Abstract
Adenosine-to-inosine (A-to-I) RNA editing was recently shown to be abundant in the human transcriptome, affecting thousands of genes. Employing a bioinformatic approach, we identified significant global hypoediting of Alu repetitive elements in brain, prostate, lung, kidney, and testis tumors. Experimental validation confirmed this finding, showing significantly reduced editing in Alu sequences within MED13 transcripts in brain tissues. Looking at editing of specific recoding and noncoding sites, including in cancer-related genes, a more complex picture emerged, with a gene-specific editing pattern in tumors vs. normal tissues. Additionally, we found reduced RNA levels of all three editing mediating enzymes, ADAR, ADARB1, and ADARB2, in brain tumors. The reduction of ADARB2 correlated with the grade of malignancy of glioblastoma multiforme, the most aggressive of brain tumors, displaying a 99% decrease in ADARB2 RNA levels. Consistently, overexpression of ADAR and ADARB1 in the U87 glioblastoma multiforme cell line resulted in decreased proliferation rate, suggesting that reduced A-to-I editing in brain tumors is involved in the pathogenesis of cancer. Altered epigenetic control was recently shown to play a central role in oncogenesis. We suggest that A-to-I RNA editing may serve as an additional epigenetic mechanism relevant to cancer development and progression.
A growing body of evidence indicates that deregulation of epigenetic mechanisms collaborates with genetic alterations in the development and progression of cancer (Bhalla 2005; Feinberg et al. 2006). DNA methylation and differences in histone modifications are the best-studied epigenetic control mechanisms shown to be altered in cancer (Feinberg and Vogelstein 1983; Feinberg et al. 2006). Recently RNA silencing mechanisms, such as RNA interference and microRNA regulation, were also linked to cancer (Lu et al. 2005; Feinberg et al. 2006).
Adenosine to inosine (A-to-I) RNA editing is a site-specific modification in stem–loop structures within precursor mRNAs, catalyzed by members of the double-stranded RNA (dsRNA)-specific ADAR (adenosine deaminase acting on RNA) family (Bass 2002). ADAR-mediated RNA editing is essential for the normal development of both invertebrates (Palladino et al. 2000) and vertebrates (Higuchi et al. 2000; Wang et al. 2000; Hartner et al. 2004). The splicing and translational machineries recognize inosine (I) as guanosine (G). Therefore the result of ADAR editing consists of genomically encoded adenosines that are read as guanosines in the RNA sequence. Conveniently, the laboratory sequencing reaction also recognizes I as G.
A number of editing sites occur in coding regions and may result in amino acid substitutions affecting the protein properties and interactions. This type of modification is usually characterized by a single site-specific editing event. Until recently, only a handful of such A-to-I editing sites were known in the human transcriptome. In the past few years, bioinformatic and experimental studies have revealed that the extent of editing is much larger, affecting at least tens of thousands of sites and >1600 different genes (Levanon et al. 2004), where the actual number of edited genes is probably much higher (Levanon et al. 2005b). These A-to-I editing events occur in noncoding repetitive sequences, mostly Alu elements, and tend to undergo multi-editing in tight clusters. Similar results were obtained by several other groups (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004). Editing in noncoding sequences was proposed to be involved in general functions such as RNA stabilization, degradation and translation, splicing, RNA interference, heterochromatic silencing, protection from retrotransposition, and destabilization of viral duplexes (Kim et al. 2004; Levanon et al. 2004; Fernandez et al. 2005; Scadden and O’Connell 2005; Nie et al. 2006). In addition, RNA editing was shown to be involved in the regulation of nuclear retention and in miRNAs biogenesis (Prasanth et al. 2005; Blow et al. 2006; Yang et al. 2006).
Altered editing patterns, mainly of coding regions, were shown to be associated with inflammation (Patterson and Samuel 1995), epilepsy (Brusa et al. 1995), depression (Gurevich et al. 2002), and amyotrophic lateral sclerosis (ALS) (Kawahara et al. 2004). Interestingly, analysis of glutamate receptor transcripts in several samples of human gliomas revealed reduced editing compared to the level in normal brain tissue (Maas et al. 2001). Here we demonstrate that RNA editing differs between cancer and normal tissues: (1) Tumors are characterized by widespread, global hypoediting of repetitive elements. (2) Specific recoding and noncoding sites, including those in cancer-related genes, exhibit a complex picture, showing a gene-specific editing pattern in tumors vs. normal tissues.
Results
Bioinformatic analysis identifies reduced A-to-I editing in brain tumors compared to normal brain tissue
The database of editing sites reported by Levanon et al. (2004) was used to search for abnormal editing patterns in cancer tissues. For this purpose, we looked at all expressed sequence tags (ESTs) which have a specific tissue and status (normal or cancer) annotation and calculated the fraction of those ESTs exhibiting editing in at least one of the editing sites identified. The most significant result was obtained for brain tissues: 74,974 normal adult brain ESTs were identified, out of which 5679 exhibit editing (7.6%). In comparison, only 624 out of 15,739 (4.0%) cancer brain ESTs show traces of editing (χ2 = 262; P < 10−58). The brain ESTs belong to 471 normal and 236 cancer libraries. The large number of ESTs and libraries in the sample reduces the probability of some auxiliary bias.
Reduced editing of Alu repeats in human brain tumors
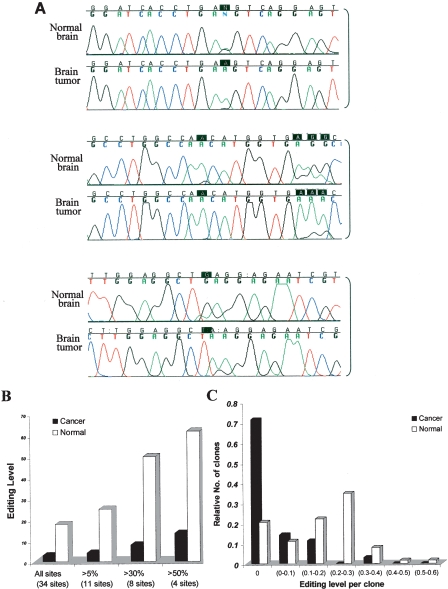
Our bioinformatic analysis suggests that Alu sequences in brain tumors undergo less editing compared to normal brains. In order to experimentally study how this trend is manifested in specific targets, Alu repeats within the gene encoding for the thyroid hormone receptor associated protein 1 (MED13, previously known as THRAP1) were analyzed. MED13 interacts with the thyroid receptor in activating transcription. Editing within this gene occurs in clusters located in Alu repeats. Specifically, we tested the Alu sequence located in intron 9 of the gene, by direct sequencing of five normal and seven cancerous (six glioblastoma multiforme [GBM], one astrocytoma) samples. All the tested sites in the normal samples showed higher averaged editing levels relative to the tumor tissues (Supplemental Data Set S4). In order to assess the accuracy of the editing quantification through direct sequencing, and to further characterize the changes in editing patterns between the tissues, we cloned the PCR product of one normal sample and one cancerous sample and sequenced multiple individual transcripts. We obtained 46 cDNA clones of this Alu sequence from astrocytoma tissue and 75 cDNA clones from normal brain tissue. The results are summarized in Figure 1B and agree well with the editing levels estimated by direct sequencing.
Figure 1.
Reduced editing levels at Alu sequences in a human brain tumor. (A) Three representative clusters from the direct sequencing of an Alu sequence located in intron 9 of the MED13 gene of normal and tumor brain (low-grade glioma) tissues are illustrated. Black boxes indicate the editing sites. (B) Alu editing in individually cloned transcripts from normal and malignant brain tissues. PCR products derived from the normal and astrocytoma samples (shown in panel A) were cloned and sequenced. One-hundred-twenty-one clones were sequenced (46 clones from astrocytoma and 75 clones from normal brain tissue). Thirty-four “As” were shown to be edited at least in one of the clones. The percent of editing was analyzed in all sites and is displayed according to cutoff values of >5%, 30%, and 50% editing. White bars represent transcripts from normal brain tissue, and black bars represent clones from brain tumor. (C) Analysis of editing level of Alu sequences in intron 9 of MED13 in individual clones. MED13 transcripts from both normal and malignant brain samples were cloned and sequenced (same samples as in panel A); 71% of the cancer clones, compared to only 21% of the normal clones, do not undergo editing at any site.
There are 48 adenosines in the examined genomic segment at chromosome 17:57,437,140–57,437,328 (all genomic coordinates follow UCSC genome browser coordinates of human genome, March 2006). Alignment of cloned cDNA sequences, representing individual transcripts, identified 34 adenosines that are read as guanosines at least once. The relative level of editing of specific sites within the sequences obtained from normal tissues correlated with that in malignant tissues: the most edited sites in normal brain were also the most edited sites in cancer. However, the overall number of editing events was significantly higher in the normal tissues (Fig. 1; and Supplemental Data Set S1–3). In total, normal tissue had 2009 “A” and 436 “G” (17.8%), while in the tumor we found 1396 “A” and only 42 “G” (2.9%) (χ2 = 186.5; P < 10−42). In about one-third (11 sites) of the editing sites in the normal sample, we found only a modest level of editing (<5% editing level) that may result from residual activity of the ADAR enzymes. In the cancer tissue, however, 30 out of 34 editing sites showed <5% editing level. A similar ratio was found at sites where the editing level is >5%: in normal tissues there were 1255 “A” and 416 “G” (24.9%), while in cancer we found 936 “A” and only 40 “G” (4.1%) (χ2 = 186.8; P < 10−42). Four sites with an editing level >50% were identified: in normal tissue there were 113 “A” and 183 “G” (61.8%) in these sites, while in cancer cells there were 148 “A” and 22 “G” (13.5%) (χ2 = 104.7; P < 10−24).
The total number of edited sites per individual MED13 Alu clone also differed significantly between tumors and normal brain tissue (Fig. 1C). For example, the majority of the tumor-derived clones (25 out the 35, 71%) were not edited at any of the tested sites, while in the clones derived from the normal brain, only 21% (13 out the 63) were not edited at all. Of the tumor clones, 97% (34/35) were edited at les than seven sites, while only 54% (34/63) of the normal clones were edited at that low level.
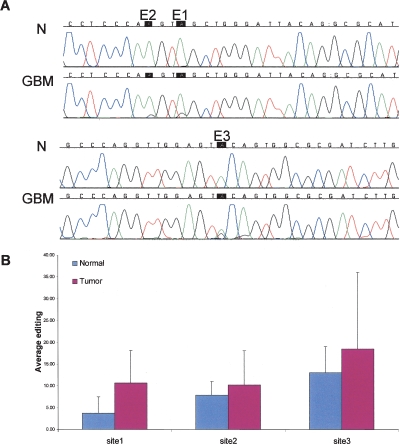
In addition to MED13, which was selected randomly from the edited targets, we repeated the analysis for three high-level (>10%) editing sites within the second intron of BRCA1, a cancer-related gene (chr17:38,523,170, chr17:38,523,173, and chr17:38,523,246). Figure 2 presents the average editing levels at these sites in normal compared to cancerous brain samples. A significantly elevated editing level was found in tumors (2.9-, 1.3-, and 1.4-fold higher editing in cancer tissues; P-values < 3 × 10−2, 4.2 × 10−2, and 3.75 × 10−2 for sites 1, 2, and 3, respectively, using MW test).
Figure 2.
Elevated editing levels in BRCA1 transcript in brain tumor vs. normal tissues; editing sites in an Alu element in the second intron of the BRCA1 gene. (A) Direct sequencing of the BRCA1 transcripts from representative samples taken from normal (N) and glioblastoma multiforme (GBM) brain tissues. A-to-I editing is detected as guanosine trace and indicated by a black box. (B) Average editing levels in BRCA1 transcripts in normal (n = 9) and brain tumor (n = 29) tissues. Editing levels at site 1 are 2.9-fold higher (P < 0.04) in the cancerous tissues relative to the normal control (10.7% vs. 3.7%). Site 2 shows 1.3-fold higher editing efficiency (10.2% editing in brain tumors compared to 7.9% in the normal brains; P < 0.04). Similarly, site 3 presents 1.4-fold elevated editing level in the tumoral tissue (18.5% compared to 13.0%; P = of 0.04).
Bioinformatic analysis of RNA sequences reveals reduced A-to-I editing levels in various human cancers
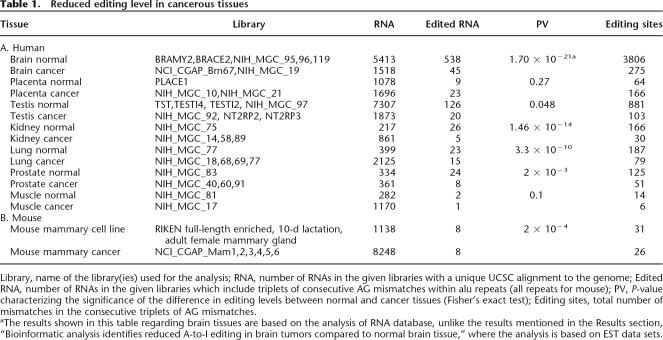
In order to test whether the reduced level of editing in tumors occurs in other tissues as well, we compared the editing level in additional types of malignancy using RNA sequence data, following the method of Neeman et al. (2006). A significantly lower editing level was observed in prostate, lung, kidney and testis tumors when compared to their normal counterparts (P-values of 2 × 10−3, 3.3 × 10−10, 1.46 × 10−14, and 4.8 × 10−2, respectively, using Fisher’s exact test), while no significant difference in editing levels were found when placenta and muscle tissues were analyzed (see Table 1A and Supplemental Material for more details). Additional tissues were not tested due to the limited number of available annotated RNAs coming from both normal and cancer tissues. While the above analysis suggests that the reduced editing level in cancer is not limited to brain tumors, the effect seems to be most significant in the brain.
Table 1.
Reduced editing level in cancerous tissues
Library, name of the library(ies) used for the analysis; RNA, number of RNAs in the given libraries with a unique UCSC alignment to the genome; Edited RNA, number of RNAs in the given libraries which include triplets of consecutive AG mismatches within alu repeats (all repeats for mouse); PV, P-value characterizing the significance of the difference in editing levels between normal and cancer tissues (Fisher’s exact test); Editing sites, total number of mismatches in the consecutive triplets of AG mismatches.
aThe results shown in this table regarding brain tissues are based on the analysis of RNA database, unlike the results mentioned in the Results section, “Bioinformatic analysis identifies reduced A-to-I editing in brain tumors compared to normal brain tissue,” where the analysis is based on EST data sets.
Lower editing levels in mice cancer tissues
Mouse has a much lower editing level than human (Eisenberg et al. 2005). Consequently, only tissues with very large libraries and sufficient numbers of RNAs allow for similar analysis. In addition, in contrast with human libraries that are dominated by cancer tissues, most mouse RNAs come from normal tissues. Yet, we found a satisfactory number of normal and cancer mammary gland RNAs available to allow for the above comparison. Here again, the editing level was significantly lower in the cancer tissue (P = 2 × 10−4). These results support our findings regarding human cancer and extend previous reports (Kim et al. 2004).
Comparison of editing in coding regions of malignant and normal tissues
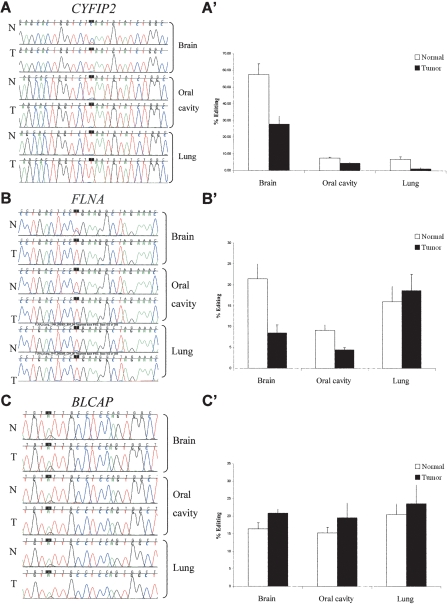
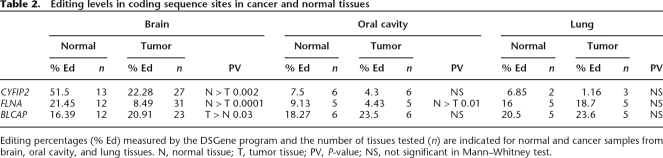
While editing is found predominantly in noncoding regions, editing sites are also found in coding sequences, where they are expected to alter the gene product. Previous bioinformatic analysis identified editing sites in an evolutionarily conserved region in the coding sequence of the CYFIP2, FLNA, and BLCAP transcripts (Levanon et al. 2005a). The editing levels of these sites were analyzed in three different types of human tumors and their normal counterparts: brain, oral cavity, and lung.
CYFIP2 (cytoplasmic FMR1 interacting protein 2) is an evolutionarily conserved gene, found in both invertebrates and vertebrates, encoding for a 1253-amino-acid protein in humans. CYFIP2 interacts with FMR1 and the FMR-related proteins, FXR1/2 (Schenck et al. 2003), and is expressed predominantly in brain tissues, white blood cells, and kidneys (Su et al. 2004). CYFIP2 transcripts undergo A→I editing at chromosome 5:156,669,386, resulting in K→E substitution at amino acid 320 (Levanon et al. 2005a). Direct sequencing analysis of the CYFIP2 editing site in 13 normal and 27 tumor samples revealed an average editing level of 52% in normal brain tissues, whereas only 22% of the sequences were edited in brain tumors (0.42-fold change, P < 2 × 10−3 by MW test) (Fig. 3A, A′; Table 2). Again, the direct sequencing results were verified by sequencing of multiple individual clones of one normal and one tumor sample. Six of 12 clones derived from normal sample exhibited “A” and six showed “G” at position chr5:156,669,386 (50%, consistent with the 56% editing found by direct sequencing of this specific sample). Only one out of 19 clones read “G” in the tumor sample (5%, in good correlation with direct sequencing data of 7% in the specific tumor tested).
Figure 3.
Site-specific editing at coding sequences in normal vs. tumor tissues; direct sequencing of the CYFIP2, FLNA, and BLCAP genes (A–C) from representative samples taken from normal and cancer tissues (brain, oral cavity, and lung). Editing position is indicated by a black box. A-to-I editing is detected as guanosine trace when products were sequenced with the forward primer and as cytidine trace when reverse primer was used for sequencing. Paired normal and tumor tissues from lung and oral cavity were taken from the same individual. (A′–C′) Average editing levels of CYFIP2, FLNA, and BLCAP in normal and cancer tissues. Normal and tumor samples are indicated by white and black bars, respectively. Error bars represent SEM (A–A′) CYFIP2 shows the highest editing percentages in brain tissues, with pronounced 2.4-fold higher editing levels in normal vs. tumor brains (51.5% [n = 13] and 22.3% [n = 27], respectively; P = 0.002). (B–B′) Highest editing percentages of FLNA were detected in the brain tissue, with a significant 2.4-fold higher editing in normal vs. tumor brains (21.4% [n = 13] and 8.8% [n = 31], respectively; P = 0.0001). (C–C′) Minor increase in editing level of BLCAP is detected in different types of tumors, with 1.3-fold (P = 0.03) in the brain (20.9% [n = 24] and 16.4% [n = 12], respectively; P = 0.03).
Table 2.
Editing levels in coding sequence sites in cancer and normal tissues
Editing percentages (% Ed) measured by the DSGene program and the number of tissues tested (n) are indicated for normal and cancer samples from brain, oral cavity, and lung tissues. N, normal tissue; T, tumor tissue; PV, P-value; NS, not significant in Mann–Whitney test.
The editing ratio was also tested in malignant and normal lung and oral cavity tissues (Table 2). In all these cases, the normal tissue was obtained from a distal, non-involved region in the same individual from whom the tumor was taken. The overall level of editing of CYFIP2 was significantly lower in these tissues, compared to the brain tissues. The average editing percentage in six oral cavity tumor samples was 4.3% compared to 7.5% in six normal controls. In lung cancer samples, the average editing percentage was 1.16% in tumors (three samples) compared to 6.85% in two normal samples. Due to the small number of oral cavity and lung samples, these differences are not statistically significant; however, the trend obtained is similar to that found in the brain tissues.
FLNA (filamin A, alpha [actin binding protein 280]) is a 2647-amino-acid ubiquitously expressed protein. It functions in F-actin, cross-linking in the cytoplasm and in anchoring membrane proteins to the cytoskeletal elements. In our previous work we identified a conserved A→I editing site in the FLNA transcript (chromosome X:153,233,144), resulting in a Q→R substitution at amino acid 2341 in the human protein (Levanon et al. 2005a). We tested this editing site by direct sequencing of normal and brain tumor samples (Fig. 3B, B′; Table 2). Normal brain samples displayed an average of 21.5% editing, while in the malignant samples, an average of 8.5% editing was found (0.39-fold change, P = 10−4, MW test).
FLNA editing frequency was also tested in paired tumor and normal tissues from lung and oral cavity. On average, the editing incidence was twofold higher in normal oral cavity tissues than in the tumor samples (9.13% and 4.43%, respectively, P = 0.01, MW test). In paired lung samples, the editing levels were similar in normal and cancer tissues (see Table 2). Again, the reliability and sensitivity of the direct sequencing methodology was validated by cloning FLNA transcripts from a single pair of malignant and normal oral cavity tissues of the same individual. Seven out of 99 clones from the normal sample exhibited “G” at the editing site (7.1%; consistent with the 8.5% editing found by direct sequencing of this specific sample), and only three out of 92 clones read “G” in the oral cavity tumor sample (3.3%, consistent with the direct sequencing data of 5.6% in this tumoral tissue). The agreement of the results obtained by direct sequencing with the multiple individual clone analysis suggests that, for large-scale analyses of editing, direct sequencing can be confidently used even for measuring low editing ratios.
BLCAP (bladder cancer-associated protein) encodes an 87-amino-acid protein, expressed predominantly in the brain and B lymphocytes (Su et al. 2004). Few editing sites were identified in the BLCAP transcript. These occur in a tight cluster, containing several A→I substitutions (most of them in the intron and three in evolutionarily conserved sites in the coding sequence). Here we focused on the A-to-I editing event (Clutterbuck et al. 2005; Levanon et al. 2005a) in the nucleotide encoding the second codon, located at chromosome 20:35,580,986, resulting in a Y→C substitution. The mean editing level at this site in normal tissues was 16%, while tumor specimens had an average value of 21%, representing a marginal 1.27-fold higher editing level (P = 0.03, MW test) in the tumor tissues (Fig. 3C, C′ Table 2). Lung and oral cavity samples have shown no significant difference in editing levels between normal and cancer tissues (Table 2). Thus, minor elevation of editing in this site was noted in malignant tissues of the three sources (1.27-, 1.15-, and 1.28-fold in brain, lung, and oral cavity, respectively, only the first being statistically significant).
Reduced expression of ADAR enzymes in human brain tumors
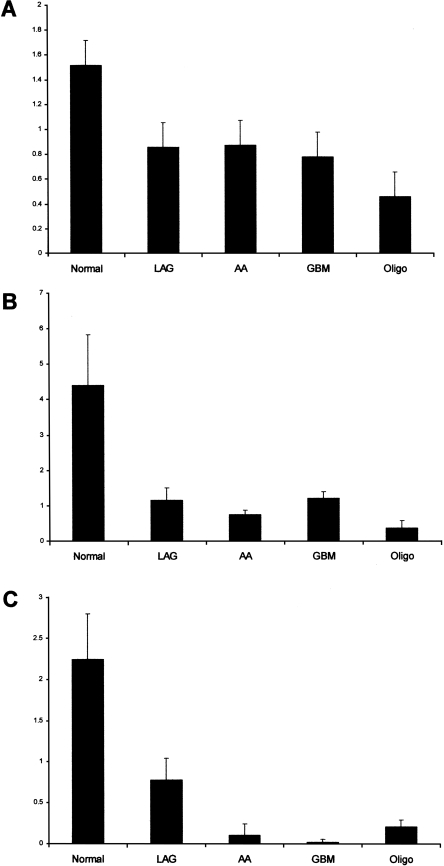
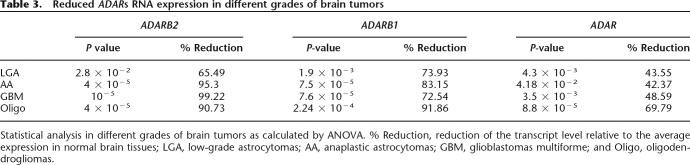
To investigate the mechanism responsible for the differences in editing levels between the normal and malignant brain tissues, we assessed the expression level of the editing enzymes. Using quantitative real-time PCR we found elevated transcript levels of ADAR, ADARB1, and ADARB2 in normal brain tissues compared to brain tumors representing different grades of gliomas. Analysis of 12 normal brain samples and 18 brain tumors revealed global reduction in ADAR and ADARB1 transcripts (Supplemental Fig. S2). Following this result, we further checked the ADARs expression in a larger cohort of well-characterized brain tumors of different histopathologies, including low-grade glioma (LGA), anaplastic astrocytoma (AA), glioblastoma multiforme (GBM), and oligodendroglioma (Oligo). Figure 4 summarizes the RNA expression levels in each tumor relative to normal brain samples. Our results demonstrate a prominent overall reduction in ADAR, ADARB1, and ADARB2 RNA levels in brain tumors compared to normal brain tissues (Fig. 4; Table 3). ADARB2 RNA levels were significantly reduced in direct correlation with the degree of malignancy of the astrocytic tumors: the less aggressive LGA displayed a 65% reduction relative to normal brain while the more aggressive AA displayed a 95.3% reduction and the most aggressive GBM displayed a 99.2% reduction.
Figure 4.
Reduced RNA level of the ADAR family is detected in human brain tumors. Real-time PCR analysis was performed in order to quantify the relative RNA levels of ADAR, ADARB1, and ADARB2 in normal and in different grades and types (according to WHO classification) of brain tumors. Values were normalized using S12, and the ΔΔCt method was applied. The y axis represents the relative ADARs expression level of (A) ADAR, (B) ADARB1, and (C) ADARB2. Error bars represent SEM. LGA, low-grade astrocytomas; AA, anaplastic astrocytomas; GBM, glioblastomas multiforme; Oligo, oligodendrogliomas.
Table 3.
Reduced ADARs RNA expression in different grades of brain tumors
Statistical analysis in different grades of brain tumors as calculated by ANOVA. % Reduction, reduction of the transcript level relative to the average expression in normal brain tissues; LGA, low-grade astrocytomas; AA, anaplastic astrocytomas; GBM, glioblastomas multiforme; and Oligo, oligodendrogliomas.
Comparison of ADARB2 transcript levels between the different grades revealed significant and gradual reduction between LGA and AA (P = 10−6), GBM and AA (P = 0.03). The most pronounced difference was seen in comparison of LGA with GBM (P = 5 × 10−5). A significant difference was also evident between GBM and Oligo (P = 6 × 10−3). Oligodendrogliomas displayed a significant reduction of 70%–90% in the RNA level of all three ADAR family members (see Table 3).
No consistent correlation found between editing efficiency and transcript level
Higher or lower editing may result from elevated transcription levels of the editing targets. Alternatively, editing efficiency may affect transcript stability. We thus looked for a possible correlation between editing levels and RNA expression, comparing the editing levels (detected by direct sequencing) and the RNA expression levels (determined by quantitative real-time PCR). Supplemental Figure S1 summarizes the results of average fold of editing and average fold of RNA expression levels in CYFIP2, FLNA, and BLCAP transcripts in normal brain tissues versus brain tumors. The results, as detailed in the legend to Supplemental Figure S1, were not consistent with a simple correlation between editing and expression levels.
Overexpression of ADAR and ADARB1 results in decreased proliferation rate
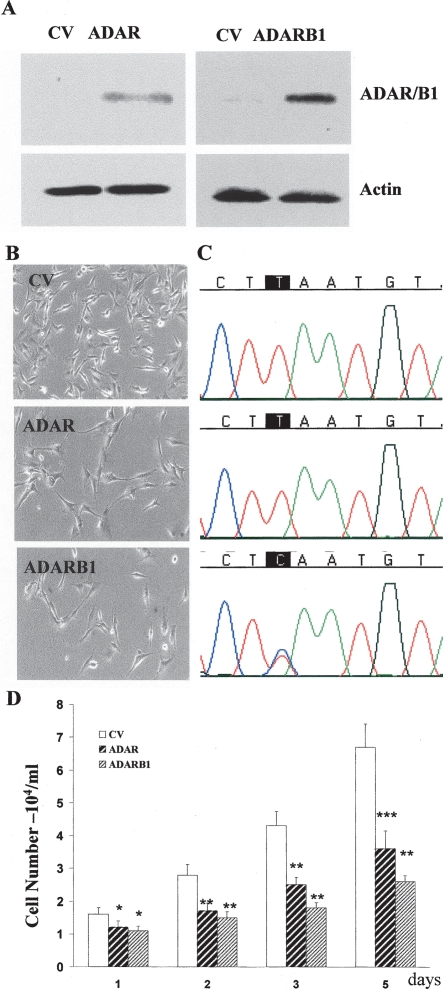
Gliomas and glioma cell lines express very low levels of the ADAR proteins. To examine the role of the ADAR proteins in glioma cells, we overexpressed ADAR and ADARB1 in the U87 glioblastoma multiforme cell line, which expresses low levels of these proteins. For these experiments, cells were transiently transfected with control vector (CV), ADAR, and ADARB1, and the morphology and cell growth were then determined. As demonstrated in Figure 5A, transfection of the cells with the different ADAR expression vectors resulted in high levels of both ADAR and ADARB1 as compared to CV-transfected cells. The elevated level of ADAR and ADARB1 was accompanied by significant alteration of editing levels in 72 sites in six different genes representing both coding and noncoding regions (Fig. 5C; Supplemental Table 1). Interestingly, U87 cells overexpressing ADAR and ADARB1 exhibited significantly lower rates of cell proliferation (Fig. 5D) and different morphologies, with larger cell bodies as compared to CV cells (Fig. 5B).
Figure 5.
Overexpression of ADAR and ADARB1 decreases glioma cell proliferation. U87 cells were transfected with ADAR, ADARB1, and a control empty expression vector (CV). After 24 h, expression of ADAR proteins was examined using Western blot analysis (A). Cells were plated (5 × 104/mL) on plastic dishes. Morphology of the cells was assessed after 24 h (B) using a phase-contrast microscope (original magnification, ×20). Direct sequencing of CYFIP2 editing site in U87 transfected cells (C). Cell number was monitored at 24-h intervals (D). Results represent the means ± SE of three independent experiments. Asterisks represent P values: *P < 0.01, **P < 10−3, ***P < 2 × 10−3.
Discussion
Using both bioinformatic and experimental approaches, we found that the A-to-I editing pattern is significantly modified in cancer compared to normal tissues. The first signs for this phenomenon were given by Mass et al., who showed that the glutamate receptor (ionotropic, AMPA 2 [GRIA2]), virtually 100% edited in normal samples, was hypoedited in a small number of glioblastoma multiforme (69%–88% editing) and low-grade astrocytoma (90% editing) samples (Maas et al. 2001). In the present work, we examined the editing level in 31 different human brain tumors and evaluated editing levels in three coding sequence targets (CYFIP2, BLCAP, and FLNA) and two clusters of noncoding editing sites. We found that the extent of both global noncoding clustered editing and gene-specific coding and noncoding regions editing differ between normal and cancer tissues. The most significant differences in editing between normal and malignant tissues were noted in brain samples, known from previous studies to be mostly affected by the editing modification. Alterations of editing levels were also demonstrated when other normal and cancer tissues were studied. The observation applies also to the mice tissues we screened, strengthening the significance of our findings.
In addition, we also demonstrated that RNA levels of the three ADAR family members (ADAR, ADARB1, and ADARB2) are significantly reduced in 106 brain tumors. The most impressive reduction of expression was found in ADARB2, for which the level of expression was shown to correlate with the tumor grade. Notably, 99% reduction in ADARB2 expression level was detected in the most aggressive type of brain tumors, glioblastoma multiforme. Expression of ADAR, ADARB1, and ADARB2 in oligodendrogliomas was reduced by 69%, 92%, and 91%, respectively. Reduction of ADARB1 activity was previously reported in tumor samples vs. normal human brain tissue (Maas et al. 2001). No direct editing activity was shown for this protein; however, its binding activity to both dsRNA and ssRNA was demonstrated (Chen et al. 2000). Possibly, the effect of the reduced expression of the ADARs is not limited to editing but extends to other activities such as inhibition of the kinase activity of the dsRNA-activated protein kinase PKR, and suppression of eIF2a phosphorylation by ADAR through a mechanism independent of dsRNA editing (Nie et al. 2006). Rather than acting as adenosine deaminase, ADARB2 could have other essential roles associated with its ssRNAs and dsRNA binding modules. It could also compete with the other editing enzymes or with other dsRNA-binding protein.
Diagnostic accuracy and reproducibility of gliomas are compromised by the subjective histologic criteria currently used to classify and grade brain tumors and by high inter-observer discrepancy (Coons et al. 1997). Our findings suggest that the analysis of ADARs levels and activity, in particular of ADARB2, may provide an efficient diagnostic tool for a better classification of brain tumors. ADARB2 is the least-studied editing enzyme.
The functional connection between carcinogenesis and RNA editing is yet to be discovered. It is not clear yet whether decreased ADAR expression/activity can serve as a cause for cancer, or just a consequence of it. However, our ADAR and ADARB1 overexpression experiments, where inhibition of GBM proliferation was correlated with increased enzyme expression, provide tantalizing hints for a possible role of altered editing in the malignant process. It can be suggested that the level of gene-specific editing is regulated by a selection process during cancer evolution and progression as editing may affect RNA level, RNA localization, alternative splicing, translation efficiency, and protein structure and function. We have previously shown that the vast majority of A-to-I editing events are expected to modify the predicted dsRNA stem and loop stability (Levanon et al. 2004). Therefore, editing is expected to affect all mechanisms depending on the dsRNA structure, such as RNA interference and miRNA activity. Indeed, it was shown that RNA editing and RNA interference are involved in the same pathway in C. elegans (Tonkin and Bass 2003) and that the biogenesis of miRNA is regulated by editing, with several miRNAs shown to be targets for editing (Luciano et al. 2004; Blow et al. 2006; Yang et al. 2006). Furthermore, a recent bioinformatic work predicts that Alu sequences are potential targets for a group of miRNAs (Smalheiser and Torvik 2006). Recently miRNAs were shown to be involved in carcinogenesis acting as both oncogenes and tumor suppressor genes (Lu et al. 2005). Thus, it can be speculated that alterations of RNA editing may be involved in cancer development and evolution by modulation of RNA interference and miRNA activities. Finally, a role for RNA editing in determining heterochromatin formation and gene silencing was shown and can also contribute to the regulation of expression of genes involved in the malignant process.
It is tempting to speculate that some of the editing alterations in the specific genes analyzed may be relevant to the malignant phenotype. This assumption is enforced by the facts connecting each of the tested genes to cancer: (1) BLCAP was first identified in a screen for genes contributing to the invasive phenotype of bladder cancer (Rae et al. 2000; Gromova et al. 2002). Later it was suggested as a novel tumor suppressor gene due to the observation that its overexpression resulted in growth inhibition, S phase arrest, and apoptosis (Yao et al. 2007). Additional work points to a correlation between BLCAP expression and aggressiveness of osteosarcoma (Su et al. 2003). (2) The well-characterized tumor suppressor gene BRCA1 appears to have elevated editing levels in specific sites within an Alu sequence. BRCA1 was shown to be important for DNA repair, cell cycle progression, ubiquitylation, and transcriptional regulation (Welcsh and King 2001; Boulton 2006). (3) The MED13 gene is located in a genomic cluster (17q23) known to be amplified in breast cancer. It was reported to be overexpressed in a breast cancer cell line as well as in primary breast tumors (Monni et al. 2001). (4) CYFIP2 is down-regulated by the tumor suppressor gene TP53 and was identified recently as a prognostic marker for some tumors, such as sarcoma and leukemia (Ceballos et al. 2005). (5) FLNA was reported to act as a transcriptional inhibitor of the androgen receptor, thus responsible for down-regulating the increased activity of the androgen receptor, associated with prostate cancer (Loy et al. 2003).
The results presented here reveal that global hypoediting in repetitive elements in cancer tissues is accompanied by regulation of site-specific editing in coding and noncoding sequences that seems to function in a gene-specific manner. Editing levels are elevated (like BLCAP and BRCA1) or reduced (e.g., FLNA and CYFIP2) in tumors, possibly depending on the growth and survival advantages provided by the specific gene product. This emerging pattern of global tumor hypoediting of repetitive sequences and gene-specific regulation in the coding sequence parallels the well-studied epigenetic alteration of DNA methylation in cancer. Generally, tumors are characterized by widespread, global hypomethylation, mainly of repetitive elements. However, hypermethylation of CpG islands of a number of “housekeeping” genes and tumor suppressor genes and hypermethylation of promoters of genes conferring growth advantage, such as oncogenes, occur in a gene-specific manner in many tumors (Sardi et al. 1997; Cheng et al. 2005; Feinberg et al. 2006). Increased expression of oncogenes and decreased expression of tumor suppressor genes may be mediated by RNA editing. Both DNA methylation and RNA editing may therefore contribute to tumor evolution in similar, complementary ways.
Similar to our findings regarding altered A-to-I editing in cancer, other types of nucleic acid editing, namely, conversion of cytidine to uridine in RNA by APOBEC1 (apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1) and DNA editing by the AICDA enzyme (activation-induced cytidine deaminase), were shown in animal models to be associated with protection from hepatocellular tumors and retrovirally induced leukemia respectively (Yamanaka et al. 1995; Gourzi et al. 2006). Taken together, the modification of RNA and DNA by editing enzymes is unraveled as an important epigenetic control mechanism that is perturbed in cancer.
In summary, we have shown that RNA editing pattern, as well as expression level of the RNA editing enzymes, is globally modified in tumors. Overexpression experiments have demonstrated that these changes lead to increased cell proliferation, suggesting a possible role for altered editing in tumorigenesis. Taken together, these findings seem to establish a link between the RNA editing and cancer and call for more experimental work trying to reveal the nature of this relation. Analysis of many editing sites in various cancer types is expected to provide new diagnostic and prognostic markers and might contribute to early detection of cancer, to the monitoring of response to therapy, and to the detection of minimal residual disease.
Methods
Tumor and control tissues
Human brain, lung, and oral cavity tissues were frozen immediately after their removal at surgery and kept at −70°C until further use. One-hundred-nineteen human brain tumors (including low-grade astrocytomas [LGA], anaplastic astrocytomas [AA], glioblastomas [GBM], and oligodendrogliomas [Oligo]) and 20 normal brain samples were obtained from M.D. Anderson Cancer Center, Tel Aviv Sourasky Medical Center, or Henry Ford Hospital (Detroit, MI), or were purchased (Stratagene, Clontech, and Biochain).
Malignant and normal samples from oral cavity (six subjects) or lung (five subjects) were collected at the Chaim Sheba Medical Center. All studies were carried out in accordance with the IRB of the respective institutes.
The number of samples tested in each assay differed due to the limited amount of RNA that was available.
RNA purification and reverse transcription (RT)
Total RNA was isolated using TRIzol reagent (Invitrogen) or the RNeasy Midi kit (QIAGEN) according to the manufacturer’s instructions. Random-primed cDNA synthesis was done on 2 μg of total RNA using M-MLV reverse transcriptase (Invitrogen) according to manufacturer’s instructions
Polymerase chain reaction (PCR), gel extraction, and sequencing
The amplified regions were selected to be specific and unique. PCR amplification used Ready Mix PCR Master Mix (ABgene). The primer sequences are listed in Supplemental Table S2. All PCR reactions were carried out at an annealing temperature of 60°C. We aimed for high sequence quality and thus amplified short (∼200 bp) sequences. The resulting PCR-fragments were purified using QiaQuick gel extraction kit (QIAGEN) cloned into pGEM-T Easy (Promega) and sequenced using ABI Prism 3100 genetic analyzer (Applied Biosystems).
Analysis of RNA editing
The level of editing, assessed by direct sequencing, was quantified by the Discovery Studio Gene (DSGene) 1.5 program (Accelrys Inc.).
The reliability of this method was further verified by cloning of individual sequences. PCR products were ligated into pGem-T Easy (Promega). After transformation of Escherichia coli, DNA was extracted using QIAprep Spin (QIAGEN), and individual plasmids were sequenced. The percentage of the edited clones was determined and compared to the DSGene quantification. This approach confirmed that the quantification was reliable even at low editing ratios of ∼5%, representing the transcript repertoire and not background noise.
Real-time quantitative PCR analysis
RQ-PCR was performed in order to determine the levels of mRNA expression of ADAR, ADARB1, ADARB2, CYFIP2, FLNA, and BLCAP. Primers (Sigma-Aldrich) were designed according to Primer-Express software guidelines (Applied Biosystems). Forward and reverse primers were designed from different exons to eliminate possible DNA contamination. The RQ-PCR reactions were run on ABI 7900HT or ABI Prism 7000 sequence detection systems utilizing SDS 2.1 Software (Applied Biosystems). All reactions were run in duplicate or triplicate. Transcripts were detected using SYBR Green I PCR Master Mix (Applied Biosystems) or 2× SYBR Green Master Mix (Invitrogen) according to manufacturer’s instructions and were normalized to beta-actin or S12. Reactions were done in a total volume of 20 μL or 50 μL, containing cDNA equivalent to 30–100 ng of RNA from each sample. The PCR conditions consisted of 4 min at 95°C and 40 cycles of 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min.
To statistically compare between different experiments we used the ΔΔCt method. Fold of changes from normal samples were calculated using the formula X = 2−ΔΔCt. ΔCt represents the difference between the Ct values of the tested gene and the reference gene, beta-actin, or S12. ΔΔCt is the difference between the ΔCt value of the samples for each target and the mean ΔCt of the calibrator.
Cell transfection
U87 cells were transfected either with the control vector (CV) or with the different ADAR expression vectors by electroporation using the Nucleofector device, protocol number A29 (Amaxa Biosystems). Transfection efficiency using nucleofection is between 70% and 90%.
Quantitation of cell growth
Cells (2 × 104) were seeded in triplicate in tissue culture dishes and were harvested at 24-h intervals using trypsin. Cells were washed in PBS and were then counted using a phase microscope. Cell numbers were then calculated per milliliter of the original dilution. Each assay was performed in quadruplets.
Western blot analysis
Cells pellets (106 cells/mL) were resuspended in 100 μL of lysis buffer (25 mM Tris-HCl at pH 7.4, 50 mM NaCl, 0.5% Na deoxycholate; 2% NP-40; 0.2% SDS; 1 mM PMSF; 50 μg/mL aprotinin; 50 μM leupeptin; 0.5 mM Na3VO4) on ice for 15 min; 2× sample buffer was then added, and the samples were boiled for 5 min.
Lysates (30 μg of protein) were resolved by SDS-PAGE and were transferred to nitrocellulose membranes. Following blocking with 5% dry milk in phosphate-buffered saline (PBS), the membranes were stained with the respective anti-ADAR antibody or with anti-actin as an equal loading control. Polyclonal anti-ADAR (176) and anti-ADARB1 (H-90) antibodies were obtained from Santa Cruz Biotechnology. ADAR (variant 1) and ADARB1 expression vectors were obtained from Origene. Specific reactive bands were detected using a goat anti-rabbit or goat anti-mouse IgG conjugated to horseradish peroxidase (Bio-Rad), and the immunoreactive bands were visualized by the ECL Western blotting detection kit (Amersham).
Acknowledgments
We thank S. Nemzer and M. Olshansky for their help and Y. Caspi for critical reading of the manuscript. We also thank the Kahn Family Foundation for supporting our research. This research was supported by the Paamei Tikva Fund, the Flight Attendants Medical Research Institute (FAMRI), and the Israel Cancer Association. G.R. holds the Djerassi Chair in Oncology at the Sackler Faculty of Medicine, Tel Aviv University, Israel. E.E. is supported by an Alon fellowship at Tel Aviv University. E.Y.L is supported by a Rothschild Fellowship. The work of N.P. and Z.S.B. was done in partial fulfillment of the requirements for a Ph.D. degree from the Sackler Faculty of Medicine, Tel Aviv University, Israel.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6493107
References
- Athanasiadis A., Rich A., Maas S., Rich A., Maas S., Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla K.N. Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies. J. Clin. Oncol. 2005;23:3971–3993. doi: 10.1200/JCO.2005.16.600. [DOI] [PubMed] [Google Scholar]
- Blow M., Futreal P.A., Wooster R., Stratton M.R., Futreal P.A., Wooster R., Stratton M.R., Wooster R., Stratton M.R., Stratton M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M.J., Grocock R.J., van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M.R., Grocock R.J., van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M.R., van Dongen S., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M.R., Enright A.J., Dicks E., Futreal P.A., Wooster R., Stratton M.R., Dicks E., Futreal P.A., Wooster R., Stratton M.R., Futreal P.A., Wooster R., Stratton M.R., Wooster R., Stratton M.R., Stratton M.R. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S.J. BRCA1-mediated ubiquitylation. Cell Cycle. 2006;5:1481–1486. doi: 10.4161/cc.5.14.2930. [DOI] [PubMed] [Google Scholar]
- Brusa R., Zimmermann F., Koh D.S., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R., Zimmermann F., Koh D.S., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R., Koh D.S., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R., Feldmeyer D., Gass P., Seeburg P.H., Sprengel R., Gass P., Seeburg P.H., Sprengel R., Seeburg P.H., Sprengel R., Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Ceballos E., Munoz-Alonso M.J., Berwanger B., Acosta J.C., Hernandez R., Krause M., Hartmann O., Eilers M., Leon J., Munoz-Alonso M.J., Berwanger B., Acosta J.C., Hernandez R., Krause M., Hartmann O., Eilers M., Leon J., Berwanger B., Acosta J.C., Hernandez R., Krause M., Hartmann O., Eilers M., Leon J., Acosta J.C., Hernandez R., Krause M., Hartmann O., Eilers M., Leon J., Hernandez R., Krause M., Hartmann O., Eilers M., Leon J., Krause M., Hartmann O., Eilers M., Leon J., Hartmann O., Eilers M., Leon J., Eilers M., Leon J., Leon J. Inhibitory effect of c-Myc on p53-induced apoptosis in leukemia cells. Microarray analysis reveals defective induction of p53 target genes and upregulation of chaperone genes. Oncogene. 2005;24:4559–4571. doi: 10.1038/sj.onc.1208652. [DOI] [PubMed] [Google Scholar]
- Chen C.X., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K., Cho D.S., Wang Q., Lai F., Carter K.C., Nishikura K., Wang Q., Lai F., Carter K.C., Nishikura K., Lai F., Carter K.C., Nishikura K., Carter K.C., Nishikura K., Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.K., Chow L.W., Loo W.T., Chan T.K., Chan V., Chow L.W., Loo W.T., Chan T.K., Chan V., Loo W.T., Chan T.K., Chan V., Chan T.K., Chan V., Chan V. The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res. 2005;65:8646–8654. doi: 10.1158/0008-5472.CAN-04-4243. [DOI] [PubMed] [Google Scholar]
- Clutterbuck D.R., Leroy A., O’Connell M.A., Semple C.A., Leroy A., O’Connell M.A., Semple C.A., O’Connell M.A., Semple C.A., Semple C.A. A bioinformatic screen for novel A-I RNA editing sites reveals recoding editing in BC10. Bioinformatics. 2005;21:2590–2595. doi: 10.1093/bioinformatics/bti411. [DOI] [PubMed] [Google Scholar]
- Coons S.W., Johnson P.C., Scheithauer B.W., Yates A.J., Pearl D.K., Johnson P.C., Scheithauer B.W., Yates A.J., Pearl D.K., Scheithauer B.W., Yates A.J., Pearl D.K., Yates A.J., Pearl D.K., Pearl D.K. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79:1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Eisenberg E., Nemzer S., Kinar Y., Sorek R., Rechavi G., Levanon E.Y., Nemzer S., Kinar Y., Sorek R., Rechavi G., Levanon E.Y., Kinar Y., Sorek R., Rechavi G., Levanon E.Y., Sorek R., Rechavi G., Levanon E.Y., Rechavi G., Levanon E.Y., Levanon E.Y. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Vogelstein B., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P., Ohlsson R., Henikoff S., Ohlsson R., Henikoff S., Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Fernandez H.R., Kavi H.H., Xie W., Birchler J.A., Kavi H.H., Xie W., Birchler J.A., Xie W., Birchler J.A., Birchler J.A. Heterochromatin: On the ADAR radar? Curr. Biol. 2005;15:R132–R134. doi: 10.1016/j.cub.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Gourzi P., Leonova T., Papavasiliou F.N., Leonova T., Papavasiliou F.N., Papavasiliou F.N. A role for activation-induced cytidine deaminase in the host response against a transforming retrovirus. Immunity. 2006;24:779–786. doi: 10.1016/j.immuni.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Gromova I., Gromov P., Celis J.E., Gromov P., Celis J.E., Celis J.E. bc10: A novel human bladder cancer-associated protein with a conserved genomic structure downregulated in invasive cancer. Int. J. Cancer. 2002;98:539–546. doi: 10.1002/ijc.10244. [DOI] [PubMed] [Google Scholar]
- Gurevich I., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C., Tamir H., Arango V., Dwork A.J., Mann J.J., Schmauss C., Arango V., Dwork A.J., Mann J.J., Schmauss C., Dwork A.J., Mann J.J., Schmauss C., Mann J.J., Schmauss C., Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/s0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- Hartner J.C., Schmittwolf C., Kispert A., Muller A.M., Higuchi M., Seeburg P.H., Schmittwolf C., Kispert A., Muller A.M., Higuchi M., Seeburg P.H., Kispert A., Muller A.M., Higuchi M., Seeburg P.H., Muller A.M., Higuchi M., Seeburg P.H., Higuchi M., Seeburg P.H., Seeburg P.H. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H., Feldmeyer D., Sprengel R., Seeburg P.H., Sprengel R., Seeburg P.H., Seeburg P.H. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S., Sun H., Aizawa H., Kanazawa I., Kwak S., Aizawa H., Kanazawa I., Kwak S., Kanazawa I., Kwak S., Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A., Kobayashi Y., Matise T.C., Buyske S., Gabriel A., Matise T.C., Buyske S., Gabriel A., Buyske S., Gabriel A., Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon E.Y., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Eisenberg E., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Yelin R., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Nemzer S., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Hallegger M., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Shemesh R., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Fligelman Z.Y., Shoshan A., Pollock S.R., Sztybel D., Shoshan A., Pollock S.R., Sztybel D., Pollock S.R., Sztybel D., Sztybel D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Levanon E.Y., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E., Hallegger M., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E., Kinar Y., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E., Shemesh R., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E., Djinovic-Carugo K., Rechavi G., Jantsch M.F., Eisenberg E., Rechavi G., Jantsch M.F., Eisenberg E., Jantsch M.F., Eisenberg E., Eisenberg E. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005a;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon K., Eisenberg E., Rechavi G., Levanon E.Y., Eisenberg E., Rechavi G., Levanon E.Y., Rechavi G., Levanon E.Y., Levanon E.Y. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 2005b;6:831–835. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy C.J., Sim K.S., Yong E.L., Sim K.S., Yong E.L., Yong E.L. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc. Natl. Acad. Sci. 2003;100:4562–4567. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., Ebert B.L., Mak R.H., Ferrando A.A., Mak R.H., Ferrando A.A., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Luciano D.J., Mirsky H., Vendetti N.J., Maas S., Mirsky H., Vendetti N.J., Maas S., Vendetti N.J., Maas S., Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas S., Patt S., Schrey M., Rich A., Patt S., Schrey M., Rich A., Schrey M., Rich A., Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc. Natl. Acad. Sci. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monni O., Barlund M., Mousses S., Kononen J., Sauter G., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M.L., Barlund M., Mousses S., Kononen J., Sauter G., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M.L., Mousses S., Kononen J., Sauter G., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M.L., Kononen J., Sauter G., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M.L., Sauter G., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M.L., Heiskanen M., Paavola P., Avela K., Chen Y., Bittner M.L., Paavola P., Avela K., Chen Y., Bittner M.L., Avela K., Chen Y., Bittner M.L., Chen Y., Bittner M.L., Bittner M.L., et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc. Natl. Acad. Sci. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeman Y., Levanon E.Y., Jantsch M.F., Eisenberg E., Levanon E.Y., Jantsch M.F., Eisenberg E., Jantsch M.F., Eisenberg E., Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y., Hammond G.L., Yang J.H., Hammond G.L., Yang J.H., Yang J.H. Double stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 2006;81:917–923. doi: 10.1128/JVI.01527-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino M.J., Keegan L.P., O’Connell M.A., Reenan R.A., Keegan L.P., O’Connell M.A., Reenan R.A., O’Connell M.A., Reenan R.A., Reenan R.A. A-to-I pre-mRNA editing in Drosophila is primarily involved in adult nervous system function and integrity. Cell. 2000;102:437–449. doi: 10.1016/s0092-8674(00)00049-0. [DOI] [PubMed] [Google Scholar]
- Patterson J.B., Samuel C.E., Samuel C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L., Bennett C.F., Zhang M.Q., Spector D.L., Zhang M.Q., Spector D.L., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Rae F.K., Stephenson S.A., Nicol D.L., Clements J.A., Stephenson S.A., Nicol D.L., Clements J.A., Nicol D.L., Clements J.A., Clements J.A. Novel association of a diverse range of genes with renal cell carcinoma as identified by differential display. Int. J. Cancer. 2000;88:726–732. doi: 10.1002/1097-0215(20001201)88:5<726::aid-ijc7>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Sardi I., Dal Canto M., Bartoletti R., Montali E., Dal Canto M., Bartoletti R., Montali E., Bartoletti R., Montali E., Montali E. Abnormal c-myc oncogene DNA methylation in human bladder cancer: Possible role in tumor progression. Eur. Urol. 1997;31:224–230. doi: 10.1159/000474455. [DOI] [PubMed] [Google Scholar]
- Scadden A.D.J., O’Connell M.A., O’Connell M.A. Cleavage of dsRNAs hyper-edited by ADARs occurs at preferred editing sites. Nucleic Acids Res. 2005;33:5954–5964. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A., Bardoni B., Langmann C., Harden N., Mandel J.L., Giangrande A., Bardoni B., Langmann C., Harden N., Mandel J.L., Giangrande A., Langmann C., Harden N., Mandel J.L., Giangrande A., Harden N., Mandel J.L., Giangrande A., Mandel J.L., Giangrande A., Giangrande A. CYFIP/Sra-1 controls neuronal connectivity in Drosophila and links the Rac1 GTPase pathway to the fragile X protein. Neuron. 2003;38:887–898. doi: 10.1016/s0896-6273(03)00354-4. [DOI] [PubMed] [Google Scholar]
- Smalheiser N.R., Torvik V.I., Torvik V.I. Alu elements within human mRNAs are probable microRNA targets. Trends Genet. 2006;22:532–536. doi: 10.1016/j.tig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Su H.C., Zhao Y.H., Fan D.G., Fan Q.Y., Zhang P., Wen Y.H., Liu Y.Y., Zhao Y.H., Fan D.G., Fan Q.Y., Zhang P., Wen Y.H., Liu Y.Y., Fan D.G., Fan Q.Y., Zhang P., Wen Y.H., Liu Y.Y., Fan Q.Y., Zhang P., Wen Y.H., Liu Y.Y., Zhang P., Wen Y.H., Liu Y.Y., Wen Y.H., Liu Y.Y., Liu Y.Y. Relationship between expression of BLCAP protein and malignancy of osteosarcoma. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2003;19:465–466. [PubMed] [Google Scholar]
- Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Batalov S., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Lapp H., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Ching K.A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Zhang J., Soden R., Hayakawa M., Kreiman G., Soden R., Hayakawa M., Kreiman G., Hayakawa M., Kreiman G., Kreiman G., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin L.A., Bass B.L., Bass B.L. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Khillan J., Gadue P., Nishikura K., Khillan J., Gadue P., Nishikura K., Gadue P., Nishikura K., Nishikura K. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- Welcsh P.L., King M.C., King M.C. BRCA1 and BRCA2 and the genetics of breast and ovarian cancer. Hum. Mol. Genet. 2001;10:705–713. doi: 10.1093/hmg/10.7.705. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Balestra M.E., Ferrell L.D., Fan J., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L., Balestra M.E., Ferrell L.D., Fan J., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L., Ferrell L.D., Fan J., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L., Fan J., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L., Arnold K.S., Taylor S., Taylor J.M., Innerarity T.L., Taylor S., Taylor J.M., Innerarity T.L., Taylor J.M., Innerarity T.L., Innerarity T.L. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc. Natl. Acad. Sci. 1995;92:8483–8487. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K., Seeburg P.H., Shiekhattar R., Nishikura K., Shiekhattar R., Nishikura K., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J., Duan L., Fan M., Yuan J., Wu X., Duan L., Fan M., Yuan J., Wu X., Fan M., Yuan J., Wu X., Yuan J., Wu X., Wu X. Overexpression of BLCAP induces S phase arrest and apoptosis independent of p53 and NF-kappaB in human tongue carcinoma: BLCAP overexpression induces S phase arrest and apoptosis. Mol. Cell. Biochem. 2007;297:81–92. doi: 10.1007/s11010-006-9332-2. [DOI] [PubMed] [Google Scholar]