Abstract
The repertoire of protein architectures in proteomes is evolutionarily conserved and capable of preserving an accurate record of genomic history. Here we use a census of protein architecture in 185 genomes that have been fully sequenced to generate genome-based phylogenies that describe the evolution of the protein world at fold (F) and fold superfamily (FSF) levels. The patterns of representation of F and FSF architectures over evolutionary history suggest three epochs in the evolution of the protein world: (1) architectural diversification, where members of an architecturally rich ancestral community diversified their protein repertoire; (2) superkingdom specification, where superkingdoms Archaea, Bacteria, and Eukarya were specified; and (3) organismal diversification, where F and FSF specific to relatively small sets of organisms appeared as the result of diversification of organismal lineages. Functional annotation of FSF along these architectural chronologies revealed patterns of discovery of biological function. Most importantly, the analysis identified an early and extensive differential loss of architectures occurring primarily in Archaea that segregates the archaeal lineage from the ancient community of organisms and establishes the first organismal divide. Reconstruction of phylogenomic trees of proteomes reflects the timeline of architectural diversification in the emerging lineages. Thus, Archaea undertook a minimalist strategy using only a small subset of the full architectural repertoire and then crystallized into a diversified superkingdom late in evolution. Our analysis also suggests a communal ancestor to all life that was molecularly complex and adopted genomic strategies currently present in Eukarya.
The repertoire of protein structures encoded in a genome delimits the cellular functions and interactions that sustain cellular life. It also serves as an imprint of genomic history. While nucleic acid and protein sequence can be highly dynamic, domain structure in proteins is generally maintained for long periods of evolutionary time (Gerstein and Hegyi 1998; Chothia et al. 2003). For this reason, domains are considered not only units of structure but also units of evolution (Murzin et al. 1995; Orengo et al. 1997; Riley and Labedan 1997). In particular, the discovery of an architectural design, that is, an orderly and unique arrangement of protein components in three-dimensional (3D) space (herein referred to as an “architecture”), constitutes an important and rare event in protein evolution that adds new functions to the protein world. In fact, there have been very few of these finds in the history of life on earth. The number of fold (F) architectures discovered so far amount to only ∼1000, the number of fold superfamilies (FSF) to ∼1500, and the number of fold families (FF) to ∼2500, according to one classification (Murzin et al. 1995; Andreeva et al. 2004). F and FSF architectures are highly conserved in nature. FSF are composed of protein molecules with low sequence identity but with structures and functions indicative of a probable common evolutionary origin (they group one or more sequence-related FF). F group FSF with secondary structures that are similarly arranged in 3D space but that may not necessarily be evolutionarily related. The vast majority of F and FSF represent highly successful architectural discoveries that have accumulated and dispersed throughout the 107–108 species that inhabit our planet. A delicate balance of survival and extinction of structural discoveries probably triggered propagation, but as with Galton-Watson branching processes (Harris 1963), only successful architectures are the ones represented by the >103 proteins per genome (i.e., the complement defining a proteome) that make up the estimated ∼1010–1014 proteins in existence today. Consequently, the repertoire of architectures in proteomes can be regarded as a collection of historical imprints or molecular fossils preserved in nature by successful propagation and evolutionary “lock-in” (preservation of the original architecture by “structural canalization”) (Ancel and Fontana 2000). Indeed, the occurrence and abundance of F and FSF, and their combination in proteins, has been used successfully to build reasonable universal trees of life capable of describing the history of major organismal lineages satisfactorily (Caetano-Anollés and Caetano-Anollés 2003; Yang et al. 2005; Wang and Caetano-Anollés 2006). Furthermore, the phylogenetic analysis of the architectural repertoire can dissect deep evolutionary phenomena related to the origins of life (Caetano-Anollés and Caetano-Anollés 2003, 2005; Dupont et al. 2006; Wang et al. 2006; Caetano-Anollés et al. 2007). In this study, we take advantage of this potential.
The ancestor of all organisms alive today is at the root of the universal phylogenetic tree, and its cellular and molecular organization illuminates our understanding on how life originated and evolved (Woese 1998; Penny and Poole 1999). However, its nature has been controversial. This stems from limitations and conflict in the evolutionary signals that are embedded in the limited number of molecular or cellular features that have been analyzed. The canonical view, stemming mostly from ribosomal RNA (rRNA), elongation factors, and other molecules of the “informational” class, suggests that the ancestor was simple and prokaryotic-like (Woese et al. 1990; Woese 1998) and that horizontal gene transfer (HGT) was rampant in early evolution (Doolittle 1999). In contrast, a tracing of the origins of the tripartite world from an ancient RNA world based on DNA sequence, RNA relics, and other considerations suggests that the ancestor was eukaryotic-like and complex (Poole et al. 1998; Forterre and Philippe 1999; Penny and Poole 1999; Kurland et al. 2006). Moreover, analysis of entire genomic complements indicated that massive HGT was not warranted (e.g., Snel et al. 1999; Gough 2005) or did not impair phylogenetic reconstruction of a universal tree (Doolittle 2005). It also revealed the complexities of phylogenetic reconstruction (Delsuc et al. 2005).
Despite the promises of evolutionary genomics, the nature of the universal ancestor and the universal tree has yet to be resolved (Delsuc et al. 2005; Doolittle 2005). However, phylogenetic analyses of combined or concatenated genomic sequences (e.g., Ciccarelli et al. 2006) or genomic features describing the survey (e.g., Snel et al. 1999; Yang et al. 2005; Wang and Caetano-Anollés 2006) or arrangement (e.g., Korbel et al. 2002) of genomic component parts suggest a clear tripartite division into organismal domains Archaea, Bacteria and Eukarya (herein referred to as “superkingdoms” to avoid confusion between “domains” of organisms or molecules). We recently used a genomic census of protein architecture to generate genome-based phylogenies (phylogenomic trees) that describe the evolution of the protein world at different hierarchical levels of protein structural organization (Caetano-Anollés and Caetano-Anollés 2003, 2005; Wang et al. 2006). These trees were used to classify proteins (mostly globular), define structural transformations, and uncover evolutionary patterns in structure. We also traced patterns of organismal distribution in these trees and found that architectures at the base were omnipresent or common to all superkingdoms and that a timeline of organismal diversification could be inferred (Caetano-Anollés and Caetano-Anollés 2005; Wang et al. 2006). The diversity of ancient architectures common to superkingdoms suggested that the universal ancestor had a complex and relatively modern eukaryotic-like organization and hinted at a prokaryotic world stemming fundamentally from reductive evolutionary processes.
In this study, we embark on a systematic and global study of 185 genomes that have been fully sequenced and represent organisms from all three superkingdoms of life that exhibit free-living (FL), parasitic (P), and obligate parasitic (OP) lifestyles. We first reconstructed phylogenomic trees of F and FSF using standard phylogenetic methods. The trees uncovered congruent patterns of architectural diversification and reductive evolutionary processes. Finally, we used this information to reconstruct global trees of proteomes and to propose a scenario for the birth and diversification of the tripartite world.
Results
Patterns of F and FSF distribution in the proteome world: Three epochs in protein evolution
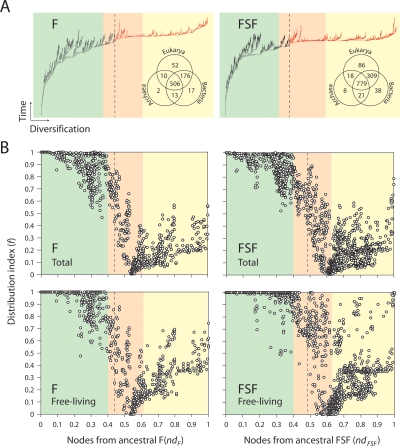
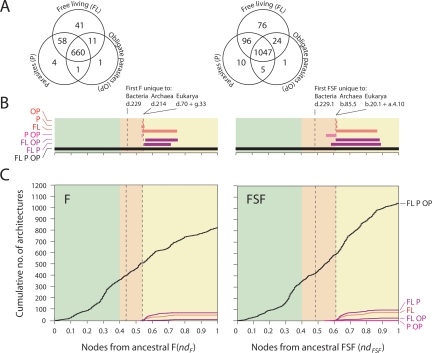
We generated intrinsically rooted trees of 776 F and 1259 FSF (Fig. 1A). Tree distribution profiles and metrics of skewness suggested strong cladistic structure (P < 0.01). The trees were well resolved, but branches were generally poorly supported by bootstrap analysis, an expected outcome with trees of this size. F and FSF trees grouped architectures into similar clades. This explains the qualitative similarity of results for F and FSF described below. To unfold the data embedded in the trees, we quantified the distribution of F and FSF among proteomes by a distribution index (f), defined as the relative number of species using each F or FSF. Figure 1B displays this index f plotted against the relative age of architectures (nd), measured on the trees as a relative distance in nodes from the hypothetical ancestor. We call these plots “architectural chronologies.” The nine most ancient F (ndF = 0–0.046) and the five most ancient FSF (ndFSF = 0–0.049) were present in all proteomes (f = 1), and representation decreased with decreasing age until f approached 0 at about ndF = 0.55 and ndFSF = 0.60 for F and FSF, respectively. At this point, a large number of architectures were clustered, each specific to a small number of organisms. Further in evolutionary time (0.55 < ndF and 0.60 < ndFSF), an opposite trend takes place, in which F and FSF increase their representation in proteomes.
Figure 1.
Architectural chronologies of (F) folds (left) and (FSF) fold superfamilies (right) suggest three evolutionary epochs in the timeline of the protein world. (A) Optimal (P < 0.01) most-parsimonious F (85,644 steps; CI = 0.043, RI = 0.770; g1 = −0.134) and FSF (118,119 steps; CI = 0.031, RI = 0.759; g1 = −0.099) trees were reconstructed from a protein domain census in 185 completely sequenced genomes. Venn diagrams show occurrence of architectures in the three superkingdoms of life, Archaea (A), Bacteria (B), and Eukarya (E). Terminal leaves were not labeled, as they would not be legible. (Red) Branches defining F and FSF that occur after the appearance of the first architecture unique to a superkingdom (B). (B) Distribution index of individual architectures (f, the number of species using an architecture/total number of species) against the age of architectures (nd, number of nodes from the root/total number of nodes in the tree) uncovers evolutionary patterns of architectural innovation and usage when studying all genomes or only those that are free-living. Based on these patterns, we propose three evolutionary epochs of the protein world: (light green) structural diversification; (salmon) superkingdom specification; (yellow) organismal diversification epochs.
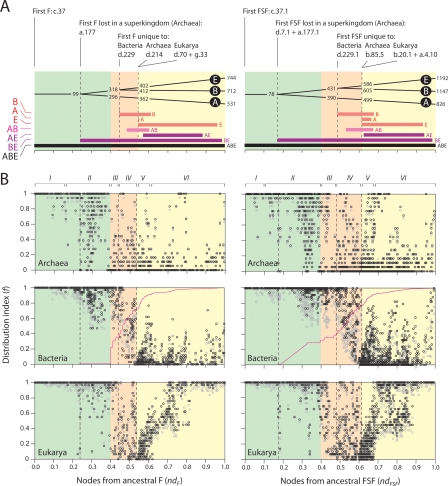
When these architectural chronologies were dissected for the three superkingdoms (Fig. 2), an additional trend became apparent: an organismal superkingdom must “lose” a significant number of architectures before “inventing” its first specific architecture (the “loser trend”). We call this a “loss,” as it usually shows as a decrease in usage (f-value) of that particular F or FSF compared to the older architectures. We hypothesize that the probability to lose an existing architecture later in evolution because of lifestyle adaptation is higher than the probability of the other lineages simultaneously discovering the same architecture at the time of its origin. In general, the higher the value of f, the higher is the probability that a few organisms lost an architecture, and the lower the probability that many organisms independently discovered the same architecture at the same time. The “loser trend” therefore signals the segregation of an emergent lineage from the pool of uniform communal ancestors, even though architectures specific to a lineage usually appear much later. As soon as the first lineage-specific architecture appears, the superkingdom is considered “specified,” and it later diversifies as superkingdom-specific architectures begin to be differentially apportioned to organisms within it.
Figure 2.
Six phases in the evolutionary timeline of the protein world based on distribution of F (left) and FSF (right) within the three superkingdoms of life. (A) Bar diagrams display ranges of age (nd) for architectures unique to superkingdoms (A, B, or E) or shared by two (AB, BE, or AE) or all (ABE) superkingdoms. Trees describe global most-parsimonious scenarios for organismal diversification of proteomes based on architectural distribution patterns. Numbers indicate the size of architectural repertoires in A, B, and E lineages at the corresponding nd values. The horizontal scale is as in B. (B) Distribution index (f) of F and FSF within the three superkingdoms for (gray) all organisms or (black) free living only against the age of the individual architectures. (Light green) Structural diversification; (salmon) superkingdom specification; (yellow) organismal diversification epochs. Roman numerals indicate the evolutionary phases of the protein world described in the text. (Red lines) Cumulative loss of BE architectures (number of architectures absent in each organism, summated over organisms, and integrated over nd); the ordinate is in logarithmic scale with units not displayed; the abscissa matches nd values.
These results suggest three epochs in protein evolution, which we then subdivide into six phases, each delimited by patterns of architectural use (elaborated in the Discussion): (1) Architectural diversification (ndF < 0.40 and ndFSF < 0.49; light green areas in Fig. 1B defining phases I and II), in which members of the ancestral community diversified their architectural repertoire through differential “loss” of architectures. (2) Superkingdom specification (0.40 <ndF < 0.618 and 0.49 < ndFSF < 0.679; salmon areas defining phases III and IV), where superkingdoms Archaea, Bacteria, and Eukarya are specified through invention of superkingdom-specific architectures. (3) Organismal diversification (0.618 < ndF and 0.674 < ndFSF; light yellow areas defining phases V and VI), where F and FSF specific to relatively small sets of organisms appear as the result of diversification of organismal lineages.
Further evidence, presented below through the analysis of architectural distribution (Fig. 2A) and representation in superkingdoms (Fig. 2B), provides support to these three evolutionary epochs.
All basal (nd < 0.1) and many of the more recent (nd < 0.4) F and FSF architectures were shared by most if not all proteomes in all superkingdoms (Figs. 1, 2). Venn diagrams of architectural use show that architectures that are common to all superkingdoms are the most abundant (Fig. 1A). Architectural diversification begins when newer architectures become differentially excluded in some species, resulting in their smaller representation (f < 1) within the organismal community. Loss of ancient architectures was mostly confined to Archaea (Fig. 2B). Very few F or FSF of ancient origin (e.g., the 53 most basal F, ndF < 0.162) were lost in bacterial and eukaryal organisms, and most of that loss occurred in organisms leading parasitic lifestyles (Fig. 1B; Fig. 2B, gray circles; Supplemental Fig. S1). Representation of ancient F in Archaea begins to drop precipitously at about ndF = 0.1, after α/β-hydrolase (c.69) and profiling-like (d.110) architectures are lost for the first time in FL archaeal species (Supplemental Fig. S1). This process becomes very extensive in the region of 0.2 < ndF < 0.4, where many F are represented only by a small fraction of archaeal organisms, and some are missing entirely (archaeal loser trend) (Fig. 2B). The sigma 2 domain of RNA polymerase sigma factors (a.177) is the first F to be lost completely in Archaea at ndF = 0.237 (BE bar in Fig. 2A). A similar trend can be seen in the representation of FSF (Fig. 2B). The LysM domain (d.7.1) and the sigma 2 domain (a.177.1) were the first to be lost in Archaea at ndFSF = 0.174 and 0.185, respectively. Such total loss of F and FSF architectures in Bacteria is delayed until ndF = 0.543 and ndFSF = 0.614, and in Eukarya until ndF = 0.439 and ndFSF = 0.489, respectively. This significant early differential loss of architectures occurring primarily in Archaea segregates them from the world of ancient organisms, establishing the first organismal divide.
Decreases in architectural representation (f-value) occurred also in Eukarya and Bacteria, but involved fewer and younger architectures. Architectural loss begins at ndF = 0.399 and ndFSF = 0.391 when the Bacteria and Eukarya (BE)-specific architectures experience a notable decrease in representation (Fig. 2B, line graph), implying that the rising architectural diversity is apportioned differently to different species (bacterial “loser trend”). This process signals the beginning of the superkingdom specification epoch, which culminates in the appearance of the first architectures unique to a superkingdom, specifically Bacteria (B bar in Fig. 2A). Those were the TilS substrate-binding domain F (d.229; ndF = 0.439) and its associated FSF (d.229.1; ndFSF = 0.489). This early start did not alter the general patterns of F and FSF representation but allowed Bacteria to acquire significant structural diversity in the 0.439 < ndF < 0.543 and 0.489 < ndFSF < 0.614 timeframe before architectures unique to other superkingdoms appeared.
The decreasing trend in architectural representation (eukaryal “loser trend”) continues until the appearance of prokaryote-specific (AB) F and FSF at ndF = 0.491 and ndFSF = 0.538 (AB bar in Fig. 2A), which coincides with the occurrence of common F and FSF (ABE) widely used in Bacteria and Eukarya (data not shown). Appearance of AB-specific architectures sets prokaryotes apart in their usage of a molecular repertoire significantly before appearance of the first Eukarya-specific (E) architectures at ndF = 0.543 and ndFSF = 0.614, the event that fully specifies Eukarya as a superkingdom. Specification of Eukarya as the last domain of life is followed by a sudden drop in representation of all subsequent architectures both in Bacteria and Eukarya until ndF = 0.601 and ndFSF = 0.674, when the last AB-specific architectures appear. We call this a “burst” of architectural innovation in Bacteria and Eukarya, as it involves a large number of architectures, all represented by a small fraction of species (f < 0.5)—evidence of organismal diversification. Here the differences between prokaryotes and eukaryotes seem to be defined, both through AB-specific and E-specific architectures (Fig. 2A) and through diversification of all three superkingdoms in their usage of the protein complement (small f). Immediately following appearance of the last AB-specific architecture, the representation strategy in Eukarya undergoes a major revision. From this point on, newer structures become more and more popular in eukaryotic genomes, until representation reaches f = 1 at ndF = 0.95. Concurrently, both Bacteria and Archaea maintain the specialization trend of small representation for almost all new F and FSF.
Evolution of cellular function
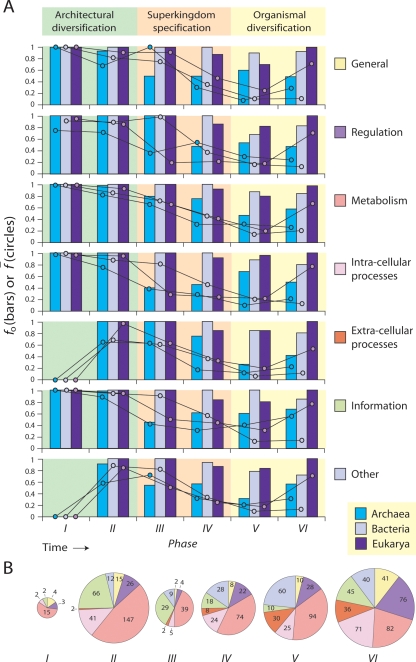
To explain the above trends from a functional perspective, we tallied the FSF participating in various cellular functions in every phase of the architectural chronology. Functions were defined using a hierarchical coarse-grained classification encompassing seven functional categories and 50 subcategories (Vogel et al. 2004a, 2005; Vogel and Chothia 2006). For each phase and category, the fraction (fo) of FSF used in each superkingdom was calculated (Fig. 3, bars). This index fo indicates what functions drop out of use in each phase and superkingdom: fo close to 1 indicates that the superkingdom in question completely lost only a few FSF of that function in that phase, whereas fo close to 0 indicates that most FSF were lost (or not gained). To aid interpretation of this index, we also calculated average f-values (f) that describe organismal FSF usage for every function, phase, and superkingdom (Fig. 3, circles). When f is close to 1, all organisms in a superkingdom use FSF for that function. When f is close to 0, most organisms fail to use them.
Figure 3.
Evolution of biological function along the six phases of the architectural chronology. (A) Bar diagrams describe the fraction of FSF corresponding to each of seven coarse-grained functional categories in each superkingdom relative to their use in all life within a particular evolutionary phase (fo), and circles describe how widely distributed these FSF are among organisms within each superkingdom, as average distribution indices (f). When bars and circles are both high or low, the relative importance of that function is either high or low, respectively—the function present in most FSF is important to most organisms in a superkingdom, or the function present in few FSF is only important to a small organismal subset. When bars are high and circles are low or when bars are low and circles are high, function in most FSF is important to small organismal subsets or function in few FSF is important to most organisms, respectively. (B) Pie charts describe FSF distribution in functional categories for every phase. The size of each pie chart is proportional to the number of FSF in each phase. Four uninformative “not annotated” FSF (d.58.45 and e.30.1 of phase V, and a.125.1 and d.46.1 of phase VI) were not included in the analysis.
Most broad functions were invented very early in phase I, and all associated FSF were necessary for cellular physiology: none of them dropped out of use (fo ∼ 1) (Fig. 3). However, the “extracellular processes” and “other” functional categories did not appear until phase II, and only 26% of functional subcategories were invented in phase I (Supplemental Fig. S2). The archaeal loser trend seems to encompass all functions approximately to the same extent (f = 0.6–0.9) in phase II, although later in phase III a large number of FSF from “intracellular processes” and “information” (fo ∼ 0.4) were completely lost in this superkingdom, most particularly protein modification and translation-related FSF—possibly the archaeal-specification event. Note that 74% of subcategories are represented in phase II (Supplemental Fig. S2). Therefore, most functions were discovered during the architectural diversification epoch.
During the superkingdom specification epoch, Bacteria became specified through the invention of several highly represented FSF corresponding to “information,” “intracellular processes,” and “regulation” functions in order of decreasing representation (f = 0.8–0.9 for functions in Bacteria, and significantly higher than those in Archaea and Eukarya, f = 0.2–0.5). Interestingly, Eukarya seem to be specified earlier than suggested by the architectural chronologies (Fig. 2). In phase II, Bacteria and Archaea substantially lost representation of FSF for “extracellular processes” (f = 0.5–0.8), whereas Eukarya retained an almost full FSF representation (f ∼ 1), corresponding mostly to cell adhesion and immune response (Supplemental Fig. S2). These include the vWA-like FSF (c.62.1) that encompasses the integrin A domain involved in cell attachment and signal transduction processes, and the 4-helical cytokine FSF (a.26.1) important for signaling glycoprotein molecules (cytokines, interferons, interleukins) that are necessary in adaptive immune responses and cellular communication.
Further in evolution, bacterial FSF invention is prominent in phase IV (fo = 1 for most functions), while Archaea and Eukarya follow the loser trend in parallel with each other. This loser trend turns into diversification, especially in phase V for all three superkingdoms, evidenced by low usage of all FSF (f close to 0) and incomplete retention of FSF invented in this phase (fo < 1). In phase VI, Eukarya retain (fo bars and f close to 1) and Bacteria diversify all functions (tall fo bars with very low f). Archaea substantially raise their usage of “information” FSF, corresponding mostly to unknown functions (Supplemental Fig. S2).
In terms of global evolutionary patterns, functions associated with “general,” “regulation,” and “intracellular processes” were abundant early and late in evolution; “metabolism” was maximal early and decreased steadily in time; “information” peaked midway (phases II and III); and “extracellular processes” and “other” were poorly represented early but increased in time (Fig. 3). Other patterns worth mentioning include the early development of translation, signal transduction, DNA binding, DNA replication/repair, chromatin structure, cell cycle/apoptosis, and cell motility functions (all during architectural diversification) and the relatively late development of photosynthesis, receptor activity, cell envelope, and proteins of viral origin (Supplemental Fig. S2).
Reconstruction of proteome trees
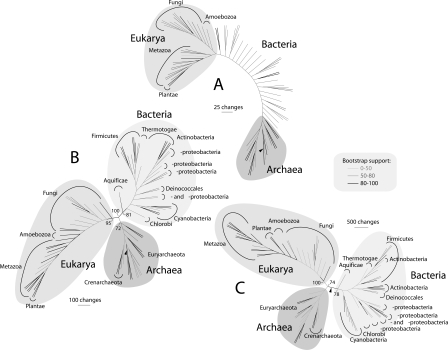
Based on previous results, we reconstructed trees of proteomes to follow the rise of three organismal superkingdoms in evolution. We excluded organisms leading parasitic lifestyles (P and OP) from further phylogenomic analysis to increase the reliability of deep branches. This decision was based on the massive loss of architectures in parasitic lifestyles (Fig. 2B; Supplemental Fig. S1), possibly causing homoplastic events frequently observed in phylogenetic trees. We built global trees using three subsets of FSF architectures (Fig. 4A–C; see also Supplemental Fig. S3) originating within different phases of the evolutionary timeline defined above, so as to follow separation of major branches through evolutionary time. The topology of rooted and unrooted trees reconstructed using polarized (directed) or nonpolarized (undirected) characters was almost identical in these studies (data not shown). A proteome tree reconstructed from ancient FSF common to all superkingdoms (ndFSF < 0.174) was rooted paraphyletically in Archaea, reflecting their early segregation through the minimalist strategy (Fig. 4A). The tree had poor resolution, likely because most architectures used were shared by all superkingdoms, but revealed clearly a monophyletic clade grouping of Eukarya. The younger architectures that appeared before the first bacterial FSF (0.174 < ndFSF < 0.489) produced a tree with three clades corresponding to the three superkingdoms rooted paraphyletically in the Archaea (Fig. 4B). It also revealed a sister-clade relationship of Eukarya and Bacteria with high confidence (100% BS). Finally, architectures contributing to the superkingdom specification and diversification epochs (0.489 < ndFSF) yielded a tree with three superkingdom clades, but it had the canonical topology with a bacterial root, reflecting the early appearance of Bacteria-specific architectures, compared to Eukarya and Archaea (Fig. 3C).
Figure 4.
Optimal most-parsimonious phylogenomic trees of proteomes from 82 free-living organisms, generated using subsets of FSF corresponding to different phases of evolutionary history. (A) Ancient FSF, ndFSF < 0.174 (6727 steps; CI = 0.232, RI = 0.687; g1 = −0.316). (B) Intermediate FSF, 0.174 < ndFSF < 0.489 (38,405 steps; CI = 0.184, RI = 0.681; g1 = −0.299). (C) Young FSF, ndFSF > 0.489 (67,555 steps; CI = 0.234, RI = 0.709; g1 = −0.576). Terminal leaves are not labeled, as they would not be legible. Individual trees with taxon labels are shown in Supplemental Figure S3. Bootstrap support (BS) levels for branches are indicated with different shades and with numbers in nodes delimiting superkingdoms.
Effect of parasitic lifestyles
Proteomes from organisms with parasitic lifestyles (both P and OP) significantly affected the distribution of protein architectures between organisms. Most prominently, parasitic organisms lack a significant number of architectures that appeared throughout evolution (depicted by gray circles in Fig. 2; see Supplemental Fig. S1). The process of architecture loss in P and OP begins very early, earlier than the appearance of the first F and FSF unique or shared by two lifestyle groups (ndF = 0.532 and ndFSF = 0.538; parasitic “loser trend”) (Fig. 5). The invention of parasite-specific architectures coincides with the rise of superkingdom-specific architectures in the superkingdom specification epoch. A Venn diagram describing the distribution of F and FSF among proteomes of organisms exhibiting FL, P, and OP lifestyles shows that most architectures were shared by all proteomes (Fig. 5): there were only 41, four, and one F and 76, 10, and one FSF unique to FL, P, and OP organisms, respectively. There was only one F and five FSF that were shared by P and OP organisms. Bar diagrams and cumulative frequency distribution plots were used to describe how F and FSF unique or shared by proteomes with different lifestyles appeared and accumulated in the course of evolution (Fig. 4). Nearly all F and FSF with restricted distribution occurred during organismal diversification.
Figure 5.
Cumulative frequency distribution of F (left) and FSF (right) along the trees of architectures that are unique or shared by organisms with (FL) free-living, (P) parasitic, or (OP) obligate parasitic lifestyles. (A) Venn and (B) bar diagrams show the distribution and range of age (nd, number of nodes from the root/total number of nodes in the tree) for architectures within one (FL, P, or OP) or more (FL-P, FL-OP, P-OP, and FL-P-OP) lifestyle categories. (C) Cumulative number of F or FSF architectures against nd.
Occurrence and abundance of architectures in proteomes
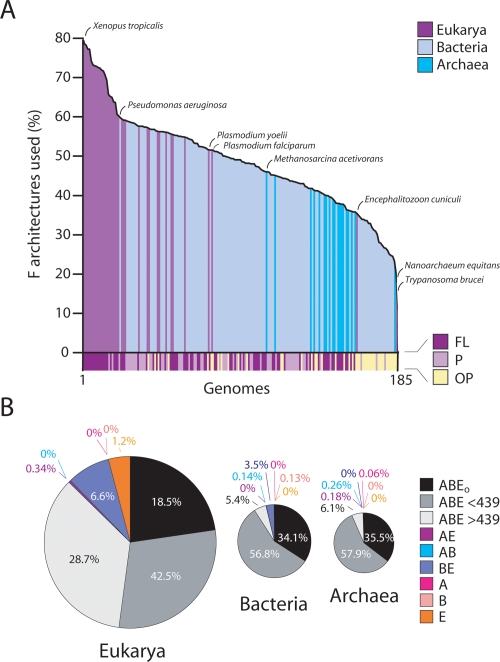
To examine the present-day outcome of the evolutionary scenario described above, we calculated the occurrence (usage) and abundance of architectures in proteomes analyzed (Fig. 6). The average percentage of F used by Eukarya, Bacteria, and Archaea was 63.0% ± 13.2 (SD)%, 46.1% ± 9.4%, and 38.4% ± 5.2%, respectively. Eukaryal genomes generally used the largest repertoire (Fig. 6A). The range of F used was also the largest in Eukarya, ranging from 9.8% in Trypanosoma brucei to 79% in Xenopus tropicalis. Archaeal genomes on average used the lowest number of architectures with F usage ranging from 20.1% in Nanoarchaeum equitans to 46% in Methanosarcina acetivorans. F usage in Bacteria was intermediate and ranged from 23.3% in Onion yellows phytoplasma to 60.3% in Pseudomonas aeuginosa. F usage in organisms with FL lifestyles accentuated these patterns; the average number of F used by free living Eukarya, Bacteria, and Archaea was 66.8% ± 8.3%, 49.7% ± 4.4%, and 39.6% ± 2.9%, respectively. Within each superkingdom, organisms with parasitic lifestyles exhibited the lowest F usage levels. Consequently, the lowest value of F usage in FL organisms increased to 54.9% (Ashbya gossypii), 39.9% (Lactobacillus johnssonii), and 35.8% (Methanocaldococcus jannaschii) for Eukarya, Bacteria, and Archaea, respectively. Overall trends in architectural abundance were similar to those of architectural occurrence, with Eukarya significantly favoring the reuse of F architectures (Fig. 6B). While ancient and common F were the most abundant in the three superkingdoms, Eukarya considerably increased the abundance of common F appearing during the superkingdom specification and diversification epochs. This includes F shared with Bacteria and those unique to Eukarya.
Figure 6.
Effect of lifestyle on use of protein F in proteomes. (A) F usage in proteomes, sorted in descending order. (FL) Free-living; (P) parasitic; (OP) obligate parasitic lifestyle. (B) Pie charts of the protein repertoire within the superkingdoms of life. The size of each pie chart is proportional to the genomic abundance of F within the respective superkingdom, and percentages represent the fraction of total abundance designated by each sector. F are identified as superkingdom-specific (A, B, or E), or shared by some (AB, BE, or AE) or by all (ABE) superkingdoms. ABE F are further divided into those that are omnipresent F shared by all organisms (ABEo) and those that appeared before (ABE < 439) or after (ABE > 439) d.229, the first F unique to Bacteria that delimits the upper bound of the organismal specification epoch at ndF = 0.439.
Discussion
Phylogenomic reconstruction of the protein world
Advances in structural bioinformatics have extended structural information deposited in the Protein Data Bank (PDB) to macromolecules encoded by more than half of gene complements identified in the >500 fully sequenced genomes published to date (Grant et al. 2004). In this study, we use information embedded in a structural genomic census of protein architecture to generate trees that describe the evolution of protein structure at F and FSF hierarchical levels (Fig. 1A). A flowchart describing the overall experimental strategy is described in Supplemental Figure S5. Our trees of F and FSF architectures are intrinsically rooted, that is, we have established evolution’s arrow without the need of outgroups. The trees were also highly unbalanced, suggesting that architectural discovery involved semipunctuated evolutionary processes, similar to those recently suggested for substitutional change at the DNA level (Pagel et al. 2006). Punctuation underscores the importance of the discovery of new architectures in evolution, as acquisition of architectural designs is rare and subject to complex processes that relate to the mapping of sequence into structure.
Our analysis does not consider the increasingly important contribution of non-coding functional RNA molecules (Eddy 2001). However, it does provide a comprehensive analysis of proteins encoded in the genomes we studied. The F and FSF examined here represent our current view of the complexity of the protein and organismal world. These architectures are associated with proteins that play diverse and fundamental functional roles in the cell, such as translational and transcriptional machinery, metabolic and signaling pathways, structural scaffolds, and many other aspects important for cellular function and interaction. The proteins themselves cannot capture adequately deep phylogenetic relationships because of the erasing effects of mutation and HGT; a comparative genomic exercise therefore reveals genomes as evolutionary mosaics of genes (Lester et al. 2005). A focus on molecular designs that are immutable for extended periods of time rather than a focus on the vagaries of gene sequence uncovers here deep historical signatures. These signatures are more successfully preserved in the architectural repertoire the older the architectures studied, because older architectures are more abundant and diverse. These ancient architectural designs provide important clues related to the molecular origins of modern life.
Thus, the conclusions of this study are independent of the outcome of major debates in the evolutionary field, including the degree of HGT in the primordial and diversifying world (Kurland 2005), the origin of the eukaryotic cell (Poole and Penny 2007), and the ability of a single bifurcating tree to represent the evolution of superkingdoms of life (Doolittle and Bapteste 2007), most of which are centered on the limitations of genomic sequence evidence. Furthermore, architectural distributions reflect evolutionary and ecological pressures on the organisms, because F and FSF represent functional units of proteins, and their function is being selected for maximum survival of an organismal lineage within its environment. Consequently, architectural distributions today carry the imprint of the adaptation strategies adopted by the three superkingdoms during their evolution, and it is the evolution of those adaptations that we infer in this study. Specifically, we infer the timing of superkingdom specification and organismal diversification based on F and FSF distribution in organisms. The differences in F and FSF distribution patterns allow us to propose a timeline and mechanisms of organismal lineage segregation from the communal ancestor, as discussed below.
Mechanisms of protein architecture distribution between organisms
Phylogenetic trees of architectures embed timelines of protein discovery. Along these architectural chronologies, the distribution (f) of F and FSF in the organismal world as a function of their age (nd) was variable (Figs. 1, 2). As new architectures appeared, they spread unevenly between organisms, that is, some were absent in individual proteomes causing f < 1. Several evolutionary processes can explain changes in f-values. In addition to (1) HGT and (2) vertical descent (Woese 2000) illustrated by sorting of architectures in organismal lineages, we also identified (3) genome reduction, (4) genome expansion, and (5) processes of architectural fusion and fission that result in the combination or rearrangement of architectures in proteins. Supplemental Figure S4 describes how these processes affect the rise of architectures during the emergence of lineages. Genome reduction decreases the number of genes in an organism and can simplify the architectural complement, resulting in loss of F and FSF and decrease in f-values. In contrast, genome expansion will favor retention of architectures with an opposite effect on f. In the absence of HGT, architectures can be lost in one lineage yet gained by others with the same effect on f, but independent gains become unlikely as lineages increase in number. In Supplemental Figure S4, for example, it is more likely to gain FSF b in one out of four lineages than to lose it in three out of four. Consequently, the probability of loss or gain depends on how structured or diversified is the organismal world. During lineage diversification (i.e., cladogenesis induced by reproductive isolation) in which all lineages are retained (unlikely in light of extinction), f will decrease with increasing nd by lineage sorting. For example, if an architecture appears early in a lineage, it could distribute by vertical descent in the many lineages that are splitting. Conversely, if the architecture appears late in one of the splitting lineages, it will be confined to the lineages where it occurs. Geographical or niche isolation will similarly decrease f by constraining the spread of architectures. In contrast, HGT processes homogenize the organismal world with partial (e.g., xenology) or total (e.g., synology, endosymbiosis) exchange of genetic complements (Mindell and Meyer 2001), increasing f in every instance. Finally, the modular combination and rearrangement of architectures in proteins (known as “domain combinations”) (Vogel et al. 2004b) can increase f by altering the representation of individual architectures in the protein world. Architectures are studied here individually, not in combination, yet effects of architectural fusion and fission are noticeable in the f-value. For example, an architecture can appear early before the split of a lineage as a combination of two or more architectures, but fission of its components can result in the architecture appearing later in the protein world in one or both of the resulting lineages and at different times. This kind of process will tend to increase f. In contrast, the fusion of individual architectures to form a combination may not affect f.
Here we use terminology that describes decreases in f as relative “loss” of architectures. In reality, decreases in f are solely due to changes in their representation. When a new molecular design appears, it is added to the global molecular repertoire. However, when some species fail to acquire the design, it may appear as a loss from their proteome, resulting in f < 1. We cannot distinguish this from the possibility of an original acquisition and subsequent loss of the design owing to it being unnecessary or incompatible with the lifestyle of the organism.
Proteome evolution and the birth of the three superkingdoms of life
Architectural chronologies derived from F and FSF trees revealed clear and congruent evolutionary patterns of origin and diversification of organismal groups (Figs. 1, 2). These patterns were used to formulate three epochs and six phases that describe the history of the protein and organismal world. These epochs and phases capture salient evolutionary features, but their boundaries should be considered arbitrary.
Epoch 1: Architectural diversification
Phase I: Organisms at the start of the protein world were molecularly complex and part of a rich communal world (0.000 < ndF < 0.162 and 0.000 < ndFSF < 0.092)
All proteomes in all superkingdoms shared ancient F and FSF that were basal in the trees, including even P and OP organisms whose genomes are highly reduced. The 53 most basal F probably encompass the proteome complexity of this evolutionary period of life (Supplemental Fig. S1). The mere number of shared architectures suggests that the primordial organisms were molecularly complex and largely similar to each other (Fig. 2A). We call this part of evolutionary history a “rich communal world.” These ancient architectures were probably generated when HGT events were rampant, and the distribution of all molecular modules was highly homogeneous, before significant barriers to information exchange (e.g., adaptive, reproductive, ecological) were established (Kurland 2005). It is likely that these F and FSF were retained in all modern life because of their importance in cellular function. The nine most ancient F were responsible for a metabolic “big bang” of architectural diversification that originated in nucleotide metabolism and gave rise to most metabolic subnetworks that exist today (Caetano-Anollés et al. 2007). They also encompass multifunction FSF with small molecule binding activities (Supplemental Fig. S2) that support the translation and transcription machinery, including a substantial number of structures that make up aminoacyl-tRNA-synthetases and rRNA-associated proteins (Ji and Zhang 2007; D. Caetano-Anollés, unpubl.). These and many basal architectures of this phase are also involved in functions associated with ancient genes from an extant proteome core identified by physical clustering of evolutionarily conserved genes in bacterial genomes (Danchin et al. 2007).
The data from this evolutionary phase are compatible with the concept of a communal world similar to the one proposed by Woese (1998). However, this world was molecularly rich and contained complex architectures that encompassed each and every one of the six major SCOP classes of protein structure. About 40% of F and ∼32% of FSF were in place before any superkingdom-specific architectures emerged, setting an upper bound for the architectural repertoire of the communal world. The relative richness of the architectural repertoire in the primordial organisms does not necessarily entail a large size of the proteome in comparison with modern organisms; thus the absolute size of the ancestral proteome still remains unknown.
Phase II: The first organismal divide produced archaeal-like ancestors with reduced proteomes and a minimalist strategy (0.162 < ndF < 0.399 and 0.092 < ndFSF < 0.391)
The organismal representation of architectures that occurred later in evolution was progressively smaller. The initially moderate decrease in representation (f-values high but <1) can be explained by architectural loss due to proteome reduction, not by architectural sorting processes in lineages. Additional decreases in f were likely caused by secondary adaptations that are not contemporary to this period, for example, due to organismal-dependent P and OP lifestyles (see below).
The differential loss of F and FSF was particularly extensive in Archaea—the superkingdom that was also the first to experience complete loss (or lack of appearance) of architectures. Over time, this superkingdom lost a total of 175 F and 308 FSF specific to Eukarya and Bacteria (EB), resulting in the highly compact proteomes typical of today’s Archaea (Fig. 6). Because this minimalist trend is present in Archaea throughout the evolutionary timeline, this early commitment may have been more important in segregating Archaea as a superkingdom than the appearance of Archaea-specific architectures later at ndF = 0.543 and ndFSF = 0.614. The minimalist trend suggests an early split of life into two organismal groups, an archaeal-like ancestor undergoing proteome reduction and a eukaryal-like ancestor that retained the molecular complexity of the rich communal world. This caused modern Eukarya to be more closely related to Bacteria at gene sequence, gene content (e.g., Esser et al. 2004; Lester et al. 2005), and structural levels (e.g., Wang and Caetano-Anollés 2006), despite preserving many commonalities of the ancient protein world with Archaea, such as the phylogenetically ancestral components of the translation and transcription apparatus (Walsh and Doolittle 2005).
Epoch 2: Superkingdom specification
Phase III: Reductive tendencies in the eukaryal-like ancestor led to the first superkingdom specification event and the emergence of Bacteria (0.399 < ndF < 0.439 and 0.391 < ndFSF < 0.489)
Reductive tendencies were also present in the eukaryal-like ancestor, but involved fewer and younger architectures compared to Archaea. The first superkingdom-specific architecture appeared in Bacteria, signaling the “official” start of the superkingdom specification epoch. However, the appearance of the first superkingdom-specific architecture in the trees should be regarded as upper bounds to this period. Lineage diversification in Eukarya and Bacteria may have started significantly before their specific architectures appeared, as suggested by the significant loss of earlier F and FSF in both superkingdoms.
Phase IV: Discovery of prokaryote-specific architectures and the rise of superkingdoms Eukarya and Archaea (0.439 < ndF < 0.543 and 0.489 < ndFSF < 0.614)
This evolutionary phase delimits the steady decrease of f during species diversification in Bacteria, concurrent with lineage specification in the other two superkingdoms. We propose that reduced representation of architectures among organisms at this time may have been caused by several factors, including sorting of architectures in lineages, increased fusion of domains into domain combinations (M. Wang and G. Caetano-Anollés, in prep.), and intensification of proteome reductive tendencies that started in phases II and III.
The concomitant appearance of the first F and FSF unique to Archaea and Eukarya marked the start of their specification. The late specification of Archaea contrasts with the early proteome reduction that defined the primordial archaeal-like ancestor. Perhaps the rates of processes underlying the adaptation of the archaeal-like ancestor to extreme environments were very different from those operating in the ancestors of the other superkingdoms and caused a delay of the lineage specification process. Ultimately, the timing of lineage specification follows the canonical and widely accepted topology of the universal tree of life, which is also reflected in the phylogeny from architectures arising during the superkingdom specification and diversification epochs (Fig. 4C).
Epoch 3: Organismal diversification
Phase V: A burst of architectural innovation in Bacteria and Eukarya (0.543 < ndF < 0.601 and 0.614 < ndFSF < 0.674)
During this brief period, a marked burst of F and FSF architectures with low f-values was evident in Bacteria and Eukarya, associated with proteins that establish domain combinations (M. Wang and G. Caetano-Anollés, in prep.). Many architectures that originated here are unique to Bacteria or to Eukarya. This, combined with their low representation, suggests that this was a period of “experimentation,” when organisms “searched” through the possible protein configurations for a promising beginning of stable lineages within the recently specified superkingdoms.
Phase VI: Genome expansion and homogenization of proteomes in Eukarya and genome reduction in Archaea and Bacteria (0.601 < ndF < 1.000 and 0.674 < ndFSF < 1.000)
Once commitment to archaeal, bacterial, or eukaryal lifestyle was in place, the proteomes in the three superkingdoms appeared to follow divergent evolutionary paths. While Archaea and Bacteria show signs of alternating retention and loss of architectures, architectural retention was increased in eukaryal lineages. We suggest that increases in architectural representation in Eukarya were caused by genome expansion, fission, and fusion/fission of domain combinations previously generated in the burst of phase V, endosymbiotic events mostly involving Bacteria, and HGT events, in order of decreasing importance. The process continued in Eukarya until new architectures were present in most eukaryotic genomes analyzed (f close to 1 again). This striking evolutionary path peculiar to Eukarya differs notably from mechanisms operating in Archaea and Bacteria, which seem to follow lineage sorting, genome reduction tendencies, and genome expansion due to HGT events (e.g., viral or plasmid transfer).
Ecological and functional mechanisms of superkingdom diversification
The patterns of F and FSF acquisition and retention within each superkingdom were certainly affected by the specific needs of organisms and their adaptation to the environment. The entire history of protein architectural evolution can thus be interpreted in ecological terms. As we have seen, Archaea were the first superkingdom to segregate from the rest by adopting the minimalist approach to the molecular repertoire. This early segregation of the archaeal-like ancestor from the eukaryal-like ancestor must have been compromised by HGT, as no substantial lineage splitting was evidenced by appearance of superkingdom-specific architectures at that time. Later they may have turned into ecologically more structured populations because of both natural selection (Vestigian et al. 2006) and adaptations to new environmental niches (L.S. Yafremava, J.E. Mittenthal, and G. Caetano-Anollés, in prep.). The archaeal-like ancestor may have been defined by adaptation to physical extremes, because extreme conditions, such as very high or very low pH, acidity, or pressure, may limit the number of functional protein variants, thus reducing the number of viable protein architectures in a cell (L.S. Yafremava, J.E. Mittenthal, and G. Caetano-Anollés, in prep.). For example, adaptation to extremely high temperatures is believed to cause proteins to be more compact and hydrophobic (structure-based thermostabilization) (Penny and Poole 1999; Berezovsky and Shaknovich 2005). Adaptations to possible chronic energy stress in methanogens, methane oxidizers, and nitrifiers (Valentine 2007) may also have led to a limited number of protein architectures that an organism is able to support. All these processes can impose constraints on structure that lead to a reduced and highly specialized protein repertoire, resulting, for example, in loss of FSF in all biological functions in phase III—these FSF could have been unstable in harsh environments (Fig. 3). Once these FSF, and possibly proteins that use them, were lost, a fraction of protein-modification machinery was lost too (phase III: “intracellular processes”) (Fig. 3; Supplemental Fig. S2), as being unnecessary. The archaeal translation machinery is also significantly reduced when compared to the more elaborate counterparts of Eukarya and Bacteria (phase III: “information”), suggesting a possible role of thermoadaptation or preservation of primordial translation repertoires (e.g., translation initiation) (Kyrpides and Woese 1998).
The eukaryal-like emerging lineage with its large and diverse architectural repertoire may have been better suited for K-selection by exploiting flexibility of use of environmental resources (Carlile 1982). Later, some lineages may have discovered the advantages of rapid growth in times when nutrients were accessible (possibly enabled by a DNA-binding apparatus invented in phase III and fully retained by bacteria) (Fig. 3; Supplemental Fig. S2), entering into r-selection and a competitive strategy of survival, diversification, and streamlining (Penny and Poole 1999), adopting a bacterial lifestyle. This decision encouraged genome reduction to shorten replication cycles (streamlining) and increase the variety of metabolic functions (diversification) to gain competitive advantage (note how bars for metabolic function in Bacteria are tallest in all phases with slight decreases during phase V and VI) (Fig. 3). The latter was made possible by the availability of most metabolic functions at the time of superkingdom specification, as shown by the detailed tracing of architectural ancestries linked to enzymatic functions in metabolism (Kim et al. 2006; Caetano-Anollés et al. 2007). Quick turnover of metabolites was facilitated by early appearance and complete bacterial retention of proteases in phase III (Fig. 3; Supplemental Fig. S2). Ultimately, competition among bacterial-like ancestors led to rapid increase in the number of emerging lineages, irreversible commitment to a competitive strategy by some of them, and generation of a wide diversity of proteomic complements and associated functions. Perhaps this variety was made possible by the early invention of the protein-modification machinery (phase III) (Fig. 3; Supplemental Fig. S2). At some point, reduced HGT and geographical-niche isolation allowed formation of reproductive barriers and generation of true organismal lineages in these streamlined bacterial-like organisms. The result is the evolutionary milestone of “speciation” and the rise of the superkingdom Bacteria.
The first functional specification event in Eukarya seems to occur in phase II: all the cell adhesion and immune response FSF invented in that phase were retained in all modern eukaryotic organisms—the only trend that is different in Eukarya compared to other superkingdoms. It is possible that full retention of these functions allowed Eukarya-like lineages to escape the survival struggle that necessitates quick reproduction, thereby setting up the conditions for long-term growth, storage, and multicellularity peculiar to eukaryotic organisms.
Proteome reductions triggered by parasitic lifestyle
Analysis of F and FSF specific to organisms with FL, P, and OP lifestyles showed that architectures unique to the P and OP categories and shared by them appeared concurrently with architectures specific to Archaea and Eukarya, and once the Bacterial superkingdom was in place (Fig. 5). This result is expected. Since parasitism usually involves adaptations to a particular host, the organismal world had to be fully diversified so that lineages could engage in host–parasite interactions.
In addition, we observed an expected tendency of parasitic organisms to have the smallest molecular repertoire within their respective superkingdoms. This reductive tendency significantly contributed to decreases in f throughout the evolutionary timeline until ndF = 0.757 or ndFSF = 0.886, delimiting a period of development of most P and OP interactions (Figs. 1, 2). The molecular repertoire was most limited in organisms that established obligate symbiotic or parasitic interactions and thus cannot live and reproduce without a host. They have highly reduced genomes and have discarded fundamental enzymatic and cellular machinery in exchange for resources from their hosts (Ochman and Moran 2001). For example, the bacterial endosymbiont of sap-feeding insects Carsonella ruddii, with the smallest genome to date, has only 182 putative protein-encoding genes embedded in 0.159 Mb of sequence (Nakabachi et al. 2006). The genomes of these endosymbiotic organisms sometimes show remarkable stasis, with virtually no rearrangements or inflow of genetic material occurring during millions of years (Tamas et al. 2002). Several studies of structural and functional prediction in minimal genomes suggested how coexistence of organisms has an impact on genomic repertoires (Fraser et al. 1995; Ouzounis et al. 1996; Rychlewski et al. 1998; Chandonia and Kim 2006). Comparison of proteomes of parasites and symbionts with highly reduced genomes showed that essential proteins related to transcription and translation exhibited a higher degree of conservation in F usage than proteins in other functional categories, and were over-represented in organisms with minimal genomes (Chandonia and Kim 2006). However, decreases in f throughout our evolutionary timeline suggest that secondary adaptations driven by reductive evolution have global (though mild) effects on the protein world.
Evolutionary impact on architectural repertoires of present-day organisms
Based on the above observations, we predict that genome reductive tendencies in Archaea and Bacteria must result in a substantial reduction in size of their proteomic repertoires, compared to Eukarya. The early start and protractive tendencies of architectural loss in Archaea predict that proteome reduction must be maximal in this superkingdom. Indeed, patterns of architectural occurrence and abundance in genomes (Fig. 6) show that Eukarya tend to use most of the architectures available, whereas Archaea use the smallest portion out of all free-living organisms. Bacteria seem to occupy the position in between, with many different species using a different subset of architectures. Consequently, the proteomes of organisms in superkingdoms have imprinted in them the evolutionary effects of genome reduction and expansion that were derived from our F and FSF trees and reflect the lifestyle adaptations of the three superkingdoms.
Rooting the universal phylogenomic tree
The topologies of the trees of proteomes reflect the events of the evolutionary timeline that are contemporary to the FSF architectures used in tree reconstruction and provide another tool to visualize the process of superkingdom specification and diversification, regardless of their possible ancestral relationship. Global trees of proteomes reconstructed from ancient FSF encompassing the architectural diversification epoch revealed a paraphyletic rooting in Archaea, reflecting their early segregation through the minimalist strategy. A rooting of the universal tree in Archaea supports paleobiological claims of early archaeal lipids and methanogenic activity linked to the fossil record (Chappe et al. 1979; Michaelis and Albrecht 1979; Schopf 1999) and contrasts with the canonical view of a bacterial ancestor (Woese et al. 1990). In our global trees of proteomes, ancient FSF revealed a paraphyletic rooting in Archaea and the monophyly of Eukarya, defining eukaryal-like ancestors as heirs to the rich communal world and progenitors of Eukarya and Bacteria. FSF of intermediate age revealed a strongly supported sister-clade relationship of Bacteria and Eukarya. Taken together, these trees reflected the early structuring and diversification of the communal world and the formation of archaeal-like and eukaryal-like emerging lineages during this time. In turn, a global tree reconstructed from the derived half of the FSF tree revealed the monophyletic nature of the three superkingdoms and a rooting in the Bacteria, consistent with their leading role in superkingdom specification.
The inclusion of only FL organisms in this analysis minimized historical reconstruction artifacts due to parasitic lifestyle. Exclusion of problematic taxa notably enhanced the support of basal branches in the trees and minimized inconsistent placement of taxa. Indeed, some of the excluded taxa (e.g., Trypanosoma, Encephaltozoon, Nanoarchaeum) had highly reduced proteomes, were big losers of ancient architectures, and were generally oddly placed in trees of proteomes that have been previously reconstructed (Yang et al. 2005; Wang and Caetano-Anollés 2006).
Conclusions
In this study, we use an unorthodox approach to analyze the origins of the tripartite world. This approach focuses on building trees of architectures instead of universal trees of organisms and reveals evolutionary relationships at a genomic scale. The importance of the analysis presented here is that it pinpoints a possible mechanism by which superkingdoms emerged from the communal ancestor, specifically by adopting different strategies of F and FSF usage, possibly in response to different environmental pressures. These strategies involve reduction (notable in Archaea) and expansions (Bacteria and Eukarya) of the global protein repertoire:
Our evolutionary timeline supports the existence of a universal communal ancestor that was complex and architecturally rich (Poole et al. 1998; Forterre and Philippe 1999; Penny and Poole 1999; Glansdorff 2000). It shows that a substantial number of architectures had been already discovered prior to the emergence of the first superkingdom-specific architecture, suggesting that the ancestral organisms may not have been as minimalistic as previously thought (e.g., small protein repertoires matching minimal gene sets) (Mushegian and Koonin 1996). Eukarya retained more ancestral protein architectures compared to prokaryotes. Thus, we call the process of emergence of the three superkingdoms of life “reductive evolution,” to highlight the reductive tendencies of prokaryotes relative to eukaryotes in their usage of architectures, which we think reflects their adaptation to the environment.
We provide for the first time evidence that Archaea established the first organismal divide by losing a substantial number of architectures early in evolution, reflecting the environmental pressures on protein stability and functionality in the harsh environments to which most Archaea are adapted. This important event dissects two ancient lineages, one committed to genome reduction and the other committed to genome expansion, large molecular repertoires, and notable increases in organismal size.
The ancient archaeal lineage suffered a protracted history of reductive evolution and did not “crystallize” (sensu) (Woese 1998) into a diversified superkingdom until later, concurrently with Eukarya. Subsequent genome reductions and expansions in the remaining communal genealogy results in the rise of two lineages with complex proteomic repertoires, one partitioning (Bacteria) and the other homogenizing (Eukarya) the architectural diversity within species in each superkingdom.
Methods
Genomic census
We analyzed the genome sequence of 185 organisms, including 19 Archaea (A), 129 Bacteria (B), and 37 Eukarya (E). Of these, 82, 58, and 45 had FL, P, and OP lifestyles, respectively, using the general strategy described in Supplemental Figure S5. Free-living, parasite, obligate parasite, commensal, obligate commensal, symbiotic, and other lifestyles were annotated manually using various sources of information. For convenience, we pooled genomes from organisms that established symbiotic or commensal interactions into the parasitic groups to define the FL, P, and OP lifestyles. Structural protein domains were assigned to genome-encoded proteins at FSF level using hybrid linear hidden Markov models (HMMs) for remote homology detection in SUPERFAMILY version 1.67 (Gough et al. 2001). Genome sequences were scanned against an HMM library generated using the iterative Sequence Alignment and Modeling System (SAM) method. Each model generated by SAM-T02 identified each non-identical SCOP domain. The HMM searching protocol used a probability cutoff E of 0.02; more stringent cutoff values did not alter the topologies of the reconstructed trees (Yang et al. 2005). An internal calibration of the accuracy of HMM prediction against Protein Data Bank (PDB) records in the ASTRAL compendium (Brenner et al. 2000) showed that the method correctly identified 98% of sequences analyzed (Kim et al. 2006). The structural census assigned protein domains to ∼50% of genomic sequences, ranging from 15% to 71% in individual genomes with a median of 52% (Wang et al. 2006). FSF were assigned to F using the Structural Classification of Proteins (SCOP) database release 1.67. SCOP classifies 24,037 PDB entries into 65,122 domains, which are then grouped into 2630 FF, 1447 FSF, and 887 F architectures (Murzin et al. 1995). Biological functions associated with FSF were annotated using the coarse-grained classification described in SUPERFAMILY (Vogel et al. 2004a, 2005; Vogel and Chothia 2006). Functions related to small molecule metabolism were dissected using MANET (Kim et al. 2006). Note that FSF functions were annotated with respect to their usual role in a protein or biological network, which can be a matter of debate. Moreover, while an older FSF is likely to have generated a function at an earlier time, statistical correlations between FSF ancestry and age of the function may not be necessarily valid for individual proteins because of the vagaries of recruitment in networks (e.g., Caetano-Anollés et al. 2007). For example, a younger FSF could be recruited to perform a particular function in a protein earlier than an older FSF.
Phylogenomic analysis
The frequencies with which individual protein architectures occur in an individual genome, termed GENOMIC ABUNDANCE (G), were used to describe at global levels the popularity of F and FSF architectures. For phylogenetic analysis, G values were normalized to compensate for differences in genome size and proteome representation and were subjected to logarithmic transformation to account for unequal variance (Wang et al. 2006). The gap-recoding technique of Thiele (1993) developed for the analysis of morphometric data was used in which a rescaling function rescores character information on both rank order and size of gaps between character states. Values were range standardized to a 0–20 scale, as this range is compatible with most phylogenetic analysis programs, encoded using an alphanumeric format with numbers 0–9 and letters A–K in the NEXUS format, and subjected to phylogenetic analysis using maximum parsimony (MP) as the optimality criterion in PAUP* (Swofford 2002). Phylogenomic trees of proteomes and trees of architectures analyzed at F and FSF levels of protein classification were generated using linearly ordered multistate phylogenetic characters. Characters are observable features that distinguish one object from another and constitute hypotheses of primary homology. In our case, they display multiple numerical values and frequency distribution of values called character states. The ANCSTATES command was used to polarize characters, based on two fundamental premises: (1) that protein structure is far more conserved than sequence and carries considerable phylogenetic signal, and (2) that F and FSF architectures that are successful and popular in nature are generally more ancestral. We consider that FF that originated early in evolution are prominent in genomes and that the number of FF members increases in single steps corresponding to the addition or removal of a homologous gene in the family. We assume that this process is reversible and expresses an asymmetry with gene duplication being favored over gene loss. Details and support for character argumentation and absence of circularity in assumptions have been described previously (Caetano-Anollés and Caetano-Anollés 2003, 2005; Wang et al. 2006). Because F and FSF are retained over long evolutionary times, their gain or loss constitute important evolutionary events that appear to be independent of HGT and other convergent evolutionary processes (Gough 2005). Phylogenetic reliability was evaluated by the bootstrap method in PAUP*. The structure of phylogenetic signal in the data was tested by the skewness (g1) of the length distribution of >104 random trees and permutation tail probability (PTP) tests of cladistic covariation using >103 replicates (Hillis and Huelsenbeck 1992). Ensemble consistency (CI) and retention (RI) indices were used to measure homoplasy and synapomorphy, confounding and desired phylogenetic characteristics, respectively.
Our phylogenetic analyses depend on the accuracy and balance of genomic databases, efficient and accurate assignment of structures to protein sequences, adequate structural classification schemes in SCOP, and methods of phylogenetic tree reconstruction. For example, there are biases in the detection of FSF from protein with PDB entries used as seed sequences of the HMMs and biases in the representation of sequences and genomes in the databases, favoring Bacteria over Eukarya and Archaea. The effects that these factors have on our approach have been discussed previously (Caetano-Anollés and Caetano-Anollés 2003, 2005) and have not been controlled in our experimental design. We do not expect that the operational definition of F and FSF will be seriously challenged, even though many F can be better described by continuous rather than discrete distributions in structure space (Harrison et al. 2002). Domain structures of globular proteins that have not been discovered to date are probably of low genomic abundance and are expected to be highly diverse (Gerstein and Hegyi 1998). Gene sequences with no structural assignments probably encode membrane proteins or globular proteins that are difficult to crystallize (Liu and Rost 2002). Future advances in structural genomics and bioinformatics will help fill structural “gaps,” will decrease the bias introduced by unassigned domains and structural elements, and will benefit our approach.
Organismal distribution analysis of F and FSF architectures
Protein architectures were classified into F and FSF distribution categories that describe their spread across the three superkingdoms of life. Architectures appearing in all 185 organisms analyzed were assigned to the ABE0 category; those present in at least one proteome but in all superkingdoms were assigned to the ABE category; those present in two superkingdoms were assigned to the AE, AB, and BE categories; and those present in only one superkingdom were assigned to the A, B, and E categories. A distribution index (f) describing the distribution of individual architectures among proteomes was calculated. The f index represents the fraction of proteomes harboring an architecture within a category and ranges from absence (f = 0) to presence in all proteomes considered (f = 1).
Because reconstructed trees were intrinsically rooted, we used a PERL script to establish the relative age (ancestry) of individual protein architectures by measuring a distance in nodes from the hypothetical ancestral F or FSF on a relative 0–1 scale. This node distance (nd) counts the number of nodes (cladogenic events) along a lineage in the tree of architectures, starting from the root and traveling to each terminal leaf. Consequently, the nd ancestry value is 0 for the most ancient architecture and 1 for the most derived. Since rates of genetic evolution are generally linked to speciation (Webster et al. 2003), the total genetic distance from the root to its tips (path length) will be correlated with the number of nodes and consequently with nd. The contribution of processes of gradual evolution will therefore be negligible, and nd will represent a good approximation of a path length based on character state change in individual branches.
Protein classification databases are continuously updated to include more completely sequenced genomes and newly described F and FSF architectures. We cross-checked our results for several releases of the SUPERFAMILY database and found that the main overarching conclusions of this study remain the same: F and FSF distribution between organisms is preserved. However, some of the details may change as new discoveries are made. Thus, we ask the reader to be careful in interpreting the results and focus more on the general trends in the data (reductive tendencies in Archaea vs. retention tendencies in Eukarya) as opposed to the specifics, such as the exact number of F and FSF found in each superkingdom, which is prone to change with time. Also, the exact ancestry values (nd) that we mention in this study for easier description and reference to the graphs will change in the new data sets but are not as important as the relative position of architectures on the trees of F and FSF, which will remain the same. Thus, the reader should treat nd values as relative.
Architectural use and abundance in genomes
The architectural usage in genomes, that is, percentage of architectures used in an organism, was calculated by dividing the number of F or FSF appearing in the organism by the total number appearing in all organisms. G values were used to measure architectural abundance as frequencies with which individual architectures occurred in individual genomes.
Acknowledgments
We thank Simina M. Boca for preliminary results on effects of parasitic lifestyle and Christine Vogel for pointing us to her FSF functional annotation scheme. M.W., L.Y., and G.C. conceived and designed experiments, generated and analyzed data, and produced figures, with significant input from J.E.M. D.C. contributed functional annotation. G.C. and L.Y. wrote the paper, with contributions from all authors. Research was supported in part with funds from UIUC and grants from NSF (MCB-0343126) and the C-FAR Sentinel Program to GC.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6454307
References
- Ancel L.W., Fontana W., Fontana W. Plasticity, evolvability, and modularity in RNA. J. Exp. Zool. 2000;288:242–283. doi: 10.1002/1097-010x(20001015)288:3<242::aid-jez5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Andreeva A., Howorth D., Brenner S.E., Hubbard T.J.P., Chothia C., Murzin A.G., Howorth D., Brenner S.E., Hubbard T.J.P., Chothia C., Murzin A.G., Brenner S.E., Hubbard T.J.P., Chothia C., Murzin A.G., Hubbard T.J.P., Chothia C., Murzin A.G., Chothia C., Murzin A.G., Murzin A.G. SCOP database in 2004: Refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezovsky I.N., Shaknovich E.I., Shaknovich E.I. Physics and evolution of thermophylic adaptation. Proc. Natl. Acad. Sci. 2005;102:12742–12747. doi: 10.1073/pnas.0503890102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S.E., Koehl P., Levitt M., Koehl P., Levitt M., Levitt M. The ASTRAL compendium for protein and sequence analysis. Nucleic Acids Res. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G., Caetano-Anollés D., Caetano-Anollés D. An evolutionarily structured universe of protein architecture. Genome Res. 2003;13:1563–1571. doi: 10.1101/gr.1161903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anollés G., Caetano-Anollés D., Caetano-Anollés D. Universal sharing patterns in proteomes and evolution of protein fold architecture and life. J. Mol. Evol. 2005;60:484–498. doi: 10.1007/s00239-004-0221-6. [DOI] [PubMed] [Google Scholar]
- Caetano-Anollés G., Kim H.-S., Mittenthal J.E., Kim H.-S., Mittenthal J.E., Mittenthal J.E. The origins of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc. Natl. Acad. Sci. 2007;104:9358–9363. doi: 10.1073/pnas.0701214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile M. Prokaryotes and eukaryotes: Strategies and successes. Trends Biochem. Sci. 1982;7:128–130. [Google Scholar]
- Chandonia J.M., Kim S.H., Kim S.H. Structural proteomics of minimal organisms: Conservation of protein fold usage and evolutionary implications. BMC Struct. Biol. 2006;6:7. doi: 10.1186/1472-6807-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappe B., Michaelis W., Albrecht P., Ourisson G., Michaelis W., Albrecht P., Ourisson G., Albrecht P., Ourisson G., Ourisson G. Fossil evidence for a novel series of archaebacterial lipids. Naturwissenschaften. 1979;66:522–523. [Google Scholar]
- Chothia C., Gough J., Vogel C., Teichmann S.A., Gough J., Vogel C., Teichmann S.A., Vogel C., Teichmann S.A., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- Ciccarelli F.D., Doerks T., Mering C.V., Creevey C.J., Snel B., Bork P., Doerks T., Mering C.V., Creevey C.J., Snel B., Bork P., Mering C.V., Creevey C.J., Snel B., Bork P., Creevey C.J., Snel B., Bork P., Snel B., Bork P., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- Danchin A., Fang G., Noria S., Fang G., Noria S., Noria S. The extant core bacterial proteome is an archive of the origin of life. Proteomics. 2007;7:875–889. doi: 10.1002/pmic.200600442. [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H., Philippe H., Brinkmann H., Philippe H., Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Doolittle W.F. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2129. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- Doolittle R.F. Evolutionary aspects of whole-genome biology. Curr. Opin. Struct. Biol. 2005;15:248–253. doi: 10.1016/j.sbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Doolittle W.F., Bapteste E., Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc. Natl. Acad. Sci. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C.L., Yang S., Palenik B., Bourne P.E., Yang S., Palenik B., Bourne P.E., Palenik B., Bourne P.E., Bourne P.E. Modern proteomes contain putative imprints of ancient shifts in trace metal geochemistry. Proc. Natl. Acad. Sci. 2006;103:17822–17827. doi: 10.1073/pnas.0605798103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001;2:919–929. doi: 10.1038/35103511. [DOI] [PubMed] [Google Scholar]
- Esser C., Ahmadinejad N., Wiegand C., Rotte C., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Ahmadinejad N., Wiegand C., Rotte C., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Wiegand C., Rotte C., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Rotte C., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Sebastiani F., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Gelius-Dietrich G., Henze K., Kretschmann E., Richly E., Leister D., Henze K., Kretschmann E., Richly E., Leister D., Kretschmann E., Richly E., Leister D., Richly E., Leister D., Leister D., et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- Forterre P., Philippe H., Philippe H. Where is the root of the universal tree of life? BioEssays. 1999;21:871–879. doi: 10.1002/(SICI)1521-1878(199910)21:10<871::AID-BIES10>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Fraser C.M., Gocayne J.D., White O., Adams M.D., Clayton R.A., Fleichmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., Gocayne J.D., White O., Adams M.D., Clayton R.A., Fleichmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., White O., Adams M.D., Clayton R.A., Fleichmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., Adams M.D., Clayton R.A., Fleichmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., Clayton R.A., Fleichmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., Fleichmann R.D., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., Bult C.J., Kerlavage A.R., Sutton G., Kelley J.M., Kerlavage A.R., Sutton G., Kelley J.M., Sutton G., Kelley J.M., Kelley J.M., et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Gerstein M., Hegyi H., Hegyi H. Comparing genomes in terms of protein structure: Surveys of a finite parts list. FEMS Microbiol. Rev. 1998;22:277–304. doi: 10.1111/j.1574-6976.1998.tb00371.x. [DOI] [PubMed] [Google Scholar]
- Glansdorff N. About the last common ancestor, the universal life-tree and lateral gene transfer: A reappraisal. Mol. Microbiol. 2000;38:177–185. doi: 10.1046/j.1365-2958.2000.02126.x. [DOI] [PubMed] [Google Scholar]
- Gough J. Convergent evolution of domain architectures (is rare) Bioinformatics. 2005;21:1464–1471. doi: 10.1093/bioinformatics/bti204. [DOI] [PubMed] [Google Scholar]
- Gough J., Karplus K., Hughey R., Chothia C., Karplus K., Hughey R., Chothia C., Hughey R., Chothia C., Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- Grant A., Lee D., Orengo C., Lee D., Orengo C., Orengo C. Progress towards mapping the universe of protein folds. Genome Biol. 2004;5:107. doi: 10.1186/gb-2004-5-5-107. http://genomebiology.com/2004/5/5/107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.A. The theory of branching processes. Dover Publications; New York: 1963. [Google Scholar]
- Harrison A., Pearl F., Mott R., Thornton J., Orengo C., Pearl F., Mott R., Thornton J., Orengo C., Mott R., Thornton J., Orengo C., Thornton J., Orengo C., Orengo C. Quantifying the similarities within fold space. J. Mol. Biol. 2002;323:909–926. doi: 10.1016/s0022-2836(02)00992-0. [DOI] [PubMed] [Google Scholar]
- Hillis D.M., Huelsenbeck J.P., Huelsenbeck J.P. Signal, noise, and reliability in molecular phylogenetic analysis. J. Hered. 1992;83:189–195. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- Ji H.-F., Zhang H.-Y., Zhang H.-Y. Protein architecture chronology deduced from structures of amino acid synthases. J. Biomol. Struct. Dyn. 2007;24:321–323. doi: 10.1080/07391102.2007.10507122. [DOI] [PubMed] [Google Scholar]
- Kim H.-S., Mittenthal J.E., Caetano-Anollés G., Mittenthal J.E., Caetano-Anollés G., Caetano-Anollés G. MANET: Tracing evolution of protein architecture in metabolic networks. BMC Bioinformatics. 2006;7:351. doi: 10.1186/1471-2105-7-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J.O., Snel B., Huynen M.A., Bork P., Snel B., Huynen M.A., Bork P., Huynen M.A., Bork P., Bork P. SHOT: A Web server for the construction of genome phylogenies. Trends Genet. 2002;18:158–162. doi: 10.1016/s0168-9525(01)02597-5. [DOI] [PubMed] [Google Scholar]
- Kurland C.G. What tangled web: Barriers to rampant horizontal gene transfer. Bioessays. 2005;27:741–747. doi: 10.1002/bies.20258. [DOI] [PubMed] [Google Scholar]
- Kurland C.G., Collins L.J., Penny D., Collins L.J., Penny D., Penny D. Genomics and the irreducible nature of eukaryote cells. Science. 2006;312:1011–1014. doi: 10.1126/science.1121674. [DOI] [PubMed] [Google Scholar]
- Kyrpides N.C., Woese C.R., Woese C.R. Archaeal translation initiation revisited: The initiation factor 2 and eukaryotic initiation factor 2B α-β-δ subunit families. Proc. Natl. Acad. Sci. 1998;95:3726–3730. doi: 10.1073/pnas.95.7.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester L., Meade A., Pagel M., Meade A., Pagel M., Pagel M. The slow road to the eukaryotic genome. BioEssays. 2005;28:57–64. doi: 10.1002/bies.20344. [DOI] [PubMed] [Google Scholar]
- Liu J., Rost B., Rost B. Domains, motifs and clusters in the protein universe. Curr. Opin. Chem. Biol. 2002;7:5–11. doi: 10.1016/s1367-5931(02)00003-0. [DOI] [PubMed] [Google Scholar]
- Michaelis W., Albrecht P., Albrecht P. Molecular fossils of Archaebacteria in Kerogen. Naturwissenschaften. 1979;66:420–421. [Google Scholar]
- Mindell D.P., Meyer A., Meyer A. Homology evolving. Trends Ecol. Evol. 2001;16:434–440. [Google Scholar]
- Murzin A., Brenner S.E., Hubbard T., Clothia C., Brenner S.E., Hubbard T., Clothia C., Hubbard T., Clothia C., Clothia C. SCOP: A structural classification of proteins for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Mushegian A.R., Koonin E.V., Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M., Yamashita A., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M., Toh H., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M., Ishikawa H., Dunbar H.E., Moran N.A., Hattori M., Dunbar H.E., Moran N.A., Hattori M., Moran N.A., Hattori M., Hattori M. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Ochman H., Moran N.A., Moran N.A. Genes lost and genes found: Evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1098. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- Orengo C.A., Michie A.D., Jones S., Jones D.J., Swindells M.B., Thornton J.M., Michie A.D., Jones S., Jones D.J., Swindells M.B., Thornton J.M., Jones S., Jones D.J., Swindells M.B., Thornton J.M., Jones D.J., Swindells M.B., Thornton J.M., Swindells M.B., Thornton J.M., Thornton J.M. CATH: A hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Ouzounis C., Casari G., Valencia A., Sander C., Casari G., Valencia A., Sander C., Valencia A., Sander C., Sander C. Novelties from the complete genome of Mycoplasma genitalium. Mol. Microbiol. 1996;20:898–900. doi: 10.1111/j.1365-2958.1996.tb02529.x. [DOI] [PubMed] [Google Scholar]
- Pagel M., Venditti C., Meade A., Venditti C., Meade A., Meade A. Large punctuational contribution of speciation to evolutionary divergence at the molecular level. Science. 2006;314:119–121. doi: 10.1126/science.1129647. [DOI] [PubMed] [Google Scholar]
- Penny D., Poole A., Poole A. The nature of the last universal common ancestor. Curr. Opin. Genet. Dev. 1999;9:672–677. doi: 10.1016/s0959-437x(99)00020-9. [DOI] [PubMed] [Google Scholar]
- Poole A., Penny D., Penny D. Evaluating hypotheses for the origin of eukaryotes. Bioessays. 2007;29:74–84. doi: 10.1002/bies.20516. [DOI] [PubMed] [Google Scholar]
- Poole A., Jeffares D.C., Penny D., Jeffares D.C., Penny D., Penny D. The path from the RNA World. J. Mol. Evol. 1998;46:1–17. doi: 10.1007/pl00006275. [DOI] [PubMed] [Google Scholar]
- Riley M., Labedan B., Labedan B. Protein evolution viewed through Escherichia coli protein sequences: Introducing the notion of a structural segment of homology, the module. J. Mol. Biol. 1997;268:857–868. doi: 10.1006/jmbi.1997.1003. [DOI] [PubMed] [Google Scholar]
- Rychlewski L., Zhang B., Godzik A., Zhang B., Godzik A., Godzik A. Fold and function predictions for Mycoplasma genitalium proteins. Fold. Des. 1998;3:229–238. doi: 10.1016/S1359-0278(98)00034-0. [DOI] [PubMed] [Google Scholar]
- Schopf J.W. Deep divisions in the tree of life—What does the fossil record reveal? Biol. Bull. 1999;196:351–355. doi: 10.2307/1542968. [DOI] [PubMed] [Google Scholar]
- Snel B., Bork P., Huynen M.A., Bork P., Huynen M.A., Huynen M.A. Genome phylogeny based on gene content. Nat. Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- Swofford D.L. Phylogenetic analysis using parsimony and other programs (PAUP*) Sinauer Associates; Sunderland, MA: 2002. version 4. [Google Scholar]
- Tamas I., Klasson L., Canbäck B., Näslund K., Eriksson A.-S., Sandström J., Wernegreen J., Moran N.A., Andersson S.G.E., Klasson L., Canbäck B., Näslund K., Eriksson A.-S., Sandström J., Wernegreen J., Moran N.A., Andersson S.G.E., Canbäck B., Näslund K., Eriksson A.-S., Sandström J., Wernegreen J., Moran N.A., Andersson S.G.E., Näslund K., Eriksson A.-S., Sandström J., Wernegreen J., Moran N.A., Andersson S.G.E., Eriksson A.-S., Sandström J., Wernegreen J., Moran N.A., Andersson S.G.E., Sandström J., Wernegreen J., Moran N.A., Andersson S.G.E., Wernegreen J., Moran N.A., Andersson S.G.E., Moran N.A., Andersson S.G.E., Andersson S.G.E. 50 million years on genomic stasis in endosymbiotic bacteria. Science. 2002;196:2376–2379. doi: 10.1126/science.1071278. [DOI] [PubMed] [Google Scholar]
- Thiele K. The holy grail of the perfect character: The cladistic treatment of morphometric data. Cladistics. 1993;9:275–304. doi: 10.1111/j.1096-0031.1993.tb00226.x. [DOI] [PubMed] [Google Scholar]
- Valentine D.L. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nature. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- Vestigian K., Woese C., Goldenfeld N., Woese C., Goldenfeld N., Goldenfeld N. Collective evolution and the genetic code. Proc. Natl. Acad. Sci. 2006;103:10696–10701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Chothia C., Chothia C. Protein family expansions and biological complexity. PLoS Comput. Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C., Berzuini C., Bashton M., Gough J., Teichmann S.A., Berzuini C., Bashton M., Gough J., Teichmann S.A., Bashton M., Gough J., Teichmann S.A., Gough J., Teichmann S.A., Teichmann S.A. Supra-domains—Evolutionary units larger than single protein domains. J. Mol. Biol. 2004a;336:809–823. doi: 10.1016/j.jmb.2003.12.026. [DOI] [PubMed] [Google Scholar]
- Vogel C., Bashton M., Kerrison N.D., Chothia C., Teichmann S.A., Bashton M., Kerrison N.D., Chothia C., Teichmann S.A., Kerrison N.D., Chothia C., Teichmann S.A., Chothia C., Teichmann S.A., Teichmann S.A. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 2004b;14:208–216. doi: 10.1016/j.sbi.2004.03.011. [DOI] [PubMed] [Google Scholar]
- Vogel C., Teichmann S.A., Pereira-Leal J.B., Teichmann S.A., Pereira-Leal J.B., Pereira-Leal J.B. The relationship between domain duplication and recombination. J. Mol. Biol. 2005;346:355–365. doi: 10.1016/j.jmb.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Walsh D.A., Doolittle W.F., Doolittle W.F. The real ‘domains’ of life. Curr. Biol. 2005;15:R237–R240. doi: 10.1016/j.cub.2005.03.034. [DOI] [PubMed] [Google Scholar]
- Wang M., Caetano-Anollés G., Caetano-Anollés G. Evolution inferred from domain combination in proteins. Mol. Biol. Evol. 2006;23:2444–2454. doi: 10.1093/molbev/msl117. [DOI] [PubMed] [Google Scholar]
- Wang M., Boca S.M., Kalelkar R., Mittenthal J.E., Caetano-Anollés G., Boca S.M., Kalelkar R., Mittenthal J.E., Caetano-Anollés G., Kalelkar R., Mittenthal J.E., Caetano-Anollés G., Mittenthal J.E., Caetano-Anollés G., Caetano-Anollés G. A phylogenomic reconstruction of the protein world based on a genomic census of protein fold architecture. Complexity. 2006;12:27–40. [Google Scholar]
- Webster A.J., Payne R.J.H., Pagel M., Payne R.J.H., Pagel M., Pagel M. Molecular phylogenies link rates of evolution and speciation. Science. 2003;301:478. doi: 10.1126/science.1083202. [DOI] [PubMed] [Google Scholar]
- Woese C.R. The universal ancestor. Proc. Natl. Acad. Sci. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R. Interpreting the universal phylogenetic tree. Proc. Natl. Acad. Sci. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R., Kandler O., Wheelis M.L., Kandler O., Wheelis M.L., Wheelis M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria and Eukarya. Proc. Natl. Acad. Sci. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Doolittle R.F., Bourne P.E., Doolittle R.F., Bourne P.E., Bourne P.E. Phylogeny determined by protein domain content. Proc. Natl. Acad. Sci. 2005;102:373–378. doi: 10.1073/pnas.0408810102. [DOI] [PMC free article] [PubMed] [Google Scholar]