Abstract
Using ChIP–chip assays (employing ENCODE arrays and core promoter arrays), we examined the binding patterns of three members of the E2F family in five cell types. We determined that most E2F1, E2F4, and E2F6 binding sites are located within 2 kb of a transcription start site, in both normal and tumor cells. In fact, the majority of promoters that are active (as defined by TAF1 or POLR2A binding) in GM06990 B lymphocytes and Ntera2 carcinoma cells were also bound by an E2F. This very close relationship between E2F binding sites and binding sites for general transcription factors in both normal and tumor cells suggests that a chromatin-bound E2F may be a signpost for active transcription initiation complexes. In general, we found that several E2Fs bind to a given promoter and that there is only modest cell type specificity of the E2F family. Thus, it is difficult to assess the role of any particular E2F in transcriptional regulation, due to extreme redundancy of target promoters. However, Ntera2 carcinoma cells were exceptional in that a large set of promoters were bound by E2F6, but not by E2F1 or E2F4. It has been proposed that E2F6 contributes to gene silencing by recruiting enzymes involved in methylating histone H3. To test this hypothesis, we created Ntera2 cell lines harboring shRNAs to E2F6. We found that reduction of E2F6 only induced minimal alteration of the transcriptome of Ntera2 transcriptome. Our results support the concept of functional redundancy in the E2F family and suggest that E2F6 is not critical for histone methylation.
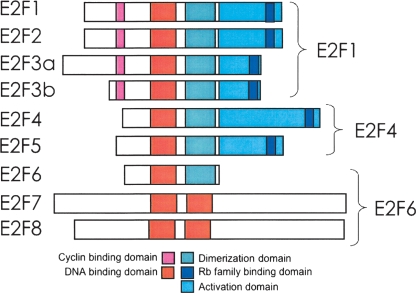
The E2F family has been implicated in controlling a myriad of critical cellular (entrance into S phase, regulation of mitosis, apoptosis, DNA repair, and DNA damage checkpoint control) and organismal (regulation of differentiation and development and tumorigenesis) functions (Slansky and Farnham 1996; Dimova and Dyson 2005; Kong et al. 2007). In general, the ability of E2F family members to regulate these processes has been linked to their ability to modulate the transcriptional output of target promoters. There are eight genes for E2F family members encoded in the human genome, with one family member (E2F3) encoding two proteins through the use of alternative promoters (for a recent review of the E2F family, see DeGregori and Johnson 2006). The highest degree of homology among the E2F family members is in their DNA binding domains, which is consistent with the finding that in vitro they can all bind to the same consensus motif of TTTSSCGC (where S is either a G or a C). However, the E2Fs can be divided into two classes based on whether they function as hetero- or homodimers. E2Fs 1–6 require dimerization with a DP family member (DP1 or DP2) to bind in vitro to the consensus site, whereas E2F7 and E2F8 bind in vitro to the consensus site as homodimers. The E2F family members have also been divided into several subclasses based on the patterns of their expression and their transcriptional regulatory properties in reporter gene assays (Fig. 1). E2F1, E2F2, and E2F3a are often considered to be activators and display maximal expression during S phase of the cell cycle. Members of a second class of E2F proteins, E2F3b, E2F4, and E2F5, are expressed throughout the cell cycle and are often referred to as repressors because they are thought to function mainly in combination with members of the retinoblastoma (RB1) family of corepressors. E2F6, E2F7, and E2F8 are also classified as transcriptional repressors, but they function independently of the RB1 family. However, there is little biological evidence to support the classification of the different E2Fs as activators or repressors. For example, it is important to note that the transactivation domain and the RB1 family binding domain overlap and therefore E2Fs 1–5 have the potential to function as both activators and repressors. Most studies have relied on artificial reporter assays to define the role of a specific E2F in the regulation of a particular gene. It is more likely that it is the assembly of proteins at a particular promoter and the specific cellular environment that determines if a specific E2F family member activates or represses transcription (Fry and Farnham 1999).
Figure 1.
The E2F family. Shown is a schematic comparing the domain structure of E2Fs 1–8; not shown are the lower abundance isoforms of E2F6 (Kherrouche et al. 2004). We have chosen E2F1 as a representative of the class of E2F family members that contains the N-terminal cyclin binding domain and the C-terminal RB1 family binding domain, E2F4 as a representative of the class of E2F family members that lacks the cyclin binding domain but contains the RB1 family member binding domain, and E2F6 as a representative of the class of E2F family members that are thought to function as repressors in an RB1-independent manner.
Knockout experiments have, in general, revealed fairly modest effects of loss of a single E2F, suggesting redundancy in their function. However, some phenotypic changes have been observed upon loss of an individual E2F (Dimova and Dyson 2005). For example, loss of E2F1 results in decreased T-cell apoptosis, testicular atrophy, and tumor formation (Field et al. 1996); E2F2 mutant mice show defects in the proliferative properties of hematopoietic cells (Murga et al. 2001; Zhu et al. 2001); E2F3 mutants display partially penetrant embryonic lethality and heart defects (Humbert et al. 2000); E2F4 mutants are runted and have craniofacial, hematopoietic, and intestinal defects (Humbert et al. 2000; Rempel et al. 2000); mice lacking E2F5 develop hydrocephalus and are perinatal lethal (Lindeman et al. 1998); and E2F6 knockout mice display modest homeotic transformations of the axial skeleton (Storre et al. 2002). E2F7 and E2F8 knockout mice have not yet been reported. Greater effects have been observed when the levels of various combinations of E2Fs have been reduced. For example, E2F1, E2F2, and E2F3 must be knocked out simultaneously to see an inhibitory effect on proliferation (Wu et al. 2001). Also, a double knockout of both E2F4 and E2F5 results in embryonic lethality (Gaubatz et al. 2000). Interestingly, cells derived from the E2F4−/−/E2F5−/− embryos showed normal cell cycle regulation. One issue that may complicate attempts to understand the role of the individual E2Fs is that loss of one family member may lead to compensation by another, either as a result of increased levels of other family members or replacement of one family member for the other at particular promoters. In fact, a recent study has suggested that long-term loss of certain E2Fs may lead to compensatory function by the others to ensure cell viability and/or survival of the organism (Kong et al. 2007).
As noted in a recent review (Dimova and Dyson 2005), a complete understanding of the role of the different E2Fs requires identification of the full range of their target genes. Therefore, to gain further understanding of the role of the different E2Fs, we have taken the unbiased approach of ChIP–chip that allows the identification of in vivo binding sites for a particular E2F without artificially altering the ratios of the E2Fs to each other. For these studies, we have chosen E2F1 as the representative member of the class of E2Fs that have an N-terminal extension that contains a cyclin A-interaction domain, E2F4 as the representative member of the class of E2Fs that lacks the N-terminal extension but shares very similar transactivation and RB1 family binding domains with E2Fs 1–3, and E2F6 as the representative member of the class of E2Fs that lacks a transactivation and an RB1 family binding domain. To ensure that the results we obtained were not unique to a specific cell type, we have performed these assays in the context of five different cell types, including both normal and tumor cells.
Results
E2F family members localize to core promoter regions in both normal and tumor cells
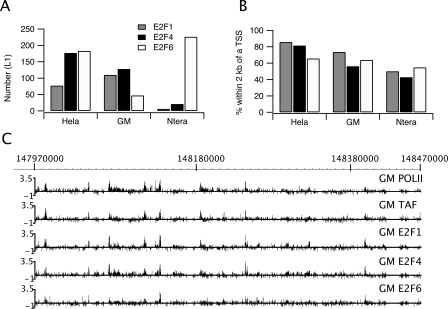
Using ChIP–chip analysis and genomic tiling arrays, we had previously shown that E2F1 binds predominantly within 2 kb of transcription start sites in HeLa cells (Bieda et al. 2006). This striking localization pattern was quite distinct from certain other transcription factors that bind predominantly to regions outside of core promoters (Cawley et al. 2004; Carroll et al. 2005; Jin et al. 2007). To determine whether other E2F family members show a similar binding pattern, we performed ChIP–chip analysis of E2F1, E2F4, and E2F6. For these experiments, we performed ChIP assays using three independent cultures of HeLa cells, prepared amplicons, and probed ENCODE arrays (which contain ∼400 genes and include 44 regions ranging from 500 kb to 2 Mbp). We used the Tamalpais peak-calling program (Bieda et al. 2006) to identify E2F1, E2F4, and E2F6 binding sites for each of the three arrays and then selected all sites that were identified in at least two of the three experiments as the set of binding sites for that specific E2F family member (Fig. 2A). A list of all the arrays for this paper, including the cell type, array platform, and array identification number, is provided in Supplemental Table S1. The number of called peaks for each ENCODE array and the overlap of binding sites for each of the triplicate experiments are detailed in Supplemental Table S2. As can be seen in Figure 2B, in HeLa cells all three E2F family members localized within 2 kb of the start site of transcription. Thus, the binding pattern that we had previously reported for E2F1 was not unique for this family member.
Figure 2.
E2F family members bind near start sites in both normal and tumor cells. ChIP–chip assays were performed using three independent cultures for each of three different cell types (HeLa, GM06990, or Ntera2 cells) and antibodies to E2F1, E2F4, or E2F6. The samples were analyzed on ENCODE arrays (see Methods) and peaks were called for each array using the L1 category of peaks called by the Tamalpais program (Bieda et al. 2006). A binding site was identified if a site was enriched in at least two of the biological replicate experiments for a particular antibody/cell type combination. (A) Shown are the number of E2F1, E2F4, or E2F6 binding sites identified in the ENCODE regions for the three different cell types. Because the E2Fs bind to many of the same regions, the total number of E2F binding sites for a given cell type is less than the sum of the E2F1, E2F4, and E2F6 sites; there were 270 distinct E2F binding sites for HeLa cells, 187 for GM06990 cells, and 232 for Ntera2 cells (see Supplemental Table S2). (B) Shown is the % of each set of E2F1, E2F4, and E2F6 binding sites for each cell type that is within 2 kb of a transcription start site. (C) ChIP–chip binding patterns for a region of chromosome 1 are shown for RNA polymerase II (POLII), TAF1 (TAF), E2F1, E2F4, and E2F6 in GM06990 (GM) cells. The Y-axis indicates fold enrichment of the ChIP sample.
It was possible that the binding pattern we observed was specific to a particular cell type. In our previous studies (e.g., Bieda et al. 2006) and in the experiments reported above, we used HeLa cells, a cervical cancer cell line that has been grown in culture for decades. To determine the binding patterns of E2F family members in other cell types, we next used the GM06990 cell line, which is derived from normal B lymphocytes, and Ntera2 cells, which are derived from a testicular embryonal carcinoma. Again, ChIP–chip experiments using antibodies to E2F1, E2F4, and E2F6 were performed in triplicate using three independent cultures of GM06990 or Ntera2 cells, peaks were called, and binding sites were identified as those peaks that were common to at least two of the three experiments. We found that the number of E2F targets in the 1% of the genome represented by the ENCODE regions for each cell type is fairly similar (270 in HeLa, 232 in Ntera2, and 187 in GM06990). After merging all the targets for all three E2F family members in all three cell lines, the 689 possible sites collapsed to 480, because many of sites were occupied by an E2F in more than one cell type. We also noted that the 480 sites collapse to 387 1-kb regions, indicating that about 20% of the sites are near other sites. As noted above, the total number of E2F binding sites that we identified in GM06990 and Ntera2 cells was fairly similar (187 vs. 232; see Supplemental Table S2). However, we found that there was a very different distribution of the binding sites relative to the different E2F family members (Fig. 2A). Specifically, there was a similar number of E2F1 and E2F4 binding sites in GM06990 cells, but the number of E2F6 sites was quite small. In contrast, almost all of the E2F binding sites in Ntera2 cells were due to binding of E2F6. Importantly, although the number of binding sites for each E2F family member varied in the different cells (Fig. 2A), in all cases there was strong preference for binding near core promoter regions (Fig. 2B). In fact, the binding pattern of the E2Fs was very similar to the binding patterns of POLR2A and TAF1 (Fig. 2C). Using a simple analysis (see Methods), we formed a rough and conservative (i.e., high) estimate that ∼14% of peaks would randomly be found within 2 kb of a transcription start site. Hence, in all cases, there is a large enrichment of E2F sites near the transcription start site over the random expectation.
De novo motif analysis of E2F family members
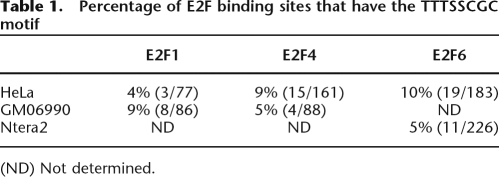
In our previous study (Bieda et al. 2006), we found that the classical E2F consensus (TTTSSCGC) previously identified by in vitro experiments (Tao et al. 1997) was found at only ∼12% of E2F1 binding sites identified by in vivo HeLa cell ChIP–chip experiments. This suggests that an in vivo binding motif may be different than a motif developed using in vitro binding data. To determine an in vivo binding motif for E2F1, we applied the ChIPMotifs approach (Jin et al. 2006) to train different data sets obtained from the ChIP–chip experiments performed using ENCODE arrays. Briefly, the ChIPMotifs approach incorporates a statistical bootstrap resampling method to identify the top motifs detected from a set of ChIP–chip training data using ab initio motif-finding programs such as Weeder (Pavesi et al. 2004) and MEME (Bailey and Gribskov 1997). The strength of our ChIPMotifs approach is that it takes advantage of the important discriminating features embedded in ChIP–chip data that identify the enriched binding data as a positive data set and the nonenriched binding data as a negative data set. Our approach not only identifies a consensus-like motif, but it also provides a cutoff threshold for the position weight matrix (PWM) of that motif.
We began by choosing the sites bound by E2F1, E2F4, and E2F6 in HeLa cells as a positive training set and a set of regions not bound by these factors (each region having an average of length of 500 bp and within 10 kb of a promoter region of a known gene) as a negative data set (see Supplemental Table S3). After performing the de novo motif analysis (see Methods for details), we identified motif logos and cutoff thresholds for the PWMs for E2F1, E2F4, and E2F6 (Supplemental Fig. S1). Our results clearly show that different E2F family members share the same core consensus STTTS binding site in HeLa cells. Using the cutoff scores shown in Supplemental Figure S1, more than 50% of the HeLa cell E2F binding sites identified from the ChIP–chip experiments included the identified motif, whereas more than 80% of the sites in the negative control data set lacked the identified motif. Although the identified E2F motifs partially resemble the previously defined E2F consensus motif TTTSSCGC identified using in vitro casting experiments (Tao et al. 1997), it was surprising that the GC-rich core SSCGC was not the predominant motif in the set of in vivo E2F binding sites, because this GC-rich segment appears important for in vitro binding in a structural study (Zheng et al. 1999). As a further examination of E2F binding sites, we performed the ChIPMotifs analysis using the E2F1 and E2F4 binding sites from GM06990 cells (there were not enough E2F6 binding sites in these cells for analysis) and the E2F6 binding sites from Ntera2 cells (there were not enough E2F1 or E2F4 binding sites in these cells for analysis). We found that the STTTS core motif was the predominant in vivo motif, regardless of which family member or which cell type was analyzed (Supplemental Fig. S1). It was possible that a subset of the binding sites did contain the TTTSSCGC motif but that the de novo motif identification programs could not pick these up because of their low occurrence. Therefore, we directly examined the binding sites and found that 4%–10% (depending on the specific E2F and cell type) of the in vivo binding sites contained a consensus motif (Table 1). These numbers are similar to those observed in previous studies in which pooled ChIP samples (with no amplification) were used to identify E2F4 targets on CpG arrays (Weinmann et al. 2002) or in which amplicons produced by LMPCR were used to identify E2F1 targets on ENCODE arrays (Bieda et al. 2006). Thus, multiple types of experimental samples (pooled ChIPs, amplicons produced by LMPCR, or amplicons produced by the whole genome amplification procedure used in this study) and multiple types of array platforms all identify E2F binding sites that differ from the in vitro consensus motif.
Table 1.
Percentage of E2F binding sites that have the TTTSSCGC motif
(ND) Not determined.
Comparison of binding specificity of E2F family members
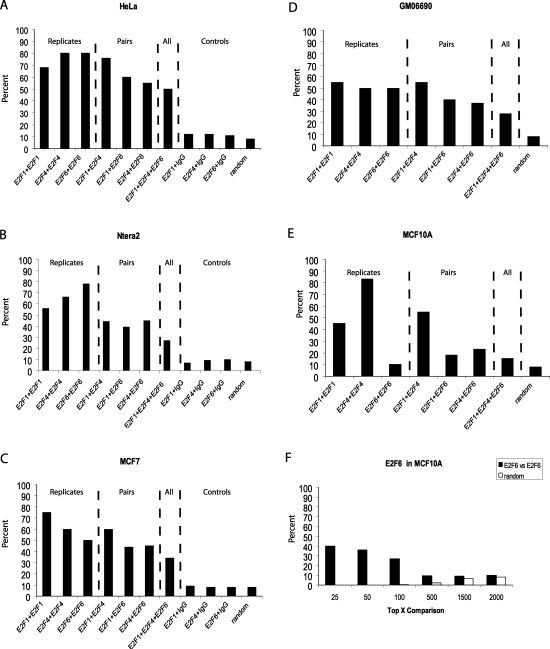
Because the experiments using ENCODE arrays demonstrated that E2F1, E2F4, and E2F6 were highly enriched at core promoter regions in all cell types analyzed, we were confident that core promoter arrays would be suitable for identifying a fairly comprehensive set of E2F target genes. For our experiments, we used arrays that contain 1.5 kb for each of 24,000 human promoters and performed two independent (from different cultures of cells) E2F1, E2F4, or E2F6 ChIP–chip assays for each of five different cell lines (see Supplemental Fig. S2 below for a Western blot analysis of the E2Fs in the different cell types). Promoters were then ranked based on the average enrichment value of the top 11 of the 15 probes for each promoter and the ranked lists were compared for each of the two experiments for a given E2F and cell line combination. The results using a particular antibody and cell type combination are very reproducible. For example, as shown in Figure 3A, 70%–80% of the promoters ranked in the top 2000 were the same for the two E2F1, E2F4, or E2F6 ChIP–chip assays in HeLa cells. For comparison, we performed duplicate ChIP–chip assays using a nonspecific rabbit IgG sample; only 10% of the top ranked targets pulled down by IgG overlapped with the E2F targets, a number approximately equal to what would be expected on a random basis. Based on the motif analysis described above indicating that at least half of the binding sites in the E2F1, E2F4, or E2F6 ENCODE data sets contained the same core motif, we would predict that at least half of the target promoters bound by the different family members should be the same in HeLa cells. In fact, we found that 55%–75% of the target promoters were the same for any pairwise comparison of E2F1, E2F4, or E2F6 and that a set of 1000 promoters (from the top 2000) were bound by all three family members in HeLa cells. In general, the same results were obtained using the other four cell lines; that is, most of the promoters that were in the top 2000 ranked list for one E2F family member were also in the top 2000 ranked list for the other E2F family members. However, the MCF10A cells gave a slightly different pattern. Although E2F1 and E2F4 bound to the same set of promoters in these cells, there was only a very small overlap between E2F6 and either E2F1 or E2F4 (Fig. 3E). We noted that the E2F6 replicates in MCF10a also showed very low overlap in the top 2000. To eliminate the possibility that one of the E2F6 experiments did not work, we performed a third E2F6 ChIP–chip assay using MCF10A cells. The same low percentage overlap in the top 2000 targets was seen when the third E2F6 experiment was compared with either the first or the second array (data not shown). Closer inspection of the E2F6 target genes in MCF10A cells revealed that a very good overlap is seen in the top 25 or top 50 promoters on each array (Fig. 3F), suggesting that there are only a few E2F6 target genes in MCF10A cells. One explanation for the small number of E2F6 target promoters in MCF10A cells could be that MCF10A cells express very little E2F6. To investigate this possibility, we performed a Western blot analysis of E2F6 using nuclear extracts (Supplemental Fig. S2). We found that the amount of E2F6 protein expressed is similar for all five cell types. Thus, the low number of E2F6 target genes in MCF10A cells is not due to a low abundance of E2F6. However, the amount of E2F4 expressed in MCF10A cells is quite high, in comparison with the other cell types. Therefore, it is possible that E2F6 is excluded from target promoters because of the abundance of E2F4.
Figure 3.
E2Fs bind to a similar set of target genes in a given cell type. Pairwise comparisons of the percentage of promoters in common in the sets of top 2000 ranked promoters in E2F1, E2F4, and E2F6 ChIP–chip assays in HeLa (A), Ntera2 (B), MCF7 (C), GM06990 (D), and MCF10A (E) cells. In A–C the overlap in top-ranked promoters after comparison of E2F arrays to IgG arrays is also shown. In A–E, the number expected by random chance to be in common when comparing the data sets is also shown. In F, the percentages of promoters in common in two independent E2F6 ChIP–chip assays in the set of the top 25, 50, 100, 500, 1500, and 2000 ranked promoters is shown for MCF10A cells. Also shown in F is the number expected by random chance to be in common when comparing the various numbers of ranked promoters.
Comparison of cell-type specificity of E2F family members
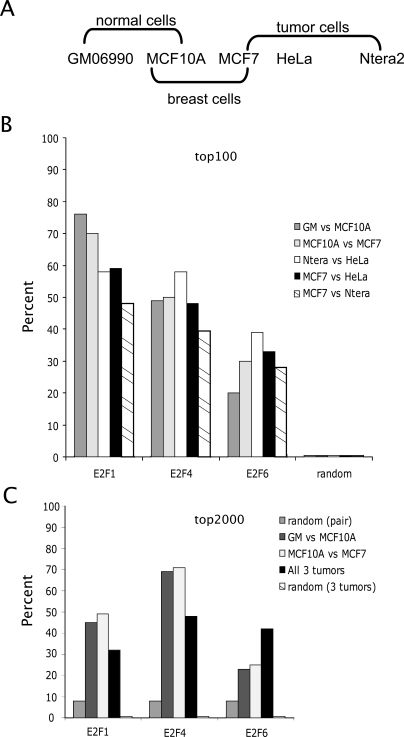
Having shown that, in general, the sets of target genes for E2F1, E2F4, and E2F6 are similar in a given cell type, we next wanted to determine whether the E2F family members regulated the same or different sets of target genes in different cell types. The five cell types that we have chosen to study include two nontumorigenic cell lines (GM06990 B lymphocytes and MCF10A breast cells) and three tumor cell lines (MCF7 breast tumor cells, HeLa cervical carcinoma cells, and Ntera2 testicular embryonal carcinoma cells). We have grouped these five cell lines into several different categories, including normal cells, normal versus tumor breast cells, and tumor cells (see Fig. 4A). We first asked whether the promoters having the highest enrichment values (the top 100 enriched promoters of the set of all 24,000 promoters on the array) for E2F1, E2F4, or E2F6 were the same in the normal cells (GM06990 vs. MCF10A), in normal versus tumor breast cells, or in any pairwise comparison of the three different tumor cell lines (Fig. 4B). We found that many of the highest enriched E2F1 and E2F4 target genes were the same in all the cell types, indicating that stable binding of E2F1 or E2F4 is not heavily influenced by cell type-specific factors. In particular, the set of promoters in the top 100 bound by E2F1 in the two normal cell lines was 75% identical, a large enrichment over the random expectation value of <1%. We also compared a larger set of target genes for each factor in the different cell types. As shown in Figure 4C, a large percentage of the top 2000 promoters bound by the E2F family members was similar in the different cell types. For example, the set of top 2000 E2F4 target genes was about 70% identical in the two normal cell lines and in the two breast cell lines, much above the random expectation value of ∼8%. The E2F6 target genes varied more in the different cell types, due in large part to the very low number of E2F6 target genes in MCF10A cells. We merged the list of promoters ranked in the top 100 for all E2Fs and all cell types (3 E2Fs × 5 cell types × 100 promoters = 1500 total) and produced a list of ∼450 unique promoters (Supplemental Table S4).
Figure 4.
E2F family members show little cell type specificity in binding. (A) Shown are the five cell types used in this study, with an indication of how they are organized into different comparison sets. (B) A comparison of the top 100 E2F1, E2F4, or E2F6 target promoters in both normal cells (GM06990 vs. MCF10A), in the normal vs. tumor breast cells (MCF10A vs. MCF7), and in all pairwise comparisons of the three tumor cell lines is shown; a combined list of the names for the top 100 E2F target promoters in the five cell types is provided as Supplemental Table S4. (C) A comparison of the top 2000 E2F1, E2F4, or E2F6 target promoters in both normal cells (GM06990 vs. MCF10A), in the normal vs. tumor breast cells (MCF10A vs. MCF7), and in all three tumor cell lines is shown.
Functional analysis of E2F target genes in Ntera2 cells
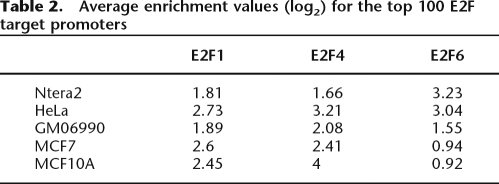
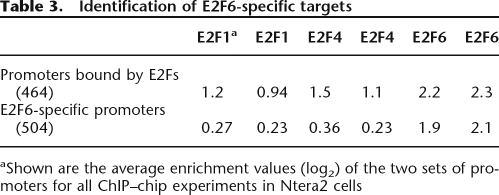
The L1 set of peaks from ENCODE ChIP–chip data represents the most stable binding sites. As seen in Figure 2A, in Ntera2 cells there are many more robust E2F6 sites than E2F1 or E2F4 sites in the ENCODE regions. Similarly, the promoter arrays (Table 2) show that the enrichment values for the E2F6 target promoters in Ntera2 cells are considerably higher than the enrichment values for the top E2F1 and E2F4 target promoters (unlike HeLa cells, which show approximately equal enrichments at E2F1, E2F4, and E2F6 target promoters). These values suggested that perhaps E2F6 has a relatively greater influence on gene expression in Ntera2 cells than in the other cell types. To investigate the role of E2F6 in Ntera2 cells further, we first identified promoters bound in common by E2F1, E2F4, and E2F6; this set is composed of 500 promoters that were in the top 2000 targets on all six arrays of the duplicate experiments for E2F1, E2F4, and E2F6. Although the log2 enrichment values for E2F6 are higher than for the other E2Fs, these promoters were also enriched approximately twofold by the antibodies to the other E2Fs (Table 3). We next determined whether we could identify any targets that are bound by E2F6 but not by the other E2Fs. As seen in Table 3, we identified about 500 promoters that were in the top 2000 on both E2F6 arrays but were not bound by E2F1 or E2F4 (we could not identify any targets that were bound by E2F1 or E2F4 and not by E2F6). Thus, it appears as if E2F6 might play two roles in Ntera2 cells, perhaps functioning in opposition to the activating E2Fs and also playing another role that is independent of the other E2Fs.
Table 2.
Average enrichment values (log2) for the top 100 E2F target promoters
Table 3.
Identification of E2F6-specific targets
aShown are the average enrichment values (log2) of the two sets of promoters for all ChIP–chip experiments in Ntera2 cells
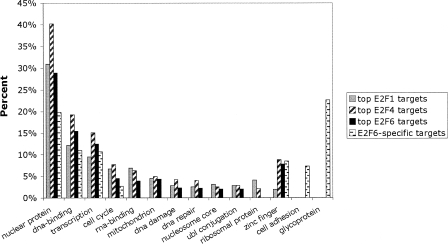
To determine whether the types of genes bound by E2F6 alone are different than the types of genes bound by E2F6 plus the other E2Fs, we performed a functional annotation using the analysis program DAVID. To select the set of E2F1 targets, we compared the promoters in the top 2000 set for both arrays probed with E2F1 samples. Similarly, we obtained a set of E2F4 and a set of E2F6 targets. Because of the overlap in binding specificity, many of the promoters in these three lists are the same. Not surprisingly, many of the promoters bound by the E2F family members are in the class of transcriptional regulators or involved in cell cycle control, DNA damage, or DNA repair. Then, we analyzed the set of genes bound only by E2F6. Interestingly, the promoters bound only by E2F6, and not by E2F1 or E2F4, included transcriptional regulators but did not include genes involved in cell cycle control, DNA damage, or DNA repair. Rather, the E2F6-specific targets were highly enriched for zinc finger genes, glycoproteins, and genes involved in cell adhesion (Fig. 5; Supplemental Table S5).
Figure 5.
E2F6 has a unique set of target genes in Ntera2 cells. The top 2000 ranked promoters in the E2F1, E2F4, and E2F6 ChIP–chip assays from Ntera2 cells were analyzed using the DAVID program. The set of promoters that were in the top 2000 on both E2F6 arrays but not in the top 2000 on either of the E2F1 or E2F4 arrays was also analyzed (E2F6-specific targets). All categories shown are significantly enriched as compared with a random list of genes (see Supplemental Table S6).
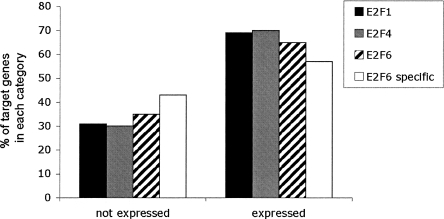
Although we have shown a strong correlation between E2F6 and binding of POLR2A, others have proposed that E2F6 functions as a transcriptional repressor. However, most studies of E2F6 activity have been performed using overexpression of E2F6 and reporter gene assays. To investigate the hypothesis that E2F6 functions as a transcriptional repressor under normal physiological conditions, we first analyzed the gene expression levels of the E2F target genes in Ntera2 cells using Illumina Sentrix Expression Beadchips. The expression levels of the promoters bound by E2F1, E2F4, or E2F6 in Ntera2 cells were divided into two categories: not expressed (P-value greater than 0; signals less than ∼300) and expressed (P-value of 0; signals greater than ∼300). We found that all three of the E2F family members bound predominantly to promoters of expressed genes. Although E2F1 is considered to be an activator and E2F6 is considered to be a repressor, the percentage of E2F1 versus E2F6 target genes having high or low expression levels was not very different (Fig. 6). Specifically, we note that there are a considerable number of genes in the E2F6-specific class that are expressed.
Figure 6.
Comparison of RNA levels of E2F target genes. The expression levels of the RNA of the top 2000 E2F1, E2F4, and E2F6 target genes were determined by analyzing RNA from Ntera2 cells using Illumina arrays. For comparison, the set of E2F6-specific genes was also analyzed. Values <300 are considered to represent genes that are not expressed, whereas values >300 represent expressed genes.
In general, it is quite difficult to determine the effects of a particular E2F on gene expression, because of the fact that multiple E2Fs bind to the same promoters. In particular, we could not identify any promoters in Ntera2 cells that were bound by E2F1 and not by E2F4 or E2F6 or any promoters bound by E2F4 and not by E2F1 or E2F6. Therefore, we cannot examine the role of E2F1 or E2F4 in Ntera2 cells. However, as noted above, we have identified a set of promoters that are specifically bound by E2F6 and not by E2F1 or E2F4. This allows us to address two related questions. First, does reduction of E2F6 affect gene expression of the E2F6-specific targets? Second, do the E2F6 promoters have a unique structure (such as a binding motif for an interacting factor) that allows E2F6, but not E2F1 or E2F4, to bind? To determine the role of E2F6 in gene regulation, we knocked down expression of E2F6 using cell lines in which we had stably transfected shRNA constructs (see Supplemental Fig. S3). Using Illumina Gene Expression Arrays, we then compared RNA levels for 48,000 transcripts in the control Ntera2 cells versus knockdown cells. We found that the levels of seven transcripts were increased at least 1.5-fold and the levels of 46 transcripts were decreased at least 1.5-fold upon reduction of E2F6. Of the seven genes whose expression increased (suggesting that E2F6 might normally repress their expression), only two of them are bound by E2F6 and of the 46 genes whose expression decreased (suggesting that E2F6 might function as an activator), only 18 are bound by E2F6. Only one of the two genes up-regulated and bound by E2F6 and five of the 18 genes down-regulated and bound by E2F6 showed a consistent change in expression in a second knockdown experiment (Supplemental Table S12). Thus, the reduction of E2F6 has essentially no effect on the overall transcriptional output of Ntera2 cells.
One reason why large effects on gene expression may not have been observed could have been that other E2Fs substituted for E2F6. To determine whether other E2Fs take the place of E2F6 in the knockdown cells, we chose 10 promoters that were bound by E2F6, but not E2F1 or E2F4, in the Ntera2 ChIP–chip assays. We then performed ChIP assays using antibodies to E2F1, E2F4, and E2F6 in Ntera2 cells stably transfected with constructs expressing control shRNAs or shRNAs specific for E2F6. The loss of signal in the E2F6 ChIP sample in the knockdown versus control cells illustrates the specificity of the E2F6 antibody (Supplemental Fig. S4); additional specificity is shown by the fact that E2F6 does not cross-react with any other protein as analyzed by Western blot (Supplemental Fig. S5). We found that at certain promoters, such as MAX, reduction of E2F6 allowed increased binding of E2F1 and E2F4 (Supplemental Fig. S4). In contrast, at other promoters (such as DYT3), reduction of E2F6 had little effect on the binding of E2F1 and E2F4. However, it remains possible that other E2F family members bind to the apparently “E2F6-specific” target promoters.
E2F6 does not affect silencing of HOX genes
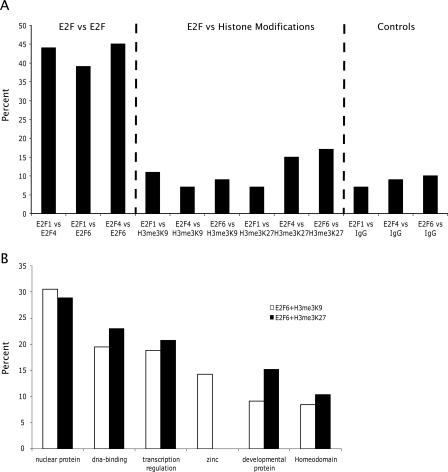
It has been proposed that E2F6 is a transcriptional repressor that mediates chromatin silencing by methylation of histone H3. For example, E2F6 has been purified in a complex containing EZH2 (Attwooll et al. 2005), which is involved in trimethylation of histone H3 at lysine 27, a modification associated with gene repression (Levine et al. 2004). Others have purified E2F6 in a complex with a histone methyltransferase that modifies lysine 9 of histone H3 (Ogawa et al. 2002), another mark associated with gene silencing. However, none of the previous studies linking E2F6 with histone methyltransferases examined histone modifications at E2F6 target genes. Therefore, we performed ChIP–chip assays using antibodies that specifically recognize histone H3 when it is trimethylated on lysine 27 or lysine 9 and compared the promoters having these modified histones with the E2F6 target promoters. To ensure that differences in cell culture conditions would not affect our results, we repeated the E2F6 ChIP–chip assays using the same set of cross-linked cells as used for analyzing the histone modifications. For comparison, we also included E2F1 and E2F4 in our analyses. As can be seen in Figure 7A, there is a high overlap in the top 2000 promoters bound by E2F1 versus E2F4, E2F1 versus E2F6, and E2F4 versus E2F6. However, there is very little overlap of any of the E2F family members with the promoters bound by H3me3K27 or H3me3K9. These results suggest that most promoters that contain these modifications are silenced by mechanisms that are not related to E2F6. However, it is possible that the few promoters that are bound by both E2F6 and H3me3K9 or E2F6 and H3me3K27 are a specific class of genes. To investigate this possibility, we performed analyses of the genes bound by E2F6 and H3me3K27 and the set of genes bound by E2F6 and H3me3K9 using the DAVID program. Not surprisingly since DNA binding proteins constitute a large portion of E2F6 target genes, most of the genes in both categories are transcription factors. However, zinc finger proteins were bound only by the combination of E2F6 and H3me3K9, whereas homeodomain proteins were bound by E2F6 plus H3me3K9 and E2F6 plus H3me3K27 (Fig. 7B).
Figure 7.
E2F6 binds to a small percentage of the H3me3K9 or H3me3K27 targets. (A) The top 2000 E2F1, E2F4, E2F6, H3me3K9 and H3me3K27 promoters were compared. As a negative control, the top 2000 promoters from a nonspecific IgG ChIP–chip assay were also compared with the E2F target promoters. (B) DAVID analysis of the set of promoters bound by both E2F6 plus H3me3K9 and by E2F6 plus H3me3K27.
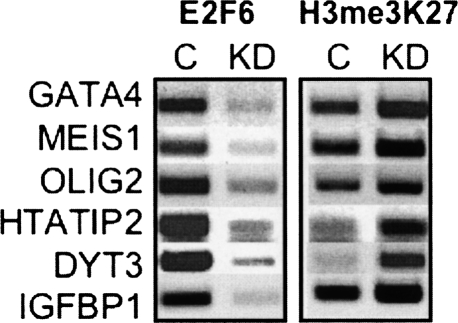
Several studies have suggested that E2F6 may play a specific role in regulating homeobox genes because an E2F6 knockout mouse shows modest homeotic transformations (Storre et al. 2002). Also, E2F6 has been shown to interact with PcG proteins, which are known to regulate HOX genes. For example, E2F6 has been shown to interact with Polyhomeotic-like (PHC3) (Isono et al. 2005; Deshpande et al. 2007) and with BMI1 (Trimarchi et al. 2001), two members of the hPRC-H family of PcG proteins. One study copurified E2F6 with EZH2 (Attwooll et al. 2005) and we have previously shown that Ezh2 binds to the promoters of hundreds of homeobox genes (Squazzo et al. 2006). Finally, Ogawa purified E2F6 with several PcG proteins, including RNF1, RNF2, h-l(3)mbt-like protein, and YAF2 (Ogawa et al. 2002). Therefore, we examined in more detail the relationship between the E2F family and the HOX A-D clusters of homeobox genes. As seen in Supplemental Table S13, E2F1 and E2F4 do not bind to the promoters of HOX genes but some of the HOX promoters are highly enriched in the E2F6 ChIP sample; all of the HOX promoters show high levels of H3me3K27. Although the expression of these HOX genes was not changed in the E2F6 knockdown cells, it was possible that changes in histone modification did occur. To determine whether E2F6 contributes to the levels of histone modification at HOX genes, we performed ChIP assays using antibodies to E2F6 and H3me3K27 in the wild type and E2F6 knockdown cells. We found that reduction of E2F6 did not result in a loss of H3me3K27 at the HOX promoters or at other promoters bound by H3me3K27 (Fig. 8). Thus, our studies do not support the hypothesis that E2F6 plays a critical role in silencing of HOX genes or in recruitment of histone methylases to genomic targets.
Figure 8.
Reduction of E2F6 does not affect histone modifications at HOX promoters. ChIP analysis was performed using antibodies to E2F6 and H3me3K27 in control Ntera2 cells (C) and Ntera2 cells harboring a plasmid that produces shRNAs against E2F6 (KD).
Discussion
We have examined the binding patterns of three members of the E2F family in five different cell types, including both tumorigenic and nontumorigenic cells. Importantly, the close relationship of the number and position of E2F, POLR2A, and TAF1 binding sites support the conclusion that a chromatin-bound E2F may be a signpost for the presence of transcription initiation complexes. Our studies indicate that, in general, several E2F family members bind to a given promoter region and that there is only modest cell type specificity (somewhat dependent on the amount of each family member in the nucleus). Finally, our results suggest, contrary to previously proposed models, E2F6 does not play a major role in histone methylation.
The universe of E2F target genes
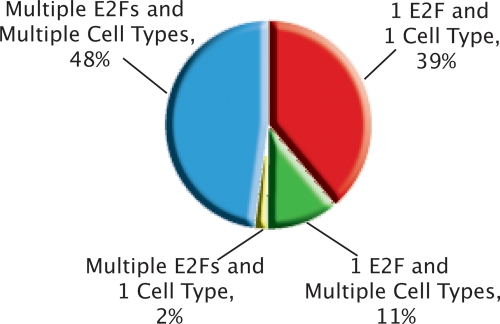
To assist in future studies of motif analyses and E2F–DNA interactions, we have provided a list of promoters that represent the very strongest E2F binding sites (identified by merging the top 100 lists for E2F1, E2F4, and E2F6 in all five cell types; ∼500 promoters, see Supplemental Table S4). However, these 500 promoters represent only a small subset of all E2F targets. By selecting all the promoters that were enriched more than twofold in the ChIP assays for the three different E2Fs in the five cell types, we have identified 8000 E2F target genes (see Supplemental Tables S7–S11 for the enrichment values for E2F1, E2F4, and E2F6 for all promoter arrays in all cell types). We note that 48% of these 8000 E2F target genes that we have identified are bound by multiple E2Fs in multiple cell types (Fig. 9). Accordingly, we have not found distinct binding motifs for the different E2Fs. As shown in Figure Supplemental S1, we identified the motif STTTS in the sets of E2F1-, E2F4-, and E2F6-bound promoters identified in the ChIP–chip studies that used ENCODE arrays. We have also analyzed the E2F1 targets identified using MCF7 cells and promoter arrays. We found that ∼20% of the promoters that were in the top 1000 ranked E2F1 targets in two independent ChIP–chip assays contain the consensus E2F site. In comparison, ∼8%–15% of promoters that are not bound by E2F1 in MCF7 cells also contain the consensus site. In contrast, ∼96% of the top 1000 ranked E2F1-bound targets identified using the promoter arrays contain the STTTS sequence. However, in vivo binding studies of site-directed mutations of the promoter regions are required before we can conclude that STTTS is critical for recruiting E2Fs to promoter regions. The difference between the previous casting experiments and the ChIP–chip experiments may be because the in vitro experiments used purified E2F and DP heterodimeric complexes, which may not represent the in vivo situation because of alterations in post-translational modifications of the E2Fs. In addition, recent data suggest that E2F family members might be recruited to promoters via interactions with factors other than, or in addition to, DP proteins (Schlisio et al. 2002; Giangrande et al. 2004; Jin et al. 2006). If so, then the in vitro studies do not represent the in vivo environment.
Figure 9.
Categorizing the universe of promoters that bind E2F. Combining the lists of highly enriched (>1.0 on the array) promoters from one array from each of the 15 conditions (3 E2Fs × 5 cell types), we found a total of exactly 8000 unique promoters. Then, we classified each promoter into the displayed categories. Almost half of the 8000 promoters were found in multiple cell types and were bound by multiple E2Fs. A large fraction of the 1 E2F in 1 Cell Type category is comprised of promoters that were only found as Ntera2 E2F6 promoters, consistent with pairwise promoter array analyses across E2Fs and cell types (see Figs. 3 and 4).
Almost all of the examples of only one E2F binding to a promoter and that binding occurring in a specific cell type are from the set E2F6 targets in Ntera2 cells; further analysis of this category of E2F targets is described below. The most enriched category of genes bound by all of the E2F family members is composed of transcriptional regulators. Although E2Fs do bind to the promoters of genes involved in DNA synthesis, cell cycle regulation, and DNA repair, these genes represent only a small percentage of the genes bound by E2F family members (5%–8%). At this point, we cannot identify characteristics of the set of promoters driving genes involved in cell cycle, DNA replication, or DNA repair that distinguish them from the thousands of other E2F-bound promoters. For example, the members of the cell cycle class of promoters do not all have a consensus E2F site nor are they overrepresented in the set of genes having the highest enrichment values. Only 6% of the set of the top 100 E2F1 targets (merged for all cell types) and 8% of the set of top 100 (merged for all cell types) E2F4 targets represent genes involved in cell cycle control.
The relationship between E2F and the transcription initiation complex
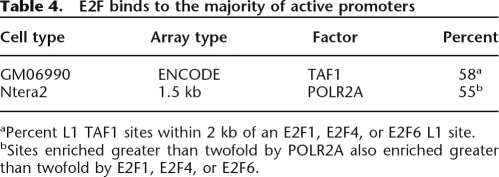
As noted above, we have identified over 8000 promoters that are bound by E2F1, E2F4, or E2F6 in at least one of five cell types. Intriguingly, we have found that most of the promoters bound by POLR2A in a given cell type are also bound by an E2F. For example, the binding pattern of the E2F family is essentially identical to that of POLR2A and TAF1, the largest subunit of TBP (Fig. 2C) and the majority of active promoters, as identified on ENCODE arrays by being bound by TAF1 in GM06990 cells or as identified on promoter arrays by being bound by POLR2A in Ntera2 cells, are also bound by E2F1, E2F4, or E2F6 (Table 4). Importantly, the GM06690 cells are nontumorigenic B lymphocytes; thus, our conclusions that a bound E2F is present at the majority of the active transcription complexes comes from cells expressing physiological levels of the E2Fs. Considering that there are five other E2Fs, our results suggest an intriguing hypothesis that a bound E2F site is the actual signal that the nearby sequences function as a promoter in that particular cell type.
Table 4.
E2F binds to the majority of active promoters
aPercent L1 TAF1 sites within 2 kb of an E2F1, E2F4, or E2F6 L1 site.
bSites enriched greater than twofold by POLR2A also enriched greater than twofold by E2F1, E2F4, or E2F6.
The role of E2F6 in transcription regulation
By investigating three E2Fs and five different cell types, we were able to identify one cell type (Ntera2 cells, derived from a testicular embryonal carcinoma) in which one E2F (E2F6) dominated the other tested family members. Initial studies of E2F6 suggested that it is a transcriptional repressor (Cartwright et al. 1998; Gaubatz et al. 1998; Trimarchi et al. 1998; Kherrouche et al. 2004). However, most of the previous work had been performed using artificial promoter constructs because of a lack of known E2F6 target genes. Within the last several years, a small set of 42 promoters has been identified as E2F6 target genes. Inspection of our ChIP–chip data indicates that almost all of these were identified as E2F6 targets in Ntera2 cells (Supplemental Table S6). Now, however, we have increased the number of E2F6 binding sites from ∼40 to >8000 (as identified by promoters that were enriched greater than 1.5-fold by ChIP–chip in Ntera2 cells). We have tested previous hypotheses that E2F6 modulates gene repression by recruiting enzymes involved in methylating histone H3 on lysine 9 or lysine 27 and have found that most promoters that are bound by histones having these modifications are not bound by E2F6. We have also reduced the levels of E2F6 and shown that levels of H3me3K27 and H3me3K9 do not decrease at the small subset of promoters bound by both E2F6 and H3me3K27 and H3me3K9. Thus, our studies do not support the hypothesis that E2F6 plays a role in silencing chromatin.
Conclusions
Perhaps the most surprising finding in our study is that although >8000 genes are bound by E2F6 in Ntera2 cells, representing over half of the promoters that are bound by POLR2A in these cells, removal of E2F6 from these promoters by expression of shRNAs does not alter the transcriptome of the cells. These findings are particularly puzzling in light of the fact that E2F6 binds at the core promoters of these 8000 target genes, not to upstream or downstream enhancer regions. A speculative hypothesis to account for the fact that E2F6 binds to a majority of the active promoters in Ntera2 cells but is not critical for transcriptional activity is that recruitment of this factor to core promoters may be a consequence, not a cause, of transcription complex formation. Recruitment of E2F6 by a general transcription factor could explain both the identical binding patterns of the factor with POLR2A and TAF1 and the finding that very few of the target promoters have an E2F consensus site. Previous studies have shown that E2F1 can interact with TBP (Emili and Ingles 1995; Pearson and Greenblatt 1997; Fry et al. 1999). It is not known whether the interactions of E2F1 with the general transcriptional machinery are conserved among the other E2Fs. Future studies will be directed at understanding in detail the mechanisms by which the different E2F family members are recruited to core promoters and the cause and effect relationships between binding of a particular E2F and the general transcriptional machinery.
Methods
Cell culture
Ntera2 and MCF-7 cells were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS, 2 mM glutamine, and 1% penicillin/streptomycin. MCF10A cells were grown in Clonetics serum-free Mammary Epithelial Growth Medium (MEGM) from Cambrex supplemented with 100 ng/mL cholera toxin from Calbiochem and all the contents of the Clonetics MEGM SingleQuots (containing BPE, hEGF, insulin, and hydrocortisone) from Cambrex except for GA-1000. HeLa-S3 cells were grown in our laboratory and by the National Cell Culture Center using Jokliks’s modified MEM supplemented with 5% NCS in spinner flask culture. GM06990 were grown by the National Cell Culture Center in Joklik’s modified MEM supplemented with 15% FBS in spinner flask culture. All cells were incubated at 37°C in a humidified 5% CO2 incubator. All cells were cross-linked for 10 min using a final concentration of 1% formaldehyde except for the HeLa-S3 and GM06990 cells obtained from the National Cell Culture Center, which were cross-linked for 20 min.
Generation of shE2F6 stable cell lines
The pRS–shE2F6 gene-specific shRNA expression cassettes, along with control shRNA plasmids including the original pRS vector (TR20003) and the pRS-shGFP vector (TR30003), were purchased from OriGene. The sequence of the E2F6-specific 29mer shRNA is ATGTGACCTATCAAGACATTCATAGCATT. Stable clones were generated by transfecting Ntera2 cells on 10-cm dishes with 1 μg of each of the shRNA plasmids using lipofectamine 2000 (Invitrogen), according to manufacturer’s recommendations. Forty-eight hours after transfection, the cells were placed under selection with 0.5 μg/mL of puromycin for a week, splitting 1:5 when the cells reached confluency. Multiple clones from the same transfection were pooled and grown under puromycin selection for ChIP assays and RNA analyses.
Western blots
Adherent cells (MCF-7, Ntera2, and MCF10A) were harvested by trypsinization, and suspension cells (HeLa-S3) were taken directly from spinner flask culture for centrifugation at 1000 rpm for 3 min. The cell pellets were washed once in PBS and processed for nuclear extraction using the NE-PER Nuclear and Cytoplasmic Extraction kit from Pierce, according to manufacturer’s instructions. Nuclear extracts (10 μg), as quantified using the BCA Protein Assay Reagents (Pierce), were separated by a 12% SDS-polyacrylamide gel and electrophoretically transferred to a nitrocellulose membrane. The membrane was blocked for 2 h in 5% nonfat dry milk (dissolved in TBST, 0.1% Tween-20) and subsequently probed with primary antibodies directed against either E2F1 (Santa Cruz Biotechnology, cat. no. sc-251), E2F4 (Santa Cruz Biotechnology, cat. no. sc-866X), E2F6 (Santa Cruz Biotechnology, cat. no. sc-22823X), or Nucleoporin p62 (Transduction Laboratories, cat. no. N43620), which was used as a loading control and to confirm the efficiency of nuclear extraction relative to whole-cell extracts and cytoplasmic extracts (data not shown). After extensive washing in TBST, the membranes were then incubated with the appropriate secondary antibody, either goat-anti mouse IgG conjugated to horseradish peroxidase (HRP) (Santa Cruz Biotechnology, cat. no. sc-2031) or goat-anti rabbit IgG HRP (Santa Cruz Biotechnology, cat. no. sc-2030) for 30 min. After extensive washing with TBST, the immunoreactive proteins were visualized using ECL Detection Reagents (GE Healthcare).
ChIP assays and amplicon preparation
ChIP assays (1 × 107 cells/assay) were performed following the protocol provided at http://genomics.ucdavis.edu/farnham and http://genomecenter.ucdavis.edu/expression_analysis. The primary antibodies used in this study were as follows: E2F1 (Upstate Cell Signaling, cat. no. 05-379), E2F4 (Santa Cruz Biotechnology, cat. no. sc-866X), E2F6 (Santa Cruz Biotechnology, cat. no. sc-22823X), H3me3K27 (Upstate Cell Signaling, cat. no. 07-449), H3me3K9 (Abcam cat. no. ab8898), POLR2A (Santa Cruz Biotechnology, cat. no. sc-899), and TAF1 (Santa Cruz Biotechnology, cat. no. sc-735). The nonspecific rabbit IgG used as a negative control in the ChIP assays was purchased from Alpha Diagnostics (cat. no. 210-561-9515 or cat. no. 20009-5). The secondary rabbit anti-mouse IgG was purchased from MP Biomedicals (cat. no. 55436). For PCR analysis of the ChIP samples before amplicon generation, QIAquick-purified immunoprecipitates were dissolved in 50 μL of water. Standard PCR reactions using 2 μL of the immunoprecipitated DNA were performed, and primer sequences are available upon request. Amplicons were prepared by adapting the standard protocol for whole genome amplification using the Sigma GenomePlex WGA kit as described in O'Geen et al. (2006). Briefly, the initial random fragmentation step was eliminated and DNA from the entire ChIP sample was used for amplification. A detailed protocol for the WGA method is provided at http://genomics.ucdavis.edu/farnham and http://genomecenter.ucdavis.edu/expression_analysis.
DNA microarrays
Amplicons were applied either to ENCODE arrays or to 1.5-kb promoter arrays (see Supplemental Table S1 and http://www.nimblegen.com for details). The labeling and hybridization of DNA samples for ChIP–chip analysis was performed by NimbleGen Systems, Inc. Briefly, each DNA sample (1 μg) was denatured in the presence of 5′-Cy3- or Cy5-labeled random nonamers (TriLink Biotechnologies) and incubated with 100 units (exo-) Klenow fragment (NEB) and dNTP mix (6 mM each in TE buffer [10 mM Tris/1 mM EDTA at pH 7.4; Invitrogen]) for 2 h at 37°C. Reactions were terminated by addition of 0.5 M EDTA (pH 8.0), precipitated with isopropanol, and resuspended in water. Then, 13 μg of the Cy5-labeled ChIP sample and 13 μg of the Cy3-labeled total sample were mixed together, dried down, and resuspended in 40 μl of NimbleGen Hybridization Buffer (NimbleGen Systems) plus 1.5 μg of human COT1 DNA. After denaturation, hybridization was carried out in a MAUI Hybridization System (BioMicro Systems) for 18 h at 42°C. The arrays were washed using NimbleGen Wash Buffer System (NimbleGen Systems), dried by centrifugation, and scanned at 5-μm resolution using the GenePix 4000B scanner (Axon Instruments). Fluorescence intensity raw data were obtained from scanned images of the arrays using NIMBLESCAN 2.0 extraction software (NimbleGen Systems). For each spot on the array, log2-ratios of the Cy5-labeled test sample versus the Cy3-labeled reference sample were calculated. Then, the biweight mean of this log2 ratio was subtracted from each point; this procedure is approximately equivalent to mean-normalization of each channel.
Data analysis
Binding sites on the ENCODE arrays were identified using the highest stringency level, L1 (six consecutive probes above the 98th percentile threshold, P < 0.0001, with S = 50 meaning that a 100 nucleotide gap was allowed between peaks), of the Tamalpais peak calling algorithm previously described (Bieda et al. 2006); see also http://genomics.ucdavis.edu/farnham. For ENCODE arrays, localization was evaluated relative to the GENCODE set of transcripts downloaded from the UCSC browser (hg17 set). If the distance from the transcription start site to the closest edge of an E2F1 binding site was <2 kb, this binding site was considered a “within 2 kb” binding site. To form a rough estimate of the number of peaks that would randomly occur within 2 kb of a transcription start site (for ENCODE-type array analyses), we calculated the total number of nucleotides that were found within 2 kb of all the transcription start sites (upstream and downstream) in the ENCODE regions, taking into account overlapping segments. This led to a total of 2080 kb. Of the 30 Mb in the ENCODE regions, ∼15 Mb are in repeat regions and not represented, with the other 15 Mb represented on the array. Hence, we find that 2.08/15 = 13.9% of nucleotides are in these regions, resulting in an ∼13.9% chance of a peak randomly being within 2 kb of a transcription start site. Because most E2F binding sites that were in the “within 2 kb” set were actually significantly closer to the TSS, and most were slightly upstream as opposed to downstream, this is a conservative (high) and very rough estimate of the random expectation. The number of genes in the ENCODE regions (433) was derived from the GENCODE set (hg17, UCSC genome browser). To derive the set of “overall nonredundant E2F family binding sites” for a single cell type, peak sets from each E2F for that cell type were combined and sites that were either overlapping or within 100 nucleotides of each other were merged. To form the overall set of nonredundant sites across cell types, we used an identical procedure, combining the predicted binding sites for each combination of E2F and cell type. All localization and set combination procedures were performed by custom Perl and bash programs under a Linux operating system. For 1.5 kb promoter array analysis, the overall enrichment value for each promoter was calculated by NimbleGen software based on the median value of the top 11 of the 15 probes for each promoter region. Comparison of sets (e.g., number in common in the top 100) was performed using the Farnham promoter array analysis website (http://chipanalysis.genomecenter.ucdavis.edu/cgi-bin/promoterarray.cgi). Functional annotations were performed using the program Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2.1 (http://david.abcc.ncifcrf.gov), as previously described (Squazzo et al. 2006). To categorize the universe of E2F genes (Fig. 9), we chose one array from each of 15 conditions (3 E2Fs × 5 cell types). To choose a single array for analysis for each of the 15 conditions, we used the array replicate that had the largest number of promoters with enrichment above 1.0. We chose promoters with values >1.0 from each of the 15 arrays and combined the lists. Each promoter was scored as to which of the 15 conditions in which it was present and results were summarized as displayed in Figure 9. Very similar results were found using the top 2000 promoters from each of the 15 arrays instead of the 1.0 cutoff criterion (data not shown).
Identification of binding site motifs
The motifs for each E2F family member in different cell types were identified using the ChIPMotifs approach developed in our previous study (Jin et al. 2007). Briefly, a set of in vivo binding sequences identified by ChIP–chip for a given factor in any cell type (also called a positive data set) was used as input for the Weeder and MEME programs. The output candidate motifs from these programs were then automatically converted to positional weight matrix (PWM) formats and subsequently tested on a set of more than ten thousand randomly rearranged sequences, generated from the same set of input sequences from our ChIP–chip experiments, at different percentile levels to determine the appropriate PWM thresholds. The latter output sequence rearrangement step was carried out to obtain randomized sequences that no longer correspond to the experimentally identified binding sites but have the same nucleotide frequencies as the experimentally identified binding sites. The Fisher test was used on the positive data set (identified by our TAM program; Bieda et al. 2006) and the negative data set (the set of regions not bound by the transcription factor of interest, where each region has an average of length of 500 bp and is within 10 kb of a promoter region of a known gene) to evaluate the best cutoff threshold for each E2F family member (see Supplemental Table S3). The similarity to the classical E2F consensus was estimated by screening against the TRANSFAC database of E2F binding sites.
RNA expression arrays
Total RNA was prepared from Ntera2 cells using the RNAeasy Kit (QIAGEN) following the manufacturer’s instructions. RNA quality was ensured using the Agilent Systems Bioanalyzer. For Illumina arrays, the Illumina TotalPrep RNA amplification kit from Ambion (AMIL1791) was used to generate biotinylated, amplified RNA for hybridization with the Illumina Sentrix Expression Beadchips, HumanRef-6 v2. The Sentrix gene expression beadchips used for this study consisted of an 8-array format comprising more than 22,000 transcripts/array from Refseq sequences (for more details, see (http://www.illumina.com/pages.ilmn?ID=197). Arrays were processed and scanned at medium PMT settings according to manufacturer’s recommendations, and analyzed using Bead Studio Software v. 2.3.41. Data was normalized using the “average” method, which simply adjusts the intensities of two populations of gene expression values such that the means of the populations become equal.
Acknowledgments
This work was supported in part by Public Health Service grant CA45250, HG003129, and DK067889. We thank the members of the Farnham laboratory for helpful discussion and data analysis. The RNA was labeled and the Illumina beadchip was processed and scanned at the UC Davis Genome Center Expression Analysis Core Facility.
Footnotes
[Supplemental material is available online at www.genome.org. The ChIP–chip data has been deposited as GEO series GSE8716.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6783507
References
- Attwooll C., Oddi S., Cartwright P., Prosperini E., Agger K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K., Oddi S., Cartwright P., Prosperini E., Agger K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K., Cartwright P., Prosperini E., Agger K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K., Prosperini E., Agger K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K., Agger K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K., Steensgaard P., Wagener C., Sardet C., Moroni M.C., Helin K., Wagener C., Sardet C., Moroni M.C., Helin K., Sardet C., Moroni M.C., Helin K., Moroni M.C., Helin K., Helin K. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation- specific manner. J. Biol. Chem. 2005;280:1199–1208. doi: 10.1074/jbc.M412509200. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Gribskov M., Gribskov M. Score distributions for simultaneous matching to multiple motifs. J. Comput. Biol. 1997;4:45–59. doi: 10.1089/cmb.1997.4.45. [DOI] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M., Green R., Farnham P.J., Xu X., Singer M., Green R., Farnham P.J., Singer M., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Unbiased location analysis of E2F1 binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J.S., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Liu X.S., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Brodsky A.S., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Li W., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Meyer C.A., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Szary A.J., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Eeckhoute J., Shao W., Hestermann E.V., Geistlinger T.R., Shao W., Hestermann E.V., Geistlinger T.R., Hestermann E.V., Geistlinger T.R., Geistlinger T.R., et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Cartwright P., Muller H., Wagener C., Holm K., Helin K., Muller H., Wagener C., Holm K., Helin K., Wagener C., Holm K., Helin K., Holm K., Helin K., Helin K. E2F-6: A novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene. 1998;17:611–623. doi: 10.1038/sj.onc.1201975. [DOI] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Johnson D.G., Johnson D.G. Distinct and overlapping roles for E2F family members in transcription, proliferation, and apoptosis. Curr. Mol. Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- Deshpande A.M., Akunowicz J.D., Reveles X.T., Patel B.B., Saria E.A., Gorlick R.G., Naylor S.L., Leach R.J., Hansen M.F., Akunowicz J.D., Reveles X.T., Patel B.B., Saria E.A., Gorlick R.G., Naylor S.L., Leach R.J., Hansen M.F., Reveles X.T., Patel B.B., Saria E.A., Gorlick R.G., Naylor S.L., Leach R.J., Hansen M.F., Patel B.B., Saria E.A., Gorlick R.G., Naylor S.L., Leach R.J., Hansen M.F., Saria E.A., Gorlick R.G., Naylor S.L., Leach R.J., Hansen M.F., Gorlick R.G., Naylor S.L., Leach R.J., Hansen M.F., Naylor S.L., Leach R.J., Hansen M.F., Leach R.J., Hansen M.F., Hansen M.F. PHC3, a component of the hPRC-H complex, associates with E2F6 during G0 and is lost in osteosarcoma tumors. Oncogene. 2007;26:1714–1722. doi: 10.1038/sj.onc.1209988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D.K., Dyson N.J., Dyson N.J. The E2F transcriptional network: Old acquaintances with new faces. Oncogene. 2005;24:2810–2826. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- Emili A., Ingles C.J., Ingles C.J. Promoter-dependent photocrosslinking of the acidic transcriptional activator E2F-1 to the TATA-binding protein. J. Biol. Chem. 1995;270:13674–13680. doi: 10.1074/jbc.270.23.13674. [DOI] [PubMed] [Google Scholar]
- Field S.J., Tsai F.-Y., Kuo F., Zubiaga A.M., Kaelin W.G., Livingston D.M., Orkin S.H., Greenberg M.E., Tsai F.-Y., Kuo F., Zubiaga A.M., Kaelin W.G., Livingston D.M., Orkin S.H., Greenberg M.E., Kuo F., Zubiaga A.M., Kaelin W.G., Livingston D.M., Orkin S.H., Greenberg M.E., Zubiaga A.M., Kaelin W.G., Livingston D.M., Orkin S.H., Greenberg M.E., Kaelin W.G., Livingston D.M., Orkin S.H., Greenberg M.E., Livingston D.M., Orkin S.H., Greenberg M.E., Orkin S.H., Greenberg M.E., Greenberg M.E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Fry C.J., Farnham P.J., Farnham P.J. Context-dependent transcriptional regulation. J. Biol. Chem. 1999;274:29583–29586. doi: 10.1074/jbc.274.42.29583. [DOI] [PubMed] [Google Scholar]
- Fry C.J., Pearson A., Malinowski E., Bartley S.M., Greenblatt J., Farnham P.J., Pearson A., Malinowski E., Bartley S.M., Greenblatt J., Farnham P.J., Malinowski E., Bartley S.M., Greenblatt J., Farnham P.J., Bartley S.M., Greenblatt J., Farnham P.J., Greenblatt J., Farnham P.J., Farnham P.J. Activation of the murine dihydrofolate reductase promoter by E2F1: A requirement for CBP recruitment. J. Biol. Chem. 1999;274:15883–15891. doi: 10.1074/jbc.274.22.15883. [DOI] [PubMed] [Google Scholar]
- Gaubatz S., Wood J.G., Livingston D.M., Wood J.G., Livingston D.M., Livingston D.M. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. 1998;95:9190–9195. doi: 10.1073/pnas.95.16.9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz S., Lindeman G.J., Ishida S., Jakoi L., Nevins J.R., Livingston D.M., Rempel R.E., Lindeman G.J., Ishida S., Jakoi L., Nevins J.R., Livingston D.M., Rempel R.E., Ishida S., Jakoi L., Nevins J.R., Livingston D.M., Rempel R.E., Jakoi L., Nevins J.R., Livingston D.M., Rempel R.E., Nevins J.R., Livingston D.M., Rempel R.E., Livingston D.M., Rempel R.E., Rempel R.E. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell. 2000;6:729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- Giangrande P.H., Zhu W., Rempel R.E., Laakso N., Nevins J.R., Zhu W., Rempel R.E., Laakso N., Nevins J.R., Rempel R.E., Laakso N., Nevins J.R., Laakso N., Nevins J.R., Nevins J.R. Combinatorial gene control involving E2F and E Box family members. EMBO J. 2004;23:1336–1347. doi: 10.1038/sj.emboj.7600134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert P.O., Rogers C., Ganiatsas S., Landsberg R.L., Trimarchi J.M., Dandapani S., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Rogers C., Ganiatsas S., Landsberg R.L., Trimarchi J.M., Dandapani S., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Ganiatsas S., Landsberg R.L., Trimarchi J.M., Dandapani S., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Landsberg R.L., Trimarchi J.M., Dandapani S., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Trimarchi J.M., Dandapani S., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Dandapani S., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Brugnara C., Erdman S., Schrenzel M., Bronson R.T., Erdman S., Schrenzel M., Bronson R.T., Schrenzel M., Bronson R.T., Bronson R.T., et al. E2F4 is essential for normal erythrocyte maturation and neonatal viability. Mol. Cell. 2000;6:281–291. doi: 10.1016/s1097-2765(00)00029-0. [DOI] [PubMed] [Google Scholar]
- Isono K.-i., Fujimura Y.-i., Shinga J., Yamaki M., Wang J.O., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Fujimura Y.-i., Shinga J., Yamaki M., Wang J.O., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Shinga J., Yamaki M., Wang J.O., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Yamaki M., Wang J.O., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Wang J.O., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Takada Y., Mizutani-Koseki Y., Koseki H., Mizutani-Koseki Y., Koseki H., Koseki H. Mammalian Polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate polycomb repression of Hox genes. Mol. Cell. Biol. 2005;25:6694–6706. doi: 10.1128/MCB.25.15.6694-6706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V., Rabinvich A., Squazzo S.L., Green R., Farnham P.J., Rabinvich A., Squazzo S.L., Green R., Farnham P.J., Squazzo S.L., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data-a case study using E2F1. Genome Res. 2006;16:1585–1595. doi: 10.1101/gr.5520206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin V.X., O'Geen H., Iyengar S., Green R., Farnham P.J., O'Geen H., Iyengar S., Green R., Farnham P.J., Iyengar S., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Identification of an OCT4 and SRY regulatory module using integrated computational and experimental genomics approaches. Genome Res. 2007;17:807–817. doi: 10.1101/gr.6006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kherrouche Z., De Launoit Y., Monte D., De Launoit Y., Monte D., Monte D. Human E2F6 is alternatively spliced to generate multiple protein isoforms. Biochem. Biophys. Res. Commun. 2004;317:749–760. doi: 10.1016/j.bbrc.2004.03.099. [DOI] [PubMed] [Google Scholar]
- Kong L.-J., Chang J.T., Bild A.H., Nevins J.R., Chang J.T., Bild A.H., Nevins J.R., Bild A.H., Nevins J.R., Nevins J.R. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- Levine S.S., King I.F.G., Kingston R.E., King I.F.G., Kingston R.E., Kingston R.E. Division of labor in polycomb group repression. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Lindeman G.J., Dagnino L., Gaubatz S., Xu Y., Bronson R.T., Warren H.B., Livingston D.M., Dagnino L., Gaubatz S., Xu Y., Bronson R.T., Warren H.B., Livingston D.M., Gaubatz S., Xu Y., Bronson R.T., Warren H.B., Livingston D.M., Xu Y., Bronson R.T., Warren H.B., Livingston D.M., Bronson R.T., Warren H.B., Livingston D.M., Warren H.B., Livingston D.M., Livingston D.M. A specific, nonproliferative role for E2F-5 in choroid plexus function revealed by gene targeting. Genes & Dev. 1998;12:1092–1098. doi: 10.1101/gad.12.8.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M., Fernandez-Capetillo O., Field S.J., Moreno B., Borlado L.R., Fujiwara Y., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Fernandez-Capetillo O., Field S.J., Moreno B., Borlado L.R., Fujiwara Y., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Field S.J., Moreno B., Borlado L.R., Fujiwara Y., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Moreno B., Borlado L.R., Fujiwara Y., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Borlado L.R., Fujiwara Y., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Fujiwara Y., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Balomenos D., Vicario A., Carrera A.C., Orkin S.H., Vicario A., Carrera A.C., Orkin S.H., Carrera A.C., Orkin S.H., Orkin S.H., et al. Mutation of E2F2 in mice causes enhanced T lymphocyte proliferation, leading to the development of autoimmunity. Immunity. 2001;15:959–970. doi: 10.1016/s1074-7613(01)00254-0. [DOI] [PubMed] [Google Scholar]
- O'Geen H., Nicolet C.M., Blahnik K., Green R., Farnham P.J., Nicolet C.M., Blahnik K., Green R., Farnham P.J., Blahnik K., Green R., Farnham P.J., Green R., Farnham P.J., Farnham P.J. Comparison of sample preparation methods for ChIP–chip assays. Biotechniques. 2006;41:577–580. doi: 10.2144/000112268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa H., Ishiguro K., Gaubatz S., Livingston D.M., Nakatani Y., Ishiguro K., Gaubatz S., Livingston D.M., Nakatani Y., Gaubatz S., Livingston D.M., Nakatani Y., Livingston D.M., Nakatani Y., Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Pavesi G., Mereghetti P., Mauri G., Pesole G., Mereghetti P., Mauri G., Pesole G., Mauri G., Pesole G., Pesole G. Weeder Web: Discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A., Greenblatt J., Greenblatt J. Modular organization of the E2F1 activation domain and its interaction with general transcription factors TBP and TFIIH. Oncogene. 1997;15:2643–2658. doi: 10.1038/sj.onc.1201451. [DOI] [PubMed] [Google Scholar]
- Rempel R.E., Saenz-Robles M.T., Storms R., Morham S., Ishida S., Engel A., Jakoi L., Melham M.F., Pipas J.M., Smith C., Saenz-Robles M.T., Storms R., Morham S., Ishida S., Engel A., Jakoi L., Melham M.F., Pipas J.M., Smith C., Storms R., Morham S., Ishida S., Engel A., Jakoi L., Melham M.F., Pipas J.M., Smith C., Morham S., Ishida S., Engel A., Jakoi L., Melham M.F., Pipas J.M., Smith C., Ishida S., Engel A., Jakoi L., Melham M.F., Pipas J.M., Smith C., Engel A., Jakoi L., Melham M.F., Pipas J.M., Smith C., Jakoi L., Melham M.F., Pipas J.M., Smith C., Melham M.F., Pipas J.M., Smith C., Pipas J.M., Smith C., Smith C., et al. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol. Cell. 2000;6:293–306. doi: 10.1016/s1097-2765(00)00030-7. [DOI] [PubMed] [Google Scholar]
- Schlisio S., Halperin T., Vidal M., Nevins J.R., Halperin T., Vidal M., Nevins J.R., Vidal M., Nevins J.R., Nevins J.R. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slansky J.E., Farnham P.J., Farnham P.J. Introduction to the E2F family: Protein structure and gene regulation. In: Farnham P.J., editor. Transcriptional control of cell growth: The E2F gene family. Springer-Verlag; New York: 1996. pp. 1–30. [DOI] [PubMed] [Google Scholar]
- Squazzo S.L., Komashko V.M., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Komashko V.M., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., O’Geen H., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Krig S., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Jin V.X., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Jang S.-W., Green R., Margueron R., Reinberg D., Farnham P.J., Green R., Margueron R., Reinberg D., Farnham P.J., Margueron R., Reinberg D., Farnham P.J., Reinberg D., Farnham P.J., Farnham P.J. Suz12 silences large regions of the genome in a cell type-specific manner. Genome Res. 2006;16:890–900. doi: 10.1101/gr.5306606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storre J., Elsasser H.P., Fuchs M., Ullmann D., Livingston D.M., Gaubatz S., Elsasser H.P., Fuchs M., Ullmann D., Livingston D.M., Gaubatz S., Fuchs M., Ullmann D., Livingston D.M., Gaubatz S., Ullmann D., Livingston D.M., Gaubatz S., Livingston D.M., Gaubatz S., Gaubatz S. Homeotic transformations of the axial skeleton that accompany a targeted deletion of E2F6. EMBO Rep. 2002;3:695–700. doi: 10.1093/embo-reports/kvf141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Kassatly R., Cress W.D., Horowitz J.M., Kassatly R., Cress W.D., Horowitz J.M., Cress W.D., Horowitz J.M., Horowitz J.M. Subunit composition determines E2F DNA-binding site specificity. Mol. Cell. Biol. 1997;17:6994–7007. doi: 10.1128/mcb.17.12.6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild B., Verona R., Moberg K., Andon N., Lees J.A., Fairchild B., Verona R., Moberg K., Andon N., Lees J.A., Verona R., Moberg K., Andon N., Lees J.A., Moberg K., Andon N., Lees J.A., Andon N., Lees J.A., Lees J.A. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl. Acad. Sci. 1998;95:2850–2855. doi: 10.1073/pnas.95.6.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi J.M., Fairchild B., Wen J., Lees J.A., Fairchild B., Wen J., Lees J.A., Wen J., Lees J.A., Lees J.A. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc. Natl. Acad. Sci. 2001;95:2850–2855. doi: 10.1073/pnas.041597698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann A.S., Yan P.S., Oberley M.J., Huang T.H.-M., Farnham P.J., Yan P.S., Oberley M.J., Huang T.H.-M., Farnham P.J., Oberley M.J., Huang T.H.-M., Farnham P.J., Huang T.H.-M., Farnham P.J., Farnham P.J. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes & Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Timmers C., Maiti B., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Timmers C., Maiti B., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Maiti B., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Saavedra H.I., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Sang L., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Chong G.T., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Nuckolls F., Giangrande P., Wright F.A., Field S.J., Giangrande P., Wright F.A., Field S.J., Wright F.A., Field S.J., Field S.J., et al. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- Zheng N., Fraenkel E., Pabo C.O., Pavletich N.P., Fraenkel E., Pabo C.O., Pavletich N.P., Pabo C.O., Pavletich N.P., Pavletich N.P. Structural basis of DNA recognition by the heterodimeric cell cycle transcription factor E2F-DP. Genes & Dev. 1999;13:666–674. doi: 10.1101/gad.13.6.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.W., Field S.J., Gore L., Thompson M., Yang H., Fujiwara Y., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Field S.J., Gore L., Thompson M., Yang H., Fujiwara Y., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Gore L., Thompson M., Yang H., Fujiwara Y., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Thompson M., Yang H., Fujiwara Y., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Yang H., Fujiwara Y., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Fujiwara Y., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Cardiff R.D., Greenberg M., Orkin S.H., DeGregori J., Greenberg M., Orkin S.H., DeGregori J., Orkin S.H., DeGregori J., DeGregori J. E2F1 and E2F2 determine thresholds for antigen-induced T-cell proliferation and suppress tumorigenesis. Mol. Cell. Biol. 2001;21:8547–8564. doi: 10.1128/MCB.21.24.8547-8564.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]