contrast, no effect was observed for the other residues we tested. The inability of Arg-298 mutants to use CCR5 was not attributed to global alteration of gp120 conformation. Neither the expression, processing, and incorporation of mutant envelope proteins into virions, nor CD4 binding were significantly affected by the mutations. This interpretation is further supported by the finding that alanine substitutions of five residues immediately adjacent to the arginine residue had no effect on CCR5 utilization. Taken together, our data strongly suggests that the highly conserved Arg-298 residue identified in the V3 of HIV-1 has a significant role in CCR5 utilization, and may represent an unusually conserved target for future anti-viral designs.
HIV type 1 (HIV-1) enters target cells through fusion of the virus envelope with the target cell membrane. Initial entry is generally believed to involve high affinity binding between the external envelope glycoprotein gp120 and CD4, the primary receptor of HIV (1). This interaction is followed by post-CD4 binding events, which involve a series of incompletely characterized processes. These events include conformational changes in gp120 and gp41, interactions with certain cellular proteins, and the insertion of the fusion peptide of the transmembrane envelope glycoprotein, gp41, into the cell membrane (2, 3). The cellular coreceptors involved in HIV entry have recently been found to belong to the seven-transmembrane (7TM) chemokine receptor family (4–9).
Chemokines are small proteins that mediate leukocyte chemotaxis. There are four types of chemokines. Those with two conserved cysteines in the N-terminal domain include two major groups, CXC—or α-chemokines—and CC—or β-chemokines—according to the position of two conserved cysteines in the N-terminal domain (10). The receptor for the α-chemokine stromal cell-derived factor 1 is CXC-chemokine receptor 4 (CXCR4), also known as LCR-1, LESTR, HUMSTR, or fusin (4, 11, 12). This receptor has been found to function as the coreceptor for T cell line tropic, syncytium-inducing (SI) viruses (4). The β-chemokine receptor CCR5, which binds RANTES, MIP-1α, and MIP-1β, is the major coreceptor for macrophage-tropic, non-SI (NSI) viruses (5–9). A few of the NSI and dual-tropic isolates can also use other β-chemokine receptors, such as CCR2B and CCR3, as the coreceptor (8, 9, 13, 14).
The importance of CCR5 as an in vivo HIV-1 entry coreceptor is supported by the observation that all primary NSI viruses tested so far use CCR5 (8, 15–20). NSI viruses are found primarily during the asymptomatic stage of infection (21), and are believed to be associated with sexual transmission (22). In addition, a homozygous defect in the CCR5 gene containing a 32-bp deletion was found to occur at a higher frequency among multiply exposed uninfected individuals (23–26) than it occurs in the general population. This suggests that defects in this gene may confer protection against infection by viruses that use CCR5 as the coreceptor.
Several studies have attempted to characterize the interaction between gp120 and CCR5. Using mouse and human CCR5, or human CCR5 and CCR2B chimeric coreceptors, it has recently been reported that several extracellular domains of CCR5 are involved in coreceptor function (27–30). Other observations suggest that hypervariable region 3 (V3) of gp120 is involved in CCR5 utilization. (i) V3 of HIV-1 gp120 is known to be a major determinant of host cell tropism (31). Chimeric viruses containing V3 of a NSI virus on a SI virus background were recently shown to utilize CCR5 (8, 30, 32). (ii) It was reported that a complex of gp120 and soluble CD4 could inhibit binding between CCR5 and MIP-1α or MIP-1β. Such inhibition can be blocked by deletion of V3 in gp120 as well as neutralizing antibodies to V3 (33–36). (iii) The NSI viruses, which use CCR5 as a coreceptor, can be inhibited by β-chemokines, RANTES, MIP-1α or MIP-1β (37, 38). The V3 loop of gp120 has been shown to be critical for such inhibition (39).
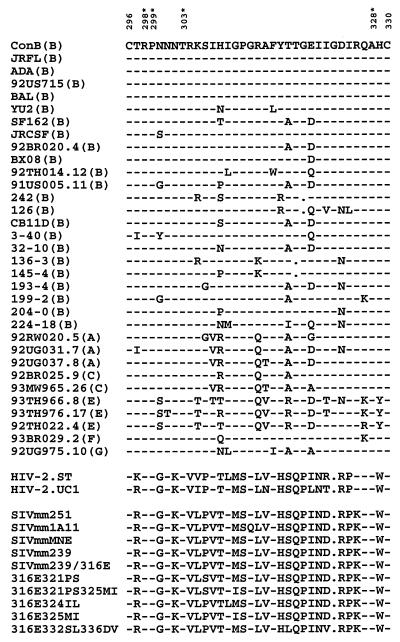
It is known that in addition to HIV-1, several simian immunodeficiency virus and HIV type 2 isolates also utilize CCR5 as a coreceptor (40–44). The high degree of sequence divergence observed within the V3 of this large family of human and nonhuman primate lentiviruses would seem to be incompatible with the functional convergence in CCR5 utilization. A closer examination of various V3 sequences, however, reveals that there are four residues highly conserved among all the viruses known to utilize CCR5 (Fig. 1). The possibility that some of these highly conserved V3 residues may mediate the observed functional convergence of CCR5 utilization was addressed in this study. We report that a highly conserved Arg-298 within V3 plays a critical role in CCR5 utilization.
Figure 1.
The V3 sequences of HIV-1, HIV-2, and SIV known to use CCR5 coreceptor. Parentheses indicate the genetic subtype of HIV-1. The numbers of two cysteine residues of V3 are shown on top. The four residues within V3, which are highly conserved among CCR5-using viruses, are numbered and marked with asterisks on top. The information was derived from references 5–9, 13, 15–20, 32, 35, 40–45. For brevity, viruses that use both CCR5 and CXCR4 are not included.
MATERIALS AND METHODS
Construction of Chimeric and Mutant Viruses.
Consensus B (ConB) virus is identical to the previously described molecular clone, HXB2RU3CI (45). The primary isolate CB11D.v was isolated from the peripheral blood mononuclear cells of an HIV-1 infected individual in Boston as described (46). CB11D.v is a macrophage tropic, NSI virus, as determined by the ability to replicate in primary macrophages but not in MT-2 cells. To generate the chimeric provirus CB11D, DNA purified from the CB11D.v-infected peripheral blood mononuclear cells was subjected to 30 cycles of PCR amplification of the envelope gene. The oligonucleotide primers −7A, 5′-GAAAGAGCAGAAGAC-AGTGG-3′ [corresponding to position 6,202–6,221 of HXB2 genome (47)], and 614B, CCAACTAGCATTCCAAAGCA-CAG-3′ (position 8,061–8,080 of HXB2), were designed to amplify a 1.87-kb env fragment containing the entire gp120 and the N-terminal 300 bp of gp41 as described (48). Amplified product was cloned into TA cloning vector PCRII (Invitrogen). DNA obtained from colonies with insert was digested with KpnI and BsmI. The 1.7-kb KpnI-BsmI fragment, which contains the 470 C-terminal residues of gp120 and 100 N-terminal residues of gp41, was cloned into a shuttle vector EX-PGEM11, which contains the 3.2-kb EcoRI-XhoI fragment of HXB2RU3. The 2.6-kb SalI-BamHI fragment containing the KpnI-BsmI insert derived from CB11D.v was cut to replace the 2.6-kb SalI-BamHI fragment of the HXB2RU3 to generate the chimeric provirus CB11D.
Oligonucleotide-directed mutagenesis was performed on the 3.2-kb EcoRI-XhoI fragment of ConB or CB11D, by the method of Kunkel (49). Mutants were identified by DNA sequencing as previously described. The 2.6-kb SalI-BamHI fragment containing the directed mutation was excised from the replicative form of each mutant and used to replace the 2.6-kb SalI-BamHI fragment of HXB2RU3. All mutations in the HXB2RU3 construct were further verified by DNA sequencing.
Virus Infection in HOS-CD4.CCR5 Cells.
HOS-CD4.CCR5, HOS-CD4.pBABE-puro, and HOS-CD4.CXCR4 were HOS-CD4 cells stably expressing CCR5, pBABE-puro, and CXCR4, respectively. They were obtained from the AIDS Research and Reference Reagent Program (National Institutes of Health, Bethesda, MD), and were propagated at 37°C in DMEM supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin-streptomycin and puromycin at 1 μg/ml as described (6). COS-7 cells were propagated in DMEM supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin. Four micrograms of wild-type or mutant DNA were transfected to 3–5 × 106 COS-7 cells by the DEAE-dextran method (48). Cell-free supernatants were collected 72 hr after transfection, filtered through 0.45 μm filters, and assayed by p24 ELISA (DuPont). Equal amounts of wild-type and mutant viruses, 10 ng of p24, were used to infect 3 × 104 HOS-CD4.CCR5, HOS-CD4.pBABE-puro, or HOS-CD4.CXCR4 cells in a 24-well plate with 1.5 ml medium per well. A half-milliliter of the culture medium was collected from each culture every 3 days for p24 analysis. The culture was monitored for 7 days.
Western Blot Analysis of Envelope Proteins in Cell Lysates.
Four micrograms of wild-type or mutant DNA were transfected to 3–5 × 106 COS-7 cells by the DEAE-dextran method (48). Mock-transfected, wild-type, and mutant-transfected COS-7 cells were collected 72 hr posttransfection, washed with PBS and subjected to centrifugation at 2,500 rpm. Cell pellets were resuspended with 100 μl RIPA lysis buffer (0.15 M NaCl/0.05 M Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and spun at 28,000 rpm (F-13 rotor, Sorvall) at 4°C for 40 min. Ten microliters of each cell lysate was electrophoresed in a SDS/10% polyacrylamide gel. A reference HIV-1 positive serum (1:200 dilution) was used for Western blot analysis as described (49).
Western Blot Analysis of Viral Lysates.
Four micrograms of wild-type or mutant DNA were transfected to 3–5 × 106 COS-7 cells by the DEAE-dextran method (48). Cell-free supernatants were collected from two transfection cultures and filtered through 0.45-μm filters (Nalgene). The supernatants were centrifuged at 20,000 rpm (Beckman, SW28 rotor) for 2 hr through a 20% (wt/vol) sucrose cushion. Virus pellets were dissolved in RIPA lysis buffer (0.15 mM NaCl/0.05 mM Tris⋅HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) and electrophoresed on a SDS/12% polyacrylamide gel. A reference HIV-1 positive serum was used for Western blot analysis as described above.
CD4 Binding Assay.
CV-1 cells were infected with recombinant vaccinia virus vCB5, a soluble CD4-expression vector, for 3 hr in cysteine-free medium and were labeled with S35-cysteine overnight. Equal amounts of supernatants (1.5 ml) were incubated with cell lysates of wild-type or mutant transfectants at 4°C for 3 hr and the mixtures were incubated with reference serum preabsorbed to protein A-Sepharose beads (50). After washing, the immunoprecipitates were eluted in a sample buffer and electrophoresed on SDS/10% polyacrylamide gel.
RESULTS
Utilization of the CCR5 Coreceptor Is V3-Dependent.
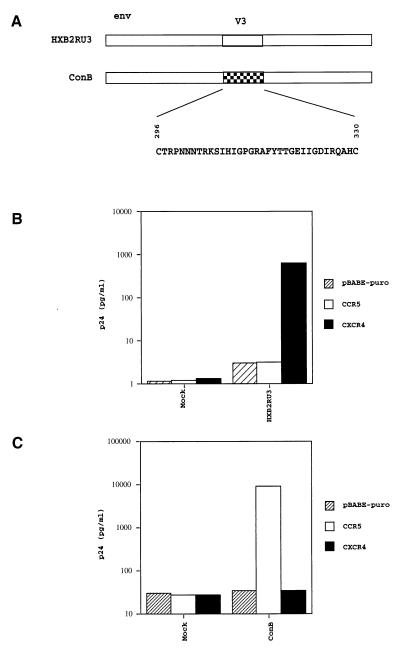
A previously described molecular clone, HXB2RU3 (45), which contains the entire coding sequences of the vpu, vpr, and nef genes was first assayed for chemokine coreceptor utilization. Cell-free viruses collected from culture supernatants of COS-7 transfectants were used to infect a human osteosarcoma cell line, HOS, stably expressing CD4 and CCR5 or CXCR4 (6). In agreement with what has been reported (4–9), this molecular clone was found to use CXCR4 (Fig. 2B). ConB used in this study is identical to the previously described molecular clone, HXB2RU3CI (Fig. 2A) (45). The only difference between ConB and HXB2RU3 is in the V3 in that ConB contains the consensus V3 sequence of HIV-1 subtype B (51). Several macrophage tropic NSI viruses, including HIV-1JRFL and HIV-1ADA (51), have a V3 sequence identical to the consensus V3 sequence of HIV-1 subtype B. As shown in Fig. 2C, cell-free ConB virus collected from culture supernatants of COS-7 transfectants was found to infect CCR5-positive cells, but neither CXCR4-positive cells nor negative control cells transfected with pBABE-puro. This finding confirms previous observations that V3 is an important determinant of chemokine coreceptor utilization (8, 30, 32). ConB was subsequently used as the parental virus to generate more mutants for the purpose of identifying V3 residues that are important for CCR5 utilization.
Figure 2.
(A) Schematic drawing of the envelope region of ConB virus and HXB2RU3. The ConB virus contains the consensus V3 sequences of HIV-1 subtype B in the backbone of molecular clone HXB2RU3. (B) Utilization of CXCR4 by HXB2RU3. (C) Utilization of CCR5 by ConB virus. Equal amounts of viruses derived from supernatants of COS-7 transfectants, as standardized by p24, were used to infect HOS-CD4 cells expressing pBABE-puro (vector control), CCR5 or CXCR4. The p24 level of culture supernatants at day 7 were shown. Mock refers to cultures infected by supernatants of mock-transfected COS-7 cultures.
Alanine Substitution of Highly Conserved V3 Residues.
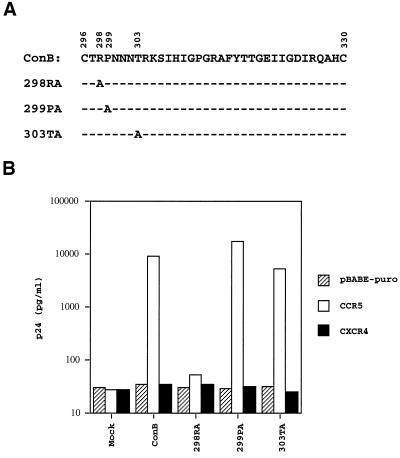
Comparing the V3 sequences of viruses known to utilize CCR5, the four highly conserved residues are Arg-298, Pro-299, Thr-303, and Ala-328 (Fig. 1). To test our hypothesis that some of these highly conserved residues may be critical for CCR5 utilization, the approach of alanine scanning was adopted. Of these four residues, the first three were individually replaced by an alanine residue by site-directed mutagenesis. The fourth residue, Ala-328, was not included in this analysis. The resulting mutant viruses of ConB were designated 298RA, 299PA, and 303TA, indicating the position of each alanine substitution (Fig. 3A). Cell-free viruses from culture supernatants of COS-7 transfectants were collected 72 hr posttransfection. Equal amounts of mutant and wild-type viruses, as standardized by p24, were used to infect HOS-CD4 cells expressing pBABE-puro, CCR5, or CXCR4. The levels of p24 in HOS-CD4.CXCR4 cells or HOS-CD4.pBABE-puro cells infected by the wild-type and mutant viruses were comparable to that of the mock-infected cells (Fig. 3B). The p24 levels of 299PA- and 303TA-infected HOS-CD4.CCR5 cells were comparable to that of ConB-infected HOS-CD4.CCR5 cells, suggesting that the conserved proline and threonine residues are not absolutely required for CCR5 utilization. In contrast, there was a significant reduction of p24 in 298RA-infected HOS-CD4.CCR5 cells, indicating that this highly conserved arginine has a critical role in CCR5 utilization.
Figure 3.
(A) The V3 sequences of ConB-derived mutants, which had alanine substitutions introduced to each of the three highly conserved V3 residues. (B) Alanine substitution of Arg-298 affects CCR5 utilization. The assay conditions were identical to those described in the legends of Fig. 2.
Mutation of the Positively Charged Arg-298 in V3.
Arginine, a positively charged residue that has been shown to be involved in protein-protein or protein-RNA interactions, contains a long side chain with two terminal amino (NH2) groups at the η position and one secondary amine (NH) at the ɛ position (52). To examine whether a positively charged residue at position 298 is essential for CCR5 utilization, the arginine residue was replaced with three different classes of amino acids. These were glutamic acid, a negatively charged residue, glutamine, a neutral residue, and leucine, a hydrophobic residue. The corresponding mutants were designated 298RE, 298RQ, and 298RL, respectively (Fig. 4A). None of these mutants infected HOS-CD4.CXCR4 cells or HOS-CD4.pBABE-puro cells (Fig. 4B). In the HOS-CD4.CCR5 cells infected by 298RE and 298RQ, a >100-fold reduction of p24 was observed when compared with cells infected by ConB. A significant reduction in p24 was also observed with 298RL-infected cells, although it was less prominent than 298RE- or 298RQ-infected cells. These findings are compatible with the interpretation that a positively charged residue at position 298 of V3 is important for CCR5 utilization. To further test this notion, mutant 298RK, which had the arginine residue replaced by another positively charged residue, lysine, was constructed. As shown in Fig. 4B, the level of p24 in 298RK-infected HOS-CD4.CCR5 cells was comparable to that of ConB-infected cells. This finding provided further evidence that a positively charged residue at position 298 of V3 appears to be required for CCR5 utilization.
Figure 4.
(A) The V3 sequences of the Arg-298 mutants 298RA, 298RK, 298RL, 298RQ, and 298RE. (B) Positively charged residue at position 298 is critical for CCR5 utilization. The assay conditions were identical to those described in the legend of Fig. 2.
Arg-298 of Another Primary Virus Linked to CCR5 Utilization.
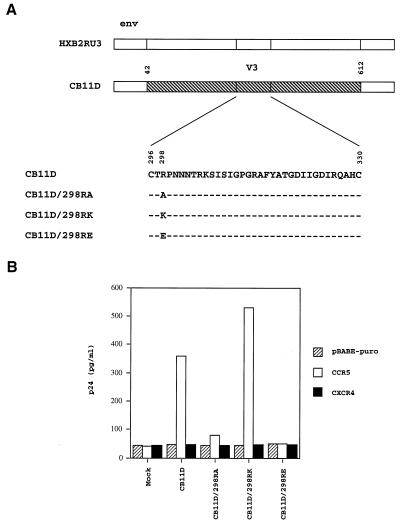
To exclude the possibility that the role Arg-298 has in CCR5 utilization is only limited to the ConB virus, the involvement of the same arginine residue of another primary virus, CB11D, in CCR5 utilization was studied (Fig. 5A). The V3 sequence of CB11D differs from that of ConB in three positions, but the arginine residue at position 298 is conserved (Figs. 1 and 5A). As shown in Fig. 5B, CB11D was able to infect HOS-CD4.CCR5 cells, but not HOS-CD4.CXCR4 or HOS-CD4.pBABE-puro cells, indicating that this virus also utilizes CCR5 as a coreceptor. Mutant viruses, designated CB11D/298RA and CB11D/298RE, were generated by substituting the arginine at position 298 of CB11D with alanine and glutamic acid, respectively. These mutations had a significant effect on virus replication in HOS-CD4.CCR5 cells (Fig. 5B). However, substitution with another positively charged residue, lysine, designated mutant CB11D/298RK, did not affect the ability of the virus to utilize CCR5 as a coreceptor. This finding lends further credence to our hypothesis that the highly conserved Arg-298 of V3 has an important role in CCR5 utilization.
Figure 5.
(A) Schematic drawing of the envelope region of CB11D and HXB2RU3. CB11D contains the KpnI-BsmI fragment (amino acids 42 to 612) of a primary isolate, CB11D.v, in the backbone of molecular clone HXB2RU3. Also shown are the V3 sequences of the CB11D and mutants, which had different type of amino acid substitutions introduced to the Arg-298. (B) Positively charged residue at position 298 is critical for CCR5 utilization by primary virus CB11D. Equal amounts of CB11D and mutant viruses, as standardized by p24, were used to infect HOS-CD4 cells expressing pBABE-puro (vector control), CCR5 or CXCR4. The p24 level of culture supernatants at day 7 were shown.
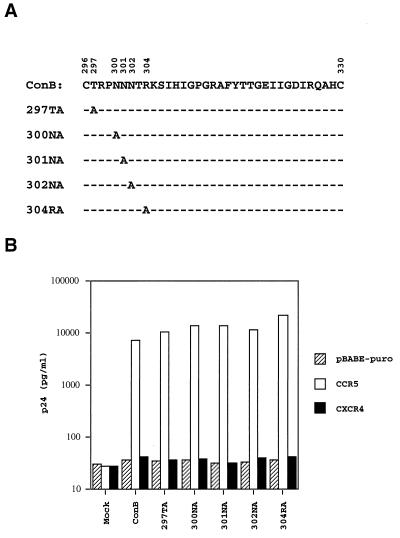
Alanine Substitution of Residues Adjacent to Arg-298.
The Arg-298 residue is located at the N-terminal base of the V3 loop, a region separated from the known β strand-β turn-β strand motif in the center of V3 (51, 53). To investigate whether residues adjacent to Arg-298 in this region may also be involved in CCR5 utilization, five more alanine substitution mutants, designated 297TA, 300NA, 301NA, 302NA, and 304RA, were constructed (Fig. 6A). As shown in Fig. 6B, the level of p24 in HOS-CD4.CCR5 cells infected by all these mutants was comparable to those infected by the wild-type virus. These results, taken together with those shown in Fig. 3B, demonstrate that of the eight consecutive residues in the N-terminal base of V3, only the highly conserved Arg-298 is indispensable for CCR5 utilization. Because seven of the eight residues in this region did not affect CCR5 utilization, it is not likely that the inability of Arg-298 mutants to utilize CCR5 are a coreceptor is caused by conformational changes in this region.
Figure 6.
(A) The V3 sequences of ConB-derived mutants, which had alanine substitutions introduced to five adjacent residues of the Arg-298. (B) Alanine substitutions introduced to five residues adjacent to the Arg-298 do not affect CCR5 utilization. The assay conditions were identical to those described in the legends of Fig. 2.
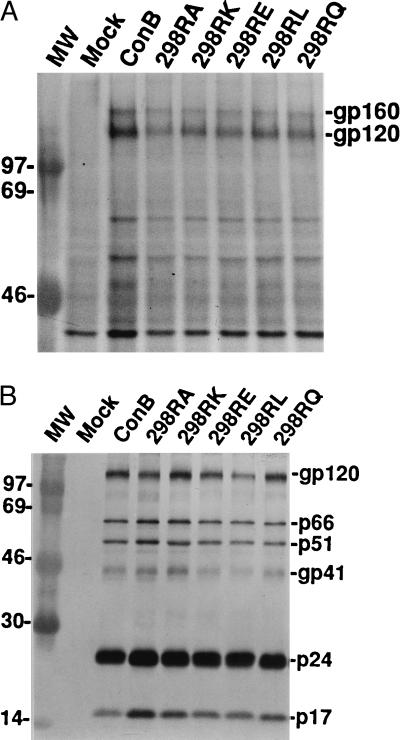
Characterization of Arg-298 Mutants.
Expression, processing, and incorporation of HIV-1 envelope proteins into virions, as well as CD4 binding, have all been used to assess if drastic changes have occurred in gp120 conformation as the result of mutation (49). To address the question of whether substitutions introduced to the Arg-298 drastically alter gp120 conformation, lysates from COS-7 cells transfected with 298RA, 298RL, 298RE, 298RQ, and 298RK were prepared for Western blot analysis. As shown in Fig. 7A, both gp160 and gp120 were readily detected by a reference HIV-1 positive human serum, indicating that neither expression nor processing of the mutant envelope protein was drastically affected by the substitutions. Western blot analysis was also performed on viral lysates derived from cell-free supernatants of COS-7 transfectants. As shown in Fig. 7B, mutant gp120 was readily detected in viral lysates and the ratio of gp120 to p24 in the viral lysates was comparable to that of the wild-type virus. Based on coprecipitation of CD4 and gp120, the amount of CD4 bound to 298 mutant proteins was found to be comparable to that of wild-type envelope protein (data not shown). This is in agreement with reports that V3 residues are not involved in CD4 binding (54, 55). Taken together, these results indicate that the inability of those 298 mutants to utilize the CCR5 receptor is unlikely to be due to global perturbation of gp120 conformation.
Figure 7.
(A) Western blot analysis of envelope proteins expressed in COS-7 cells transfected with proviral DNA of ConB or Arg-298 mutants using a HIV-1 positive human serum. Both gp160 and gp120 are indicated. (B) Western blot analysis of envelope proteins incorporated into virions of ConB and Arg-298 mutants. Viral lysates prepared from ConB, 298RA, 298RK, 298RE, 298RL, and 298RQ viruses were reacted with a HIV-1 positive human serum. Molecular mass markers are shown in lane MW.
DISCUSSION
CCR5 belongs to the superfamily of 7TM G-protein coupled receptors, which contain rhodopsin-like 7TM domains (56). The ligands for 7TM receptors cover a wide range of molecules including metal ions, monoamines, purines, peptide hormones, and large proteins (56). The interaction between 7TM receptors and their ligands follows three general patterns. The first pattern involves mainly small molecules that can penetrate the membrane and interact with residues deep in the transmembrane helices. One such example is the interaction between adrenaline and the adrenoreceptor (57). The second pattern involves peptide hormones that interact with both transmembrane and extracellular domains of the receptor. The interaction between angiotensin II, an eight-amino acid peptide, and the angiotensin AT1 receptor serves as an example. The C-terminal phenylalanine residue of angiotensin II is believed to interact with the transmembrane domain of the angiotensin AT1 receptor, whereas the N-terminal aspartic acid and arginine residues interact with the extracellular domain (58). The third pattern involves large polypeptide ligands that interact with the extracellular domain of the receptor. One such example is the interaction between interleukin 8 (IL-8) and the IL-8 type A receptor. A cluster of four residues in the N-terminal domain of IL-8 are critical for the interaction with the extracellular domain of the IL-8 type A receptor (59, 60). One of the underlying hypotheses of this study is that the interaction between gp120 and CCR5 is likely to follow the third pattern and involves only the extracellular domain of CCR5 and selective gp120 residues.
The V3 of gp120 has been found to have a significant role in coreceptor utilization (8, 30, 32–36, 39). Our finding that the chimeric virus, ConB, utilizes CCR5 as a coreceptor is in complete agreement with previous studies (8, 30, 32). Mutational analysis conducted in this study further identified an arginine residue at position 298 within the V3 of gp120 as critical for CCR5 utilization. This finding supports our hypothesis that some unusually conserved residues in the V3 loop of gp120 may be involved in CCR5 utilization.
Several lines of evidence have suggested that V3 is likely to form a structurally independent domain of gp120. (i) The entire V3 can be removed from gp120 without affecting its binding to CD4 (54, 55). (ii) Deletion of V3 does not abolish recognition of gp120 by several conformational dependent mAbs (55). (iii) Because V3 is the target of several neutralizing antibodies, it is unlikely to be buried deep inside the core of gp120 (61). In this regard, our finding that point mutations at position 298 did not significantly affect the expression and processing of mutant proteins, their incorporation into HIV-1 virions, or their ability to bind CD4, is compatible with the interpretation that the global gp120 conformation of these mutants was not drastically altered.
Although substitutions are frequently found in the V3, the length variation of V3 is rather limited due to the absence of large insertions and deletions, a feature not shared by other variable regions of gp120 (51). Based on secondary structure studies, the central part of V3 is likely to adopt a conserved motif of β strand-β turn-β strand. In contrast, the N-terminal part of V3, where the Arg-298 residue and seven other residues studied here are located, is predicted to assume a less rigid structure (51, 53). In addition to these structural features, an N-linked glycosylation site is present in the N-terminal part of the V3 of many human and nonhuman primate viruses (51). Our finding that mutations introduced to either Asn-301 or Thr-303 of the canonical N-linked glycosylation sequence had no discernible effect on CCR5 utilization suggests that this relatively conserved N-linked site is not critical for CCR5 utilization. However, this N-linked sugar may play a role in blocking the accessibility of a previously described mAb (62). Similarly, substitution of the conserved Pro-299 with an alanine residue does not affect CCR5 utilization. This highly conserved proline residue, which is located immediately adjacent to Arg-298, is unlikely to be involved in CCR5 utilization. Taken together, the observation that only substitutions of Arg-298 had an effect on CCR5 utilization, while substitutions of the seven adjacent residues did not, favors the interpretation that the involvement of the N-terminal part of V3 in CCR5 utilization is limited to the Arg-298. It is worth noting that of the four residues of IL-8 that are known to be essential for interacting with the IL-8 type A receptor, the most critical one is a positively charged arginine residue (59). The possibility that the positively charged Arg-298 residue may interact with the CCR5 coreceptor in a manner analogous to the interaction between the critical arginine residue of IL-8 and its receptor should be considered.
In addition to the Arg-298, other gp120 residues must be involved in CCR5 utilization as well. Recent studies have shown that identical HIV-1 isolates interacted with chimeras between human and murine CCR5 or with chimeras between human CCR5 and human CCR2B with distinct efficiencies (27–30). These observations suggest that the interaction between gp120 and CCR5 is likely to involve multiple extracellular domains of CCR5. Furthermore, because the Arg-298 of V3 is conserved by all the HIV-1 isolates analyzed in those studies, other gp120 residues must account for the different efficiency in CCR5 utilization. While the V3 appears to form a structurally independent domain of gp120, functional interactions between the V3 and other regions of gp120 have also been reported (63–67). In fact, a recent study reported that gp120 residues outside of the V3 might have a role in CCR5 utilization (30). Further studies are needed to address how other gp120 residues inside and outside V3 are involved in CCR5 utilization.
A more comprehensive analysis of the V3 sequence reveals that the Arg-298 is not only conserved by those viruses known to utilize CCR5, but is also conserved by the majority of human and nonhuman primate lentiviruses including those known to utilize CXCR4 as their coreceptor (51). This appears to indicate that this residue is a necessary, but not a sufficient determinant of HIV-1 for CCR5 utilization. For example, because ConB, which utilizes CCR5, and HXB2RU3, which utilizes CXCR4, share the common Arg-298 and differ only in the V3, the determinant of HIV-1 coreceptor specificity must therefore be conferred, at least in part, by V3 residues other than Arg-298.
Interacting with chemokine coreceptors through the highly conserved Arg-298 exemplifies functional convergence by the highly divergent HIV-1 envelope protein. In light of recent reports that triple therapy does not fully suppress HIV-1 in vivo (68–70), and that resistant mutants of any of the available protease inhibitors tend to be class-specific (71), it is important to continue identifying new targets for potential anti-HIV-1 strategies. The possibility that the Arg-298 represents an unusual target for antiviral therapy should be carefully considered. It is particularly intriguing that although the positively charged lysine residue appears to be interchangeable with arginine in this study, a lysine residue has not been found at the same position of Arg-298 in naturally occurring human and nonhuman primate lentiviruses. The evolutionary significance and the molecular basis of the requirement of an arginine residue in the N-terminal part of V3 remain to be explored, however, this invariable Arg-298 may represent a promising target for development of new anti-viral compounds.
Acknowledgments
We thank V. Philippon and S. Y. Chang for helpful discussions, M. Chang for technical assistance, R. Rawat for editorial assistance, and Dr. B. Moss for the soluble CD4 expression vector. The following reagents were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Disease, National Institutes of Health: HOS-CD4.CCR5, HOS-CD4.pBABE-puro, and HOS-CD4.CXCR4 cells from Dr. Nathaniel Landau. This work was supported in part by Public Health Service Grants CA-39805 from the National Institutes of Health.
ABBREVIATION
- HIV-1
HIV type 1
- V3
hypervariable region 3
- 7TM
seven transmembrane
- IL-8
interleukin 8
- SI
syncytium-inducing
- NSI
non-SI
- ConB
Consensus B
References
- 1.McDougal J S, Klatzmann D R, Maddon P J. Curr Opin Immunol. 1991;3:552–558. doi: 10.1016/0952-7915(91)90020-2. [DOI] [PubMed] [Google Scholar]
- 2.Sattentau Q J, Moore J P. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broder C C, Dimitrov D S. Pathobiology. 1996;64:171–179. doi: 10.1159/000164032. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 5.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 6.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 7.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 10.Murphy P M. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 11.Bleul C C, Farzan M, Choe H, Parolin C, Clark-Lewis I, Sodroski J, Springer T A. Nature (London) 1996;382:829–833. doi: 10.1038/382829a0. [DOI] [PubMed] [Google Scholar]
- 12.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, et al. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 13.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature (London) 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 14.Alkhatib G, Berger E A, Murphy P M, Pease J E. J Biol Chem. 1997;272:20420–20426. doi: 10.1074/jbc.272.33.20420. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L, Huang Y, He T, Cao Y, Ho D D. Nature (London) 1996;383:768. doi: 10.1038/383768a0. [DOI] [PubMed] [Google Scholar]
- 16.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dittmar M T, Simmons G, Hibbitts S, O’Hare M, Louisirirotchanakul S, Beddows S, Weber J, Clapham P R, Weiss R A. J Virol. 1997;71:8008–8013. doi: 10.1128/jvi.71.10.8008-8013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, et al. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 20.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, et al. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuitemaker H, Kootstra N A, De Goede R Y, De Wolf F, Miedema F, Tersmette M. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van’t Wout A B, Kootstra N A, Mulder-Kampinga G A, Albrecht-van Lent N, Scherpbier H J, Veenstra J, Boer K, Coutinho R A, Miedema F, Schuitemaker H. J Clin Invest. 1994;94:2060–2067. doi: 10.1172/JCI117560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 24.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, et al. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 25.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, et al. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 27.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 28.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, et al. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 29.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planelles P, Chen I S Y. In: Human Retroviruses. Cullen B R, editor. New York: Oxford Univ. Press; 1993. pp. 17–48. [Google Scholar]
- 32.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, et al. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 34.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 35.Verrier F C, Charneau P, Altmeyer R, Laurent S, Borman A M, Girard M. Proc Natl Acad Sci USA. 1997;94:9326–9331. doi: 10.1073/pnas.94.17.9326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 38.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 40.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Wu L, Newman W, Gerard N, Gerard C, Sodroski J. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kirchhoff F, Pohlmann S, Hamacher M, Means R E, Kraus T, Uberla K, Di Marzio P. J Virol. 1997;71:6509–6516. doi: 10.1128/jvi.71.9.6509-6516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z H, Clements J E, Murphey-Corb M, et al. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 45.Trujillo J R, Wang W K, Lee T H, Essex M. Virology. 1996;217:613–617. doi: 10.1006/viro.1996.0158. [DOI] [PubMed] [Google Scholar]
- 46.Wang W K, Essex M, McLane M F, Mayer K H, Hsieh C C, Brumblay H G, Seage G, Lee T H. Proc Natl Acad Sci USA. 1996;93:6693–6697. doi: 10.1073/pnas.93.13.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratner L, Haseltine W, Patarca R, Livak K J, Starcich B, Josephs S F, Doran E R, Rafalski J A, Whitehorn E A, Baumeister K, et al. Nature (London) 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 48.Wang W K, Essex M, Lee T H. J Virol. 1996;70:607–611. doi: 10.1128/jvi.70.1.607-611.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee W R, Syu W J, Bin D, Matsuda M, Tan S, Wolf A, Essex M, Lee T H. Proc Natl Acad Sci USA. 1992;89:2213–2217. doi: 10.1073/pnas.89.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee C-N, Robinson J, Mazzara G, Cheng Y-L, Essex M, Lee T-H. AIDS Res Hum Retroviruses. 1995;11:777–780. doi: 10.1089/aid.1995.11.777. [DOI] [PubMed] [Google Scholar]
- 51.Myers G, Korber B, Foley B, Jeang K-T, Mellors J W, Wain-Hobson S. Human Retroviruses and AIDS 1996: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences. Los Alamos National Laboratory, Los Alamos, NM: Theoretical Biology and Biophysics; 1996. [Google Scholar]
- 52.Calnan B J, Tidor B, Biancalana S, Hudson D, Frankel A D. Science. 1991;252:1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- 53.LaRosa G J, Davide J P, Weinhold K, Waterbury J A, Profy A T, Lewis J A, Langlois A J, Dreesman G R, Boswell R N, Shadduck P, et al. Science. 1990;249:932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- 54.Pollard S R, Rosa M D, Rosa J J, Wiley D C. EMBO J. 1992;11:585–591. doi: 10.1002/j.1460-2075.1992.tb05090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Savarese T M, Fraser C M. Biochem J. 1992;283:1–19. doi: 10.1042/bj2830001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwartz T W. Curr Opin Biotechnol. 1994;5:434–444. doi: 10.1016/0958-1669(94)90054-x. [DOI] [PubMed] [Google Scholar]
- 58.Hunyady L, Balla T, Catt K J. Trends Pharmacol Sci. 1996;17:135–140. doi: 10.1016/0165-6147(96)81588-4. [DOI] [PubMed] [Google Scholar]
- 59.Hebert C A, Vitangcol R V, Baker J B. J Biol Chem. 1991;266:18989–18994. [PubMed] [Google Scholar]
- 60.Leong S R, Kabakoff R C, Hebert C A. J Biol Chem. 1994;269:19343–19348. [PubMed] [Google Scholar]
- 61.Moore J P, Sodroski J. J Virol. 1996;70:1863–1872. doi: 10.1128/jvi.70.3.1863-1872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laman J D, Schellekens M M, Abacioglu Y H, Lewis G K, Tersmette M, Fouchier R A, Langedijk J P, Claassen E, Boersma W J. J Virol. 1992;66:1823–1831. doi: 10.1128/jvi.66.3.1823-1831.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatatos L, Cheng-Mayer C. J Virol. 1993;67:5635–5639. doi: 10.1128/jvi.67.9.5635-5639.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willey R L, Ross E K, Buckler-White A J, Theodore T S, Martin M A. J Virol. 1989;63:3595–3600. doi: 10.1128/jvi.63.9.3595-3600.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moore J P, Thali M, Jameson B A, Vignaux F, Lewis G K, Poon S W, Charles M, Fung M S, Sun B, Durda P J, et al. J Virol. 1993;67:4785–4796. doi: 10.1128/jvi.67.8.4785-4796.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wyatt R, Thali M, Tilley S, Pinter A, Posner M, Ho D, Robinson J, Sodroski J. J Virol. 1992;66:6997–7004. doi: 10.1128/jvi.66.12.6997-7004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carrillo A, Ratner L. J Virol. 1996;70:1301–1309. doi: 10.1128/jvi.70.2.1301-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 69.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Markowitz M, Ho D D, et al. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 70.Chun T W, Stuyver L, Mizell S B, Ehler L A, Mican J A M, Baseler M, Lloyd A L, Nowak M A, Fauci A S. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Condra J H, Holder D J, Schleif W A, Blahy O M, Danovich R M, Gabryelski L J, Graham D J, Laird D, Quintero J C, Rhodes A, et al. J Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]