Abstract
Bacterial trans translation is activated when translating ribosomes are unable to elongate or terminate properly. Small protein B (SmpB) and transfer-messenger RNA (tmRNA) are the two known factors required for and dedicated to trans translation. tmRNA, encoded by the ssrA gene, is a bifunctional molecule that acts both as a tRNA and as an mRNA during trans translation. The functions of tmRNA ensure that stalled ribosomes are rescued, the causative defective mRNAs are degraded, and the incomplete polypeptides are marked for targeted proteolysis. We present in vivo and in vitro evidence that demonstrates a direct role for the Lon ATP-dependent protease in the degradation of tmRNA-tagged proteins. In an endogenous protein tagging assay, lon mutants accumulated excessive levels of tmRNA-tagged proteins. In a reporter protein tagging assay with λ-CI-N, the protein product of a nonstop mRNA construct designed to activate trans translation, lon mutant cells efficiently tagged the reporter protein, but the tagged protein exhibited increased stability. Similarly, a green fluorescent protein (GFP) construct containing a hard-coded C-terminal tmRNA tag (GFP-SsrA) exhibited increased stability in lon mutant cells. Most significantly, highly purified Lon preferentially degraded the tmRNA-tagged forms of proteins compared to the untagged forms. Based on these results, we conclude that Lon protease participates directly in the degradation of tmRNA-tagged proteins.
Small protein B (SmpB) and transfer-messenger RNA (tmRNA) are essential components of a bacterial protein quality control mechanism called trans translation (12, 27, 29-31, 37). trans translation is activated when elongating ribosomes stall during translation, unable to terminate. Ribosome stalling may occur during the translation of mRNA transcripts that end without in-frame stop codons (31) or contain rare codons (39, 41) or during inefficient translation termination (10, 20, 21, 40, 48). In the trans-translation model (12, 27, 37, 53), such stalling events lead to an accumulation of trapped ribosomes and incompletely synthesized proteins, which is energetically costly and potentially toxic for the cell. SmpB and tmRNA are targeted to a stalled ribosome in a complex with elongation factor EF-Tu and GTP (5, 42). Following the recruitment of SmpB and tmRNA into the stalled ribosome, the tRNA-like domain of tmRNA donates its alanine to the nascent peptide in a transpeptidation reaction. The ribosome then cotranslationally switches from the faulty mRNA onto the open reading frame in the mRNA-like domain of tmRNA. Translation of the tmRNA message appends a degradation tag onto the C terminus of the nascent peptide. Termination occurs at the stop codon that follows the tmRNA-encoded degradation sequence, releasing recyclable ribosomal subunits and the tagged nascent peptide. The C-terminal tmRNA tag of this chimeric polypeptide targets it for degradation by the cellular proteases ClpXP, ClpAP, FtsH, and Tsp (18, 23, 31).
To identify additional elements of trans translation, we performed a genetic screen using a group of transposon insertion mutants. Our screening confirmed that smpB and ssrA are required for trans-translation function. Of the screen candidates that showed abnormal trans-translation function, we determined that three contain mutations in the gene encoding Lon protease (also known as protease La). The results of in vivo protein tagging and stability assays and in vitro protein degradation assays led us to conclude that Lon protease contributes to the cellular degradation of tmRNA-tagged proteins. Additionally, the involvement of multiple proteases in the turnover of tmRNA-tagged proteins emphasizes the physiological significance of the degradative function of the trans-translation process.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and plasmids.
Unless otherwise noted, bacterial strains were cultivated in LB medium. Antibiotics (ampicillin [100 μg/μl], kanamycin [50 μg/ml], tetracycline [24 μg/ml], or chloramphenicol [30 μg/ml]) were added to the medium when appropriate. For the in vivo protein stability assays, spectinomycin (100 μg/ml) was used to block translation and bacterial growth and to initiate a chase. The Escherichia coli K-12 derivative W3110 [F− λ− IN(rrnD-rrnE)1 rph-1] was utilized as a wild type and is the parental strain of the mutants analyzed in this study. The ssrA::Kan strain has been described previously (33). The clpA::Kan and clpP::Cm strains were prepared through P1 transduction of W3110 with P1 lysates of SG22176 and SG22174, respectively (18). The clpX::Kan strain was prepared through P1 transduction of W3110 with a P1 lysate of BW25113 ΔclpX from the Keio collection (4). λ-P22 hybrid bacteriophages λimmP22 c2-5 dis and λimmP22 c1-7 dis have been described previously (46). Plasmid pPW500 (31) encodes λ-cI-N-trpAt nonstop reporter mRNA regulated by a PTRC promoter. Plasmid pKW510 is a derivative of pPW500 that additionally encodes wild-type tmRNA regulated by the native ssrA promoter. pKW540 is a derivative of pKW510 encoding variant tmRNAH6 rather than wild-type tmRNA. The tmRNAH6 variant encodes a modified degradation tag (NH2-[A]ANDEHHHHHH-COOH) that is poorly recognized by proteases, in contrast with the wild-type tag (NH2-[A]ANDENYALAA-COOH). Green fluorescent protein (GFP) variant gfpmut3.1 was PCR amplified from pJBA27 (2), unmodified or fused with the wild-type ssrA tag, and subcloned downstream of the PBAD promoter in pBAD18-Cm (19), generating pBAD-GFP and pBAD-GFP-ssrA, respectively.
Endogenous-protein tagging assays.
Strains carrying pKW510 or pKW540 were cultivated in LB medium containing ampicillin at 37°C and 200 rpm, until the culture optical density at 600 nm (OD600) was ∼1.0. Cell harvests were normalized to 50 ml of culture with an OD600 of 1.0. Pelleted cells were resuspended in freshly prepared urea lysis buffer (8 M urea, 1% Triton X-100, 2 mM β-mercaptoethanol, 100 mM NaH2PO4, 10 mM Tris-HCl [pH 8.0]). Following 10 min of mixing at room temperature, cells were mechanically disrupted by sonication. His6-tagged proteins in clarified lysates were purified using Ni2+-nitrilotriacetic acid (NTA) chromatography. Bound proteins were eluted from Ni2+-NTA resin using freshly prepared elution buffer (8 M urea, 1% Triton X-100, 10 mM β-mercaptoethanol, 0.1 M acetic acid). Purified proteins were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (3) and Western blotting with a mouse monoclonal anti-His6-horseradish peroxidase (HRP) probe (Santa Cruz Biotechnology).
In vivo GFP-SsrA protein stability assays.
Strains carrying pBAD-GFP or pBAD-GFP-ssrA were cultivated in LB medium containing chloramphenicol at 37°C and 200 rpm until the culture OD600 was ∼0.45. Expression of GFP or GFP-SsrA was induced by addition of 0.01% arabinose to cultures. Following 1 h of induction, cells were gently harvested, washed once with warmed LB medium, resuspended in 1 culture volume of warmed LB medium containing chloramphenicol and spectinomycin, and returned to incubation at 37°C and 200 rpm. Time point samples were obtained from the cultures at 0, 10, 20, 40, 60, 90, and 120 min during the chase. The cell harvest from each time point sample was normalized to 1 ml of culture at an OD600 of 1.0. Cell pellets were resuspended and lysed in 1× SDS sample buffer (3). Total cellular protein was resolved by SDS-PAGE and analyzed by Western blotting with rabbit polyclonal anti-GFP-HRP (Santa Cruz Biotechnology).
In vivo λ-CI-N protein stability assays.
Strains carrying pPW500 were cultivated in LB medium containing ampicillin at 32°C and 200 rpm until the culture OD600 was ∼0.45. Expression of λ-cI-N-trpAt nonstop mRNA was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to cultures. Following 0.5 h of induction, cells were gently harvested, washed once with warmed LB medium, resuspended in 1 culture volume of warmed LB medium containing ampicillin and spectinomycin, and returned to incubation at 32°C and 200 rpm. Time point samples were obtained from the cultures at 0, 5, 10, 15, 20, and 30 min during the chase. The cell harvest from each time point sample was normalized to 1 ml of culture at an OD600 of 1.0. Cell pellets were resuspended and lysed in 1× Tricine sample buffer (3). Total cellular protein was resolved by Tris-Tricine-PAGE (3) and analyzed by Western blotting with mouse monoclonal anti-FLAG M2 (Scientific Imaging Systems) and goat anti-mouse IgG-HRP (Santa Cruz Biotechnology).
Quantification of reporter protein half-life in vivo.
To extrapolate half-life information for the reporter proteins studied in vivo, Western signals were quantified using a GS-710 imaging densitometer and Quantity One software (Bio-Rad). The quantified protein levels were log transformed and least-squares fit to an exponential function to obtain decay rate constants. Reporter half-lives (t1/2) were calculated based on first-order decay: t1/2 = ln(2)/k.
Protein purification.
C-terminally His6-tagged Lon was purified from E. coli strain CH1019 (a gift from the Sauer lab) carrying Lon expression plasmid pET21b-LonH6 by successive Ni2+-NTA affinity chromatography and Mono Q (Amersham Biosciences) anion-exchange chromatography. Lon-His6 expression was induced using 1 mM IPTG for 2 h at 37°C and 200 rpm. Cell pellets were resuspended and mechanically lysed in lysis buffer (1 M NH4Cl, 20 mM potassium phosphate [pH 7.4], 1 mM EDTA, 2 mM β-mercaptoethanol, 20 mM imidazole). Clarified lysate was applied to Ni2+-NTA resin and incubated at 4°C for 1 h with agitation. Lon-His6 was collected from Ni2+-NTA resin using elution buffer (100 mM KCl, 20 mM potassium phosphate [pH 7.4], 1 mM EDTA, 20 mM β-mercaptoethanol, 250 mM imidazole). Eluate containing Lon-His6 was applied to a Mono Q column in buffer A (50 mM KCl, 2 mM EDTA, 20 mM Tris-HCl [pH 7.6], 2 mM β-mercaptoethanol, 10% glycerol). Lon-His6 was eluted using a 0 to 60% linear gradient of buffer B (1 M KCl, 2 mM EDTA, 20 mM Tris-HCl [pH 7.6], 2 mM β-mercaptoethanol, 10% glycerol). The absence of contaminating proteins was verified by SDS-PAGE followed by Coomassie blue staining. Robust Lon activity was determined against fluorescein isothiocyanate-casein (type I; Sigma).
The λ-CI-N reporter protein has an internal His6 epitope that was utilized for purification by Ni2+-NTA chromatography. Untagged and tmRNA-tagged species of the λ-CI-N protein were simultaneously purified from an E. coli clpP clpX lon triple mutant expressing pPW500. Cells were grown at 37°C and 250 rpm in Terrific broth (49) containing ampicillin until the culture OD600 was ∼0.45. Reporter expression was induced for 3 h with 1 mM IPTG. Harvested cells were resuspended and mechanically lysed in lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole). Clarified lysate was applied to Ni2+-NTA resin and washed with lysis buffer containing 20 mM imidazole. Both forms of the λ-CI-N protein were eluted using elution buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 250 mM imidazole). The absence of contaminating proteins was verified by Tris-Tricine-PAGE followed by Coomassie blue staining.
GFP and GFP-SsrA were purified as described previously (56). GFP was purified from E. coli strain JM109 expressing pBAD-GFP. GFP-SsrA was purified from an E. coli clpP clpX lon triple mutant expressing pBAD-GFP-ssrA. The absence of contaminating proteins was verified by SDS-PAGE followed by Coomassie blue staining.
In vitro proteolysis assays.
All in vitro Lon proteolysis assays were carried out with a minimal activity buffer (50 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 1 mM dithiothreitol). Complete assay mixtures contained 1 μM Lon-His6, 10 μM substrate, and an ATP regeneration system (50 mM creatine phosphate, 80 μg/ml creatine kinase [Roche], 4 mM ATP). The reaction mixtures were incubated at 37°C, and analytical samples were obtained at various time points. The levels of untagged and tmRNA-tagged λ-CI-N at selected time points were analyzed by Tris-Tricine-PAGE and quantified using a GS-710 imaging densitometer and Quantity One software (Bio-Rad). The levels of GFP or GFP-SsrA at selected time points were determined from fluorescence measurements obtained using a SpectraMax M2 microplate reader (Molecular Devices) configured with empirically determined excitation and emission wavelengths of 476 and 519 nm, respectively.
RESULTS
Transposon mutagenesis screen.
We performed a transposon-based mutagenesis screen to identify additional cellular factors that might participate in trans translation. The EZ::Tn transposon has two key features that facilitated the screening process. The first feature is a kanamycin resistance gene that serves as a selectable marker for cells that carry an integrated transposon, and the second is a conditional origin of replication that allows transposon rescue cloning and mutant identification. The transposon was introduced into E. coli wild-type strain W3110 via electroporation, and LB medium containing kanamycin was used to select for transposon insertion clones. Several independent transformations were carried out to generate 20,673 insertion clones.
To facilitate mutant screening, we took advantage of a distinct bacteriophage phenotype of E. coli smpB and ssrA mutant strains. SmpB and tmRNA functions are required for lytic development of the λimmP22 c2-5 dis hybrid phage but not for lytic development of the related λimmP22 c1-7 dis phage (29, 38, 46, 52). The molecular basis for this strict dependence of λimmP22 c2-5 dis hybrid phage on SmpB and tmRNA is currently not clear. However, this dependence provided a convenient and distinguishing phenotype that could be readily incorporated into our screen. Using phage cross-streak assays, we first screened the transposon insertion clones for resistance to λimmP22 c2-5 dis phage (see Fig. S1 in the supplemental material). A total of 148 insertion clones (primary candidates) were observed to exhibit resistance to λimmP22 c2-5 dis relative to wild-type cells. Many of these clones, however, had a slightly milder phage phenotype than smpB mutant or ssrA mutant cells, which may reflect an important functional distinction for smpB and ssrA in λimmP22 c2-5 dis development.
Next, we used the hybrid phage λimmP22 c1-7 dis, which does not depend on SmpB and tmRNA functions, to screen out primary candidates that were generally resistant to phage infection rather than abnormal in some aspect of trans-translation function (see Fig. S1 in the supplemental material). Based on this difference, insertion clones that were resistant to both λimmP22 c2-5 dis and λimmP22 c1-7 dis were considered likely to contain mutations in genes that inhibit the phage infection process rather than in genes that might have a trans-translation-related function. Phage cross-streak assays using hybrid phage λimmP22 c1-7 dis narrowed the 148 primary candidates to 16 secondary candidates (see Fig. S1 in the supplemental material).
Transposons from the 16 secondary candidates were rescue cloned in order to map their integration sites. Table S1 in the supplemental material lists the transposon integration sites in the secondary candidates. Despite the small size of the smpB and ssrA genes, these genes were both independently isolated in our screen. This finding was encouraging and validated our approach for identifying genes with trans-translation-related functions. Interestingly, among these candidates were three candidates with independent insertions in the lon gene, designated lon-1, lon-2, and lon-3. The lon gene codes for the ATP-dependent protease Lon, which contains three distinct domains: an amino-terminal domain having an undefined function, a central ATPase domain crucial for substrate binding and unfolding, and a C-terminal peptidase domain. Since substrate binding, unfolding, and proteolysis are coupled functional events, it is expected that mutation of either the ATPase domain or the peptidase domain of Lon should render it nonfunctional in the cell (1, 15, 45). DNA sequencing analysis of the lon mutants revealed transposon disruptions in the region encoding the ATPase domain in the lon-1 and lon-2 mutants and in the region encoding the peptidase domain in the lon-3 mutant.
Once we identified lon multiple times in our genetic screen, we wondered why we had not identified any other major cytoplasmic protease genes, especially the clp genes. Subsequently, we tested clpA, clpP, and clpX mutants using our phage screening assays and observed that these mutants behaved like the parental wild-type strain in that they were sensitive to both λimmP22 c2-5 dis and λimmP22 c1-7 dis (data not shown). This finding effectively explained why we did not identify clp genes in our screen. The identification of smpB and ssrA in this screen suggested that perhaps one or more of the candidates, such as lon, might have a trans-translation-related function.
Assessment of trans translation in secondary candidates.
To further characterize the secondary screen candidates based on trans-translation function, we wished to determine whether the selected candidates had a defect in the SmpB-tmRNA tagging process and potential downstream functions. To this end, we utilized an endogenous protein tagging assay mediated by tmRNAH6. This assay has been used extensively and has greatly facilitated a better understanding of the trans-translation process (13, 28, 36, 40, 47). The degradation sequence encoded by wild-type tmRNA is known to target tmRNA-tagged peptides for rapid destruction by cellular proteases such as ClpXP, ClpAP, FtsH, and Tsp (18, 23, 31), thus making it difficult to evaluate defects in trans translation or to analyze the nature and identity of tagged proteins. Conversely, proteins modified by tmRNAH6, a variant of tmRNA encoding a tag that ends with a His6 epitope rather than the wild-type tag, are significantly stabilized and can be purified by Ni2+-NTA chromatography and detected by Western blot analysis (13, 28, 40, 47, 55). It should be noted that in a trans-translation-competent strain, chromosomally encoded wild-type tmRNA is able to compete with tmRNAH6, resulting in an apparent overall reduction in tmRNAH6-tagged protein levels. This effect can be observed by comparing the tagging profile of a wild-type strain to the tagging profile of an ssrA mutant (Fig. 1A, lanes 1 and 3). Our use of the endogenous protein tagging assay was thus intended to provide an approximation of candidate tagging ability relative to wild-type ability in cases where the candidate was not completely defective in trans translation. To determine the extents of trans-translation function in the secondary candidates, their levels of tmRNAH6-tagged proteins were compared to those observed in the otherwise isogenic parental wild-type strain.
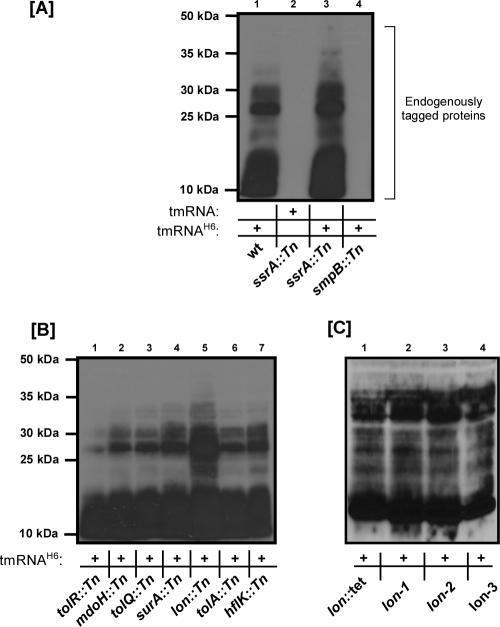
FIG. 1.
Endogenous protein tagging assays performed with selected secondary candidates and wild-type tmRNA or variant tmRNAH6. Purified His6-tagged proteins were analyzed by SDS-PAGE and Western blotting, using HRP-conjugated anti-His6 antibody. (A) The ssrA::Tn mutant had higher levels of tmRNAH6-tagged proteins than the wild-type strain due to the absence of endogenous tmRNA (lanes 1 and 3). As expected, wild-type tmRNA-tagged proteins were not detected in this assay (lane 2). The smpB::Tn mutant is completely defective in the tagging function of trans translation (lane 4). (B) Compared to the wild type, tolR::Tn, tolQ::Tn, and tolA::Tn mutants showed slightly decreased levels of tmRNAH6-tagged proteins (lanes 1, 3, and 6), while the lon::Tn mutant (lon-1) accumulated higher levels of tmRNAH6-tagged proteins (lane 5). (C) Similar endogenous tagging profiles were obtained for lon-1, lon-2, and lon-3 mutants and an otherwise isogenic, previously characterized lon::Tet mutant (from AP401). wt, wild type.
The wild-type strain carrying the tmRNAH6 variant produced a characteristic set of tmRNAH6-tagged proteins (Fig. 1A, lane 1). His6-tagged proteins were not observed when the ssrA::Tn clone was complemented with wild-type tmRNA (Fig. 1A, lane 2), confirming that the tagged proteins observed in the wild-type strain were produced via tmRNAH6 activity. As expected, complementation of the ssrA::Tn clone with tmRNAH6 resulted in the accumulation of levels of tagged proteins higher than the levels obtained with the wild-type strain due to the absence of competition (Fig. 1A, lanes 1 and 3). The smpB::Tn clone did not produce tmRNAH6-tagged proteins, as tmRNA function cannot compensate for the strict requirement for SmpB function in trans translation (Fig. 1A, lane 4).
All of the remaining secondary candidates produced tmRNAH6-tagged proteins, indicating that they were all functional in the protein tagging and ribosome rescue aspects of trans translation (Fig. 1B). However, the lon::Tn (lon-1) mutant accumulated higher levels of tmRNAH6-tagged proteins than most other clones (Fig. 1B, lane 5). Similarly, we observed increased accumulation of tmRNAH6-tagged proteins in the other two lon::Tn mutants, the lon-2 and lon-3 mutants (Fig. 1C). The results of our endogenous tagging assays emphasized that lon is a strong candidate for trans-translation-related function.
Lon protease functions in trans translation.
To confirm that the endogenous protein tagging results observed for our lon::Tn mutants were due to transposon insertion in the lon gene and not a consequence of combined multiple integration events, we took two complementary approaches. First, we complemented the lon::Tn λimmP22 c2-5 dis phage phenotype with a plasmid-borne copy of the lon gene. We cloned lon flanked by its native promoter and terminator in pBR322, to generate pLon. Complementation of all three lon::Tn mutants with the pLon plasmid restored mutant sensitivity to λimmP22 c2-5 dis phage to levels comparable to the parental wild-type strain levels (see Fig. S2 in the supplemental material). Second, we reasoned that if the endogenous tagging phenotype of our lon::Tn mutants is solely due to loss of Lon function, then we should observe the identical phenotype with an independently derived and widely used lon mutant (lon::Tet). To this end, we compared the tmRNAH6-tagging phenotype of our three lon::Tn mutants with that of the lon::Tet mutant. All four lon mutants were found to accumulate comparably high levels of tmRNAH6-tagged proteins (Fig. 1C), suggesting that the defect is a direct result of loss of Lon protease function. Since all three of our lon::Tn mutants produced matching results in our assays, we used one mutant, namely, the lon-1 mutant, for further characterization of lon in trans translation.
GFP-SsrA exhibits increased stability in Lon-deficient cells.
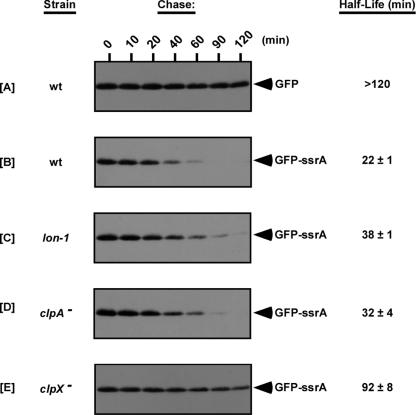
Our endogenous tagging assays clearly showed that lon mutant cells accumulate higher levels of tmRNAH6-tagged proteins. Since Lon is similar to ClpXP and ClpAP in being an ATP-dependent protease that plays an important role in cellular protein turnover, we considered whether it might also participate in the degradation of tmRNA-tagged proteins. To investigate this possibility, we assessed the stability of GFP-SsrA, a GFP variant (2) that carries a hard-coded wild-type tmRNA tag (AANDENYALAA) at its C terminus, in our lon-1 mutant. GFP-SsrA has been used extensively to study the degradation of tmRNA-tagged proteins by the ClpXP and ClpAP proteases (6, 14, 32, 44). In wild-type cells, we found that full-length untagged GFP was quite stable throughout a 2-h time course, with a half-life of more than 120 min (Fig. 2A). In contrast, GFP-SsrA was distinctly less stable (Fig. 2B), and the half-life was reduced more than fivefold to approximately 22 min. GFP-SsrA was consistently observed to be more stable in lon-1 cells than in wild-type cells, as represented by an approximately twofold increase in the half-life (Fig. 2C). These data suggest that lon participates in the turnover of tmRNA-tagged proteins.
FIG. 2.
GFP-SsrA exhibits increased stability in the lon-1 mutant. Expression of GFP or GFP-SsrA was induced using 0.01% arabinose. After removal of the inducer, protein levels were chased in medium containing spectinomycin. In vivo levels of GFP or GFP-SsrA were determined by SDS-PAGE and Western blot analysis using HRP-conjugated anti-GFP antibody. Parallel assays were performed with clpA and clpX mutants for comparative analysis. wt, wild type.
To obtain comparative information for Lon and the Clp proteases, we also studied the stability of GFP-SsrA in clpA and clpX mutants (Fig. 2D and 2E). In each of the protease mutants, GFP-SsrA was stabilized compared to the GFP-SsrA in wild-type cells. Additionally, we observed a consistent difference in GFP-SsrA stability between lon-1, clpA mutant, and clpX mutant cells. As expected, GFP-SsrA was most stable in clpX mutant cells (Fig. 2D), although it was not as stable as untagged GFP. On the other hand, GFP-SsrA was consistently more stable in lon-1 cells than in wild-type or clpA mutant cells (Fig. 2C and 2D). Our results obtained with clpA and clpX mutants are in agreement with previous reports indicating that ClpXP contributes significantly more to the in vivo turnover of tmRNA-tagged proteins than ClpAP contributes (6, 14, 18) and also suggest that Lon might play a greater role than ClpAP in the cellular degradation of tmRNA-tagged proteins.
λ-CI-N is efficiently tagged but more stable in Lon-deficient cells.
To further substantiate our endogenous tagging and GFP-SsrA data, we decided to study a reporter that specifically activates the trans-translation process by causing ribosomal stalling. The λ-cI-N-trpAt nonstop reporter mRNA encoded by pPW500 lacks an in-frame stop codon, thus promoting ribosome stalling and cotranslational addition of the tmRNA tag to the C terminus of the λ-CI-N protein. Of particular relevance is the use of λ-cI-N-trpAt to show that the reporter protein product is tagged but stabilized in clpP mutants, thus directly linking Clp proteolytic activity to trans translation (18).
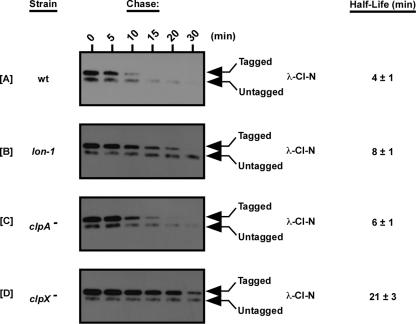
We assessed the stability of cotranslationally tagged λ-CI-N protein in wild-type, lon-1, clpA mutant, and clpX mutant cells. Consistent with our observations for GFP-SsrA, we found that cotranslationally tagged λ-CI-N was more stable in lon-1 cells than in wild-type cells (Fig. 3A and 3B). Additionally, we found that the tagged λ-CI-N protein was most stable in clpX mutant cells (Fig. 3D), moderately stable in lon-1 cells (Fig. 3B), and least stable in clpA mutant cells (Fig. 3C). In agreement with previously published reports, we found that clpA mutant cells have a quite mild defect in the turnover of tmRNA-tagged proteins (6, 14, 18). These findings further support the conclusion that the ATP-dependent protease Lon plays a role in the degradation of tmRNA-tagged proteins.
FIG. 3.
trans-translation reporter protein λ-CI-N is tagged but not efficiently degraded in the lon-1 mutant. Expression of the λ-CI-N protein from a nonstop mRNA activates trans translation, generating tmRNA-tagged λ-CI-N. Reporter expression was induced using 1 mM IPTG. After removal of the inducer, protein levels were chased in medium containing spectinomycin. In vivo levels of λ-CI-N were determined by Tris-Tricine-PAGE and Western blot analysis using anti-FLAG M2 (λ-CI-N has an internal FLAG epitope) and anti-mouse IgG-HRP antibodies. Parallel assays were performed with clpA and clpX mutants for comparative analysis. wt, wild type.
Highly purified Lon protease degrades tmRNA-tagged proteins in vitro.
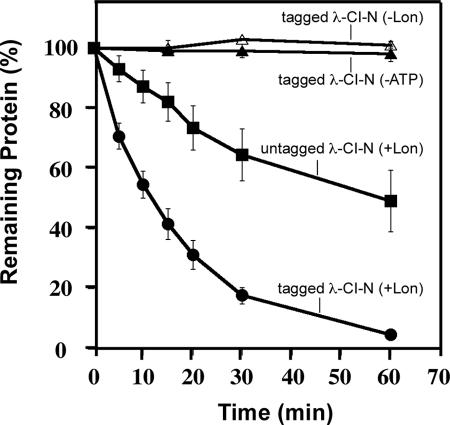
Although we had concurring data from three independent protein stability assays that supported a role for Lon protease in the degradation of tmRNA-tagged proteins, it was still possible that the phenotype that we observed for lon mutant cells resulted from some indirect effect of loss of Lon function. To verify that Lon is involved in the selective proteolysis of tmRNA-tagged proteins, we purified Lon-His6, the tagged and untagged forms of λ-CI-N protein, GFP, and GFP-SsrA. The purified Lon-His6 was fully active against a known substrate (data not shown). To directly examine the degradation of tmRNA-tagged proteins by Lon, we performed in vitro proteolysis assays with the purified Lon and the tagged and untagged forms of λ-CI-N protein (Fig. 4). We observed that Lon degraded tmRNA-tagged λ-CI-N protein much more efficiently than untagged λ-CI-N protein (Fig. 4). Previous studies had demonstrated that tagged and untagged variants of λ-CI-N are equally stable and structurally similar (18). Quantitative analysis of the degradation of tmRNA-tagged λ-CI-N confirmed that Lon protease selectively and efficiently degraded this substrate under our in vitro assay conditions (Fig. 5). Furthermore, the in vitro degradation of tagged λ-CI-N was fully dependent on the presence of both Lon and ATP, as no degradation was observed in the absence of either (Fig. 4 and 5). These data support a model in which the tmRNA-tagged form of λ-CI-N protein is preferentially recognized and degraded by Lon in an ATP-dependent manner.
FIG. 4.
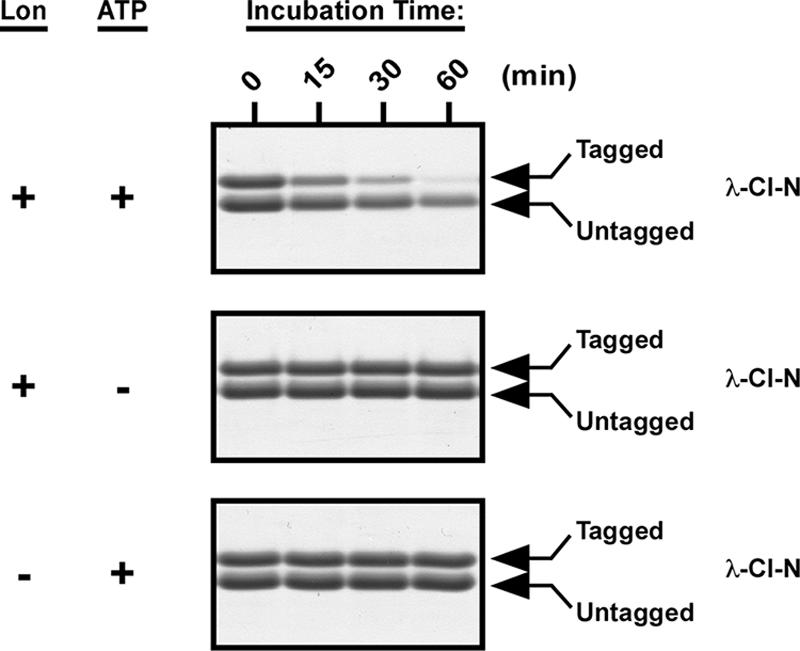
Lon preferentially degrades tmRNA-tagged λ-CI-N protein in vitro. In vitro proteolysis assays were carried out at 37°C in a minimal activity buffer containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, and 1 mM dithiothreitol. Complete reaction mixtures contained 1 μM Lon, 10 μM substrate, and an ATP regeneration system. Time point samples were taken at the indicated times and analyzed by Tris-Tricine-PAGE followed by Coomassie blue staining.
FIG. 5.
Lon preferentially degrades tmRNA-tagged λ-CI-N protein in vitro. A quantitative analysis of in vitro reactions performed with Lon and tagged and untagged forms of λ-CI-N is shown. Coomassie blue-stained λ-CI-N species were quantified using an imaging densitometer.
Next, we compared the proteolytic stabilities of purified GFP and GFP-SsrA in our in vitro degradation assay. GFP and GFP-SsrA constructs have been used extensively to examine recognition and degradation of tmRNA-tagged proteins by the ClpXP and ClpAP proteases (7, 8, 24-26, 32, 44, 51). In agreement with previous studies and our in vivo results, we found that untagged GFP was highly stable and fully resistant to degradation by Lon (Fig. 6). Addition of the tmRNA tag to the C terminus of GFP (GFP-SsrA) resulted in significant degradation of this protein by Lon, as measured by loss of the fluorescent signal of GFP (Fig. 6). Similar to our observations with tmRNA-tagged λ-CI-N, degradation of GFP-SsrA was strictly dependent on the presence of both Lon and ATP (Fig. 6). Both tagged and untagged GFP proteins were equally stable and fluorescent in the absence of Lon and ATP. These results indicated that the presence of the tmRNA tag promoted recognition and selective degradation of GFP-SsrA by Lon protease. Taken together, these data strongly support the conclusion that the ATP-dependent protease Lon participates in the cellular degradation of tmRNA-tagged proteins.
FIG. 6.
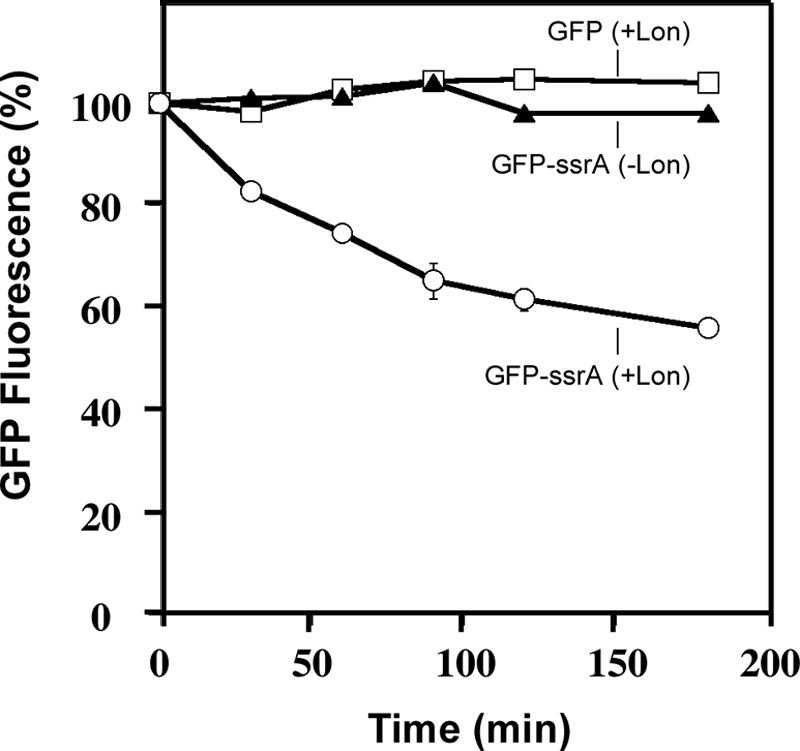
Lon degrades GFP-SsrA but not GFP in vitro. Reactions were carried out with GFP-SsrA or GFP as described in the legend to Fig. 4. The levels of GFP-SsrA and GFP at various time points were determined by microplate fluorimetry.
DISCUSSION
We have described the use of transposon mutagenesis and bacteriophage phenotype screening to identify E. coli genes that are important for trans translation. Through our screening, we confirmed the central roles of smpB and ssrA and identified several other genes as candidates for trans-translation function. The results of our endogenous tagging assays distinguished lon as our strongest candidate, and our subsequent efforts using in vivo and in vitro techniques have substantiated our initial findings. We did, however, find it interesting and perplexing that no clp genes were included in our group of secondary candidates when we were able to identify the much smaller smpB and ssrA genes. Indeed, we found that clpA, clpP, and clpX mutants exhibited sensitivity to both of our screening phages to an extent that was indistinguishable from that of wild-type cells. This effectively explained why clp genes were not identified in our screen.
While the cellular proteases ClpXP, ClpAP, FtsH, and Tsp have been shown to degrade tmRNA-tagged proteins in a tag-specific manner (18, 23, 31), this function had not been observed for Lon protease (14, 18). However, our modification of the reporter protein tagging assay for emphasis on protein decay revealed accumulation of tmRNA-tagged λ-CI-N in lon mutant cells compared to wild-type cells (Fig. 3). Similarly, we found that the GFP-SsrA reporter protein was more stable in lon mutant cells than in wild-type cells (Fig. 2). Therefore, our findings are significant because they demonstrate that lon mutants are unable to efficiently dispose of tmRNA-tagged polypeptides, despite possessing functional ClpXP and ClpAP proteases. Moreover, in vitro degradation assays with purified components clearly demonstrated that Lon has significant proteolytic activity against tmRNA-tagged λ-CI-N and GFP-SsrA, preferentially degrading these tmRNA-tagged forms over untagged controls.
We found that Lon exhibited more robust activity against tmRNA-tagged λ-CI-N than against GFP-SsrA in vitro, perhaps because GFP is intrinsically more stable than λ-CI-N. This is an interesting scenario, since in vitro studies have shown that a difference in inherent stability is not a complicating factor for ClpXP and ClpAP, given that the substrate carries the tmRNA tag (18, 32, 44). However, Lon is known to target certain proteins that are unstable (17, 50) and, therefore, might have more limited capacity in substrate unfolding, which is also the case for the ATP-dependent protease FtsH (22). Nevertheless, we have observed specific Lon activity against tmRNA-tagged proteins, indicating that the presence of the tmRNA tag stimulates proteolysis by Lon.
The C-terminal amino acids of the tmRNA tag are known to be important for targeting to ClpXP and ClpAP (16). Hence, it was intriguing when comparative endogenous tagging assays performed with wild-type, lon mutant, clpA mutant, clpP mutant, and clpX mutant cells showed that while clp mutants had levels of tmRNAH6-tagged proteins that were similar to those observed in wild-type cells, lon mutants accumulated excessive levels of these proteins (Fig. 1 and data not shown). These data suggested the possibility that Lon might recognize sequence determinants in the N-terminal region of the tmRNA tag. It is also possible that the tmRNA tag aids in the presentation of Lon substrate determinants within the tagged protein.
SspB and ClpS are the only adaptor proteins known to considerably influence the proteolysis of tmRNA-tagged peptides (11, 16, 35). Collectively, the two adaptors directly regulate the activities of ClpXP and ClpAP. While there is presently no known adaptor protein that specifically modulates Lon activity in tmRNA-tagged protein turnover, a few studies have shown that Lon proteolysis is influenced by cellular factors that function in stress response (34, 43). It is conceivable that Lon activity against tmRNA-tagged proteins may be conditionally stimulated by as-yet-unknown cellular factors. The modulation of proteolytic activity during various physiological states and responses may lead to rearrangements in the contribution of each protease to tmRNA-tagged protein turnover.
A long-standing unresolved issue has been the fate of tmRNA-tagged proteins in bacterial species that do not possess Clp proteases. Surveys of protease homologs and orthologs in Eubacteria have revealed that Lon is more strongly conserved than other bacterial energy-dependent proteases, including the Clp proteases (9, 17, 54). In contrast to the variable conservation of bacterial energy-dependent proteases, the SmpB-tmRNA system is strictly conserved and, presumably, is universally used to tag proteins for directed proteolysis. Our finding that Lon protease participates in the cellular degradation of tmRNA-tagged proteins provides a fitting resolution for this apparent paradox.
In agreement with our conclusion that Lon protease participates in the degradation of tmRNA-tagged proteins, a recent unpublished study (K. McGinness and R. Sauer, personal communications) showed that Lon protease associates specifically with tmRNA and affects the stability of tmRNA-tagged RbsK protein, a natural trans-translation substrate. The significance of Lon-tmRNA association has not been fully elucidated and requires further scrutiny. The emerging view from these studies is that Lon protease participates in the cellular turnover of tmRNA-tagged proteins, irrespective of how they are tagged. Specifically, the substrate range of Lon includes proteins that carry a C-terminal tmRNA tag encoded at the gene level (like GFP-SsrA), reporter proteins derived from mRNAs designed to activate trans translation (like tmRNA-tagged λ-CI-N), and proteins that are natural substrates of trans translation (like endogenously tagged proteins and RbsK). Based on the new information detailed in this study, we expand the current model for the degradation of tmRNA-tagged proteins to include an important role for Lon protease.
Supplementary Material
Acknowledgments
We thank Andrew Michaels for his help with the transposon mutagenesis screen. We thank Kathleen McGinness and Robert Sauer for communicating results prior to publication, insightful comments, and critical reading of the manuscript. We are grateful to members of the Center for Infectious Diseases for continued support and encouragement. We thank Daniel Dulebohn, Nihal Okan, Thomas Sundermeier, Laura Katona, and James Coleman for invaluable discussions and their help with preparation of the manuscript.
This work was supported in part by grants (to A.W.K.) from The National Institutes of Health (grants GM-65319 and AI-055621), The Northeast Biodefense Center, and The Pew Scholars Program.
Footnotes
Published ahead of print on 6 July 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Amerik, A., V. K. Antonov, A. E. Gorbalenya, S. A. Kotova, T. V. Rotanova, and E. V. Shimbarevich. 1991. Site-directed mutagenesis of La protease. A catalytically active serine residue. FEBS Lett. 287:211-214. [DOI] [PubMed] [Google Scholar]
- 2.Andersen, J. B., C. Sternberg, L. K. Poulsen, S. P. Bjorn, M. Givskov, and S. Molin. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Stuhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, NY.
- 4.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006-2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barends, S., J. Wower, and B. Kraal. 2000. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli. Biochemistry 39:2652-2658. [DOI] [PubMed] [Google Scholar]
- 6.Bohn, C., E. Binet, and P. Bouloc. 2002. Screening for stabilization of proteins with a trans-translation signature in Escherichia coli selects for inactivation of the ClpXP protease. Mol. Genet. Genomics 266:827-831. [DOI] [PubMed] [Google Scholar]
- 7.Bolon, D. N., R. A. Grant, T. A. Baker, and R. T. Sauer. 2004. Nucleotide-dependent substrate handoff from the SspB adaptor to the AAA+ ClpXP protease. Mol. Cell 16:343-350. [DOI] [PubMed] [Google Scholar]
- 8.Burton, R. E., T. A. Baker, and R. T. Sauer. 2003. Energy-dependent degradation: linkage between ClpX-catalyzed nucleotide hydrolysis and protein-substrate processing. Protein Sci. 12:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandu, D., and D. Nandi. 2004. Comparative genomics and functional roles of the ATP-dependent proteases Lon and Clp during cytosolic protein degradation. Res. Microbiol. 155:710-719. [DOI] [PubMed] [Google Scholar]
- 10.Collier, J., E. Binet, and P. Bouloc. 2002. Competition between SsrA tagging and translational termination at weak stop codons in Escherichia coli. Mol. Microbiol. 45:745-754. [DOI] [PubMed] [Google Scholar]
- 11.Dougan, D. A., B. G. Reid, A. L. Horwich, and B. Bukau. 2002. ClpS, a substrate modulator of the ClpAP machine. Mol. Cell 9:673-683. [DOI] [PubMed] [Google Scholar]
- 12.Dulebohn, D., J. Choy, T. Sundermeier, N. Okan, and A. W. Karzai. 2007. Trans-translation: the tmRNA-mediated surveillance mechanism for ribosome rescue, directed protein degradation, and nonstop mRNA decay. Biochemistry 46:4681-4693. [DOI] [PubMed] [Google Scholar]
- 13.Dulebohn, D. P., H. J. Cho, and A. W. Karzai. 2006. Role of conserved surface amino acids in binding of SmpB protein to SsrA RNA. J. Biol. Chem. 281:28536-28545. [DOI] [PubMed] [Google Scholar]
- 14.Farrell, C. M., A. D. Grossman, and R. T. Sauer. 2005. Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbiol. 57:1750-1761. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, H., and R. Glockshuber. 1994. A point mutation within the ATP-binding site inactivates both catalytic functions of the ATP-dependent protease La (Lon) from Escherichia coli. FEBS Lett. 356:101-103. [DOI] [PubMed] [Google Scholar]
- 16.Flynn, J. M., I. Levchenko, M. Seidel, S. H. Wickner, R. T. Sauer, and T. A. Baker. 2001. Overlapping recognition determinants within the ssrA degradation tag allow modulation of proteolysis. Proc. Natl. Acad. Sci. USA 98:10584-10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes, C. S., B. Bose, and R. T. Sauer. 2002. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J. Biol. Chem. 277:33825-33832. [DOI] [PubMed] [Google Scholar]
- 21.Hayes, C. S., B. Bose, and R. T. Sauer. 2002. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:3440-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herman, C., S. Prakash, C. Z. Lu, A. Matouschek, and C. A. Gross. 2003. Lack of a robust unfoldase activity confers a unique level of substrate specificity to the universal AAA protease FtsH. Mol. Cell 11:659-669. [DOI] [PubMed] [Google Scholar]
- 23.Herman, C., D. Thevenet, P. Bouloc, G. C. Walker, and R. D'Ari. 1998. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH). Genes Dev. 12:1348-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinnerwisch, J., B. G. Reid, W. A. Fenton, and A. L. Horwich. 2005. Roles of the N-domains of the ClpA unfoldase in binding substrate proteins and in stable complex formation with the ClpP protease. J. Biol. Chem. 280:40838-40844. [DOI] [PubMed] [Google Scholar]
- 25.Hoskins, J. R., S. K. Singh, M. R. Maurizi, and S. Wickner. 2000. Protein binding and unfolding by the chaperone ClpA and degradation by the protease ClpAP. Proc. Natl. Acad. Sci. USA 97:8892-8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang, S. G., J. Ortega, S. K. Singh, N. Wang, N. N. Huang, A. C. Steven, and M. R. Maurizi. 2002. Functional proteolytic complexes of the human mitochondrial ATP-dependent protease, hClpXP. J. Biol. Chem. 277:21095-21102. [DOI] [PubMed] [Google Scholar]
- 27.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 28.Karzai, A. W., and R. T. Sauer. 2001. Protein factors associated with the SsrA. SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. USA 98:3040-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karzai, A. W., M. M. Susskind, and R. T. Sauer. 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keiler, K. C. 2007. Physiology of tmRNA: what gets tagged and why? Curr. Opin. Microbiol. 10:169-175. [DOI] [PubMed] [Google Scholar]
- 31.Keiler, K. C., P. R. Waller, and R. T. Sauer. 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271:990-993. [DOI] [PubMed] [Google Scholar]
- 32.Kim, Y. I., R. E. Burton, B. M. Burton, R. T. Sauer, and T. A. Baker. 2000. Dynamics of substrate denaturation and translocation by the ClpXP degradation machine. Mol. Cell 5:639-648. [DOI] [PubMed] [Google Scholar]
- 33.Komine, Y., M. Kitabatake, T. Yokogawa, K. Nishikawa, and H. Inokuchi. 1994. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:9223-9227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 35.Levchenko, I., M. Seidel, R. T. Sauer, and T. A. Baker. 2000. A specificity-enhancing factor for the ClpXP degradation machine. Science 289:2354-2356. [DOI] [PubMed] [Google Scholar]
- 36.Moore, S. D., and R. T. Sauer. 2005. Ribosome rescue: tmRNA tagging activity and capacity in Escherichia coli. Mol. Microbiol. 58:456-466. [DOI] [PubMed] [Google Scholar]
- 37.Moore, S. D., and R. T. Sauer. 2007. The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem. 76:101-124. [DOI] [PubMed] [Google Scholar]
- 38.Retallack, D. M., L. L. Johnson, and D. I. Friedman. 1994. Role for 10Sa RNA in the growth of lambda-P22 hybrid phage. J. Bacteriol. 176:2082-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Richards, J., P. Mehta, and A. W. Karzai. 2006. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 62:1700-1712. [DOI] [PubMed] [Google Scholar]
- 40.Roche, E. D., and R. T. Sauer. 2001. Identification of endogenous SsrA-tagged proteins reveals tagging at positions corresponding to stop codons. J. Biol. Chem. 276:28509-28515. [DOI] [PubMed] [Google Scholar]
- 41.Roche, E. D., and R. T. Sauer. 1999. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18:4579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudinger-Thirion, J., R. Giege, and B. Felden. 1999. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA 5:989-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sherman, M., and A. L. Goldberg. 1992. Involvement of the chaperonin dnaK in the rapid degradation of a mutant protein in Escherichia coli. EMBO J. 11:71-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh, S. K., R. Grimaud, J. R. Hoskins, S. Wickner, and M. R. Maurizi. 2000. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proc. Natl. Acad. Sci. USA 97:8898-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starkova, N. N., E. P. Koroleva, L. D. Rumsh, L. M. Ginodman, and T. V. Rotanova. 1998. Mutations in the proteolytic domain of Escherichia coli protease Lon impair the ATPase activity of the enzyme. FEBS Lett. 422:218-220. [DOI] [PubMed] [Google Scholar]
- 46.Strauch, M. A., M. Baumann, D. I. Friedman, and L. S. Baron. 1986. Identification and characterization of mutations in Escherichia coli that selectively influence the growth of hybrid lambda bacteriophages carrying the immunity region of bacteriophage P22. J. Bacteriol. 167:191-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sundermeier, T. R., D. P. Dulebohn, H. J. Cho, and A. W. Karzai. 2005. A previously uncharacterized role for small protein B (SmpB) in transfer messenger RNA-mediated trans-translation. Proc. Natl. Acad. Sci. USA 102:2316-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sunohara, T., T. Abo, T. Inada, and H. Aiba. 2002. The C-terminal amino acid sequence of nascent peptide is a major determinant of SsrA tagging at all three stop codons. RNA 8:1416-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab Focus 9:19. [Google Scholar]
- 50.Tsilibaris, V., G. Maenhaut-Michel, and L. Van Melderen. 2006. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 157:701-713. [DOI] [PubMed] [Google Scholar]
- 51.Wah, D. A., I. Levchenko, G. E. Rieckhof, D. N. Bolon, T. A. Baker, and R. T. Sauer. 2003. Flexible linkers leash the substrate binding domain of SspB to a peptide module that stabilizes delivery complexes with the AAA+ ClpXP protease. Mol. Cell 12:355-363. [DOI] [PubMed] [Google Scholar]
- 52.Withey, J., and D. Friedman. 1999. Analysis of the role of trans-translation in the requirement of tmRNA for λimmP22 growth in Escherichia coli. J. Bacteriol. 181:2148-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Withey, J. H., and D. I. Friedman. 2003. A salvage pathway for protein synthesis: tmRNA and trans-translation. Annu. Rev. Microbiol. 57:101-123. [DOI] [PubMed] [Google Scholar]
- 54.Wong, P., and W. A. Houry. 2004. Chaperone networks in bacteria: analysis of protein homeostasis in minimal cells. J. Struct. Biol. 146:79-89. [DOI] [PubMed] [Google Scholar]
- 55.Wower, I. K., C. Zwieb, and J. Wower. 2004. Contributions of pseudoknots and protein SmpB to the structure and function of tmRNA in trans-translation. J. Biol. Chem. 279:54202-54209. [DOI] [PubMed] [Google Scholar]
- 56.Yakhnin, A. V., L. M. Vinokurov, A. K. Surin, and Y. B. Alakhov. 1998. Green fluorescent protein purification by organic extraction. Protein Expr. Purif. 14:382-386. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.