Abstract
Pseudomonas aeruginosa is an opportunistic pathogen that causes a number of infections in humans, but is best known for its association with cystic fibrosis. It is able to use a wide range of sulfur compounds as sources of sulfur for growth. Gene expression in response to changes in sulfur supply was studied in P. aeruginosa E601, a cystic fibrosis isolate that displays mucin sulfatase activity, and in P. aeruginosa PAO1. A large family of genes was found to be upregulated by sulfate limitation in both isolates, encoding sulfatases and sulfonatases, transport systems, oxidative stress proteins, and a sulfate-regulated TonB/ExbBD complex. These genes were localized in five distinct islands on the genome and encoded proteins with a significantly reduced content of cysteine and methionine. Growth of P. aeruginosa E601 with mucin as the sulfur source led not only to a sulfate starvation response but also to induction of genes involved with type III secretion systems.
Pseudomonas aeruginosa is a well-studied, opportunistic pathogen that causes a wide range of infections in humans, posing particular risk to patients with burn wounds, immune system deficiencies (e.g., AIDS), and cystic fibrosis (31, 32). It displays a high level of metabolic versatility, manifested in the presence in its genome of an unusually high number of genes encoding transcriptional regulators and transport systems for uptake of organic compounds into the cell (45). This allows the cell to make a flexible and tightly regulated response to conditions where particular nutrients become limiting. Under conditions where the supply of inorganic sulfate becomes limiting, for example, P. aeruginosa PAO1 can grow at similar rates by desulfurizing a range of organosulfur compounds such as sulfate esters and sulfonates, since it responds to inorganic sulfate limitation by synthesizing a range of proteins for uptake and utilization of these alternative sulfur sources (34). However, synthesis of these enzymes is controlled by several different nutritional constraints, and whereas expression of the arylsulfatase AtsA is controlled by sulfur supply and provides sulfur for cell growth (3, 19), the SdsA1 alkylsulfatase catalyzes the first step in mineralization of long-chain sulfate esters (14) as a carbon source, and its expression is independent of sulfate concentration. Proteomic studies have shown that sulfur supply also controls the synthesis of some of the enzymes of sulfate assimilation to cysteine in P. aeruginosa (34), and at least two corepressors are important for this control (18). Similar responses have been observed in Escherichia coli (35) and Bacillus subtilis (2), where they appear to be controlled in part by the availability of adenosine-5′-phosphosulfate and S-adenosylmethionine, respectively (2, 4). Further studies of the E. coli sulfate starvation response by microarray expression analysis (13) showed that it is dominated by increased expression of the cysteine biosynthetic pathway but is also controlled by nitrogen supply as part of an overall starvation response dependent on the RpoS protein (12).
Studies of the sulfate starvation response in P. aeruginosa have concentrated on the well-characterized isolate PAO1 (18, 34). However, P. aeruginosa strains isolated from the cystic fibrosis lung are often genetically significantly different from strain PAO1 (55). They are also exposed to a very different set of sulfur-containing molecules, since they colonize the bronchiolar lumen as microcolonies attached to mucus components such as the sulfated respiratory mucin (28, 44). A number of reports have shown that the level of mucin sulfation is enhanced in cystic fibrosis patients (7, 29, 57), and this may well be a defense response in itself, since mucin sulfation is known to prevent degradation of the mucin by bacterial enzymes (48). Mucin sulfatase activities have been found in anaerobic bacteria such as Bacteroides and Prevotella spp. (36, 48), but mucin desulfation has also been reported for several clinical isolates of P. aeruginosa (20). In the P. aeruginosa strains, mucin sulfation seems to be independent of the characterized arylsulfatase activity, suggesting that cells may react differently to exposure to mucin as a sulfur source than to sulfate limitation alone.
This study was initiated to address the questions of which sets of genes are regulated by sulfur supply in P. aeruginosa and whether these are altered in a cystic fibrosis patient isolate on exposure to mucin. Here, we report that under sulfate-limited conditions, a cystic fibrosis P. aeruginosa isolate was able to utilize mucin as the sole sulfur source, although P. aeruginosa PAO1 did not do so. Under these conditions, P. aeruginosa cells showed three groups of transcriptional responses, including a complex sulfate starvation response, a specific response to mucin as the sulfur source, and a more general response to mucin exposure.
MATERIALS AND METHODS
Bacteria and growth conditions.
Pseudomonas aeruginosa PAO1 (45) and the nonmucoid cystic fibrosis isolate P. aeruginosa E601 (20) were grown at 30°C in minimal medium with 25 mM succinate as the carbon source (24), supplemented with all proteinogenic amino acids with the exception of cysteine and methionine (40 μg/ml of each amino acid). Sulfur sources were added as described in the text, at concentrations of 500 μM for sodium sulfate or cyclamate and of 25 μg/ml for human colon cell line mucin LS174T or pig mucin. Glassware used for sulfur-limited work was washed with 3 M HCl before use. Cultures used for RNA preparation were grown as aerobic 5-ml batch cultures in 50-ml Erlenmeyer flasks on an orbital shaker (225 rpm). Growth experiments were performed with 200-μl cultures in microtiter plates with continuous shaking, using a Synergy HTi plate reader (BioTek Instruments), and growth rates were determined in the exponential growth phase by measurements of A600 taken every 10 min over 18 h.
P. aeruginosa K1040M was constructed from P. aeruginosa 1040 (met-9011 pvd-9 tonB1::Hg) (58) by replacement of the methionine auxotrophy by transduction using phage E79-tv2 grown on strain PAO1. This strain was grown in iron-supplemented medium as previously described (58). Other cystic fibrosis P. aeruginosa isolates used included three epidemic strains (C3373, LES400, and LESB58 [42]) and a selection of other isolates from the University of Liverpool laboratory collection (strains 49211, 49244, 59032, 59039, 59070, and 59079).
Preparation of mucin.
The human colon adenocarcinoma cell line LS174T, a cell type which forms well-differentiated goblet cells, was obtained from the European Collection of Animal Cell Cultures (Porton Down, United Kingdom). The presence of O-sulfate esters on LS174T secretory mucins has been demonstrated previously in [35S]sulfate incorporation studies (5). For production of human mucin LS174T, cells were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 4 mM glutamine, 10% (vol/vol) fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin in 5% CO2-95% air at 37°C. The medium was harvested 2 to 7 days after confluence was achieved, and the mucin was purified by gel filtration over Sepharose CL-2B minicolumns (5 by 1.5 cm), eluting with phosphate-buffered saline (pH 7.4) containing 0.01% (wt/vol) thimerosal. Mucin-containing fractions were desalted over PD10-Sephadex GM25 columns, lyophilized, and stored at 4°C. Pig gastric mucin (type III) was obtained from Sigma.
RNA preparation and reverse transcriptase-PCR.
Cells used for the preparation of RNA were grown to exponential phase (optical density at 600 nm, 0.2), harvested at 4°C, and immediately frozen in liquid nitrogen. Total RNA was prepared with an RNeasy mini kit and an RNase-free DNase set from QIAGEN (Hilden, Germany), by following the protocol of the manufacturer. For each growth condition, RNA was prepared from three or four independent batch cultures harvested at the same optical density at 600 nm (within 0.05 optical units).
The quality of RNA preparations was routinely checked on 1% agarose gels (39), and by reverse transcriptase-PCR of the sbp gene PA0283 (reverse primer, CGACGATCTCGAAGTTTTCC; forward primer, ATCGACGAACTGCACAAGC). Reverse transcriptase-PCR was performed by a two-step protocol, using 1 μg of RNA and the First Strand cDNA synthesis kit (MBI-Fermentas). The same PCR setup was used to check RNA preparations for the absence of contaminating DNA prior to reverse transcription.
DNA microarray hybridization.
Frozen batches of RNA were thawed and used only once. Reverse transcription of RNA, cDNA fragmentation, and hybridization followed the protocol recommended and provided by Affymetrix (Santa Clara, CA). Briefly, reverse transcription of 5 to 10 μg of P. aeruginosa RNA was performed using 750 ng of random hexamer primer mix and Superscript III transcriptase (Invitrogen). The cDNA product was purified with the QIAquick PCR purification kit (QIAGEN). Purified cDNA (3 μg) was then fragmented for 6 min at 37°C using DNase I, and the fragmented cDNA was biotin labeled and hybridized for 16 h at 50°C to an Affymetrix P. aeruginosa chip (P. aeruginosa genome array; Affymetrix). Microarrays were further processed in accordance with the recommended protocol of Affymetrix. Experiments were performed with either threefold or fourfold biological replication. Hybridization of genomic DNA was carried out similarly, after fragmentation of genomic DNA samples using the protocol provided by Affymetrix.
Analysis of microarray data.
Image analysis of the scanned arrays for the presence and absence of RNA transcripts was done using the MAS 5 software package with the following parameter settings: α1 of 0.04, α2 of 0.06, τ of 0.015, TGT value of 400, and norm factor of 1.0. Array images were checked visually for any irregularities in hybridization using dChip 1.3 (30, 59). Array data were normalized using the GC-RMA algorithm (56), and the data were then further analyzed using the microarray software package Genespring GX 7.3 (Agilent). Genes whose levels of expression were significantly influenced by sulfur source (P < 0.05) were identified with Genespring GX, using a Benjamini-Hochberg multiple-testing correction and a false detection rate of 5%. Significance analysis of the microarray results was performed using the SAM algorithm (49), with false detection rates of <5%.
Validation of microarray results with quantitative reverse transcriptase-PCR.
The accuracy of the microarray data was verified by quantitative reverse transcriptase-PCR analysis of 10 selected genes displaying high and low levels of change in the microarray analysis (see Table S1 in the supplemental material). PCR amplification was performed using an ABI Prism 7000 cycler (Applied Biosystems) with two-step TaqMan reverse transcription and a SYBR green PCR master mix kit (Applied Biosystems) and 100 ng of template RNA, in accordance with the protocol of the manufacturer. The calibration curves for each primer pair were constructed with purified PCR product as the template. Experiments were repeated three times for each of the biological triplicates of the corresponding microarrays.
Microarray data accession number.
Lists of induced and repressed genes can be found in the supplemental material. The gene expression data are available from the Gene Expression Omnibus (GEO) under accession number GSE8408.
RESULTS
Growth of P. aeruginosa with mucin as the sulfur source.
Although many Pseudomonas species express arylsulfatases (22), the ability to desulfate mucin is less common. Pseudomonas mucin sulfatase activity was first identified in the cystic fibrosis isolate P. aeruginosa E601 and is not present in the sequenced strain P. aeruginosa PAO1 (20). Growth of these two P. aeruginosa strains was tested in a succinate-based minimal medium with sulfur supplied as sulfate, cyclamate, model human mucin LS174T, pig mucin, or cyclamate mixed with human mucin LS174T (Table 1). For comparison, growth with mucin LS174T or with sulfate was also tested for a range of nine cystic fibrosis isolates selected from a previous study (42), including two different isolates of the Liverpool epidemic strain (strains LES400 and LESB58) and an isolate of the Manchester epidemic strain (strain C3373). Minimal medium containing no sulfur source was used as a negative control. Strain E601 grew with sulfate, cyclamate, human mucin, and the combination of cyclamate with mucin but not with pig mucin. A small pulse of growth (up to an A600 of 0.1) was observed in the initial part of the experiment, due to exhaustion of residual sulfate in the growth medium, and exponential growth rates (Table 1) were determined after exhaustion of this contaminating sulfate. Growth was fastest with sulfate, as a preferred sulfur source (23). Although growth with mucin as the sulfur source was 35-fold slower than with sulfate, it was consistently above the slow growth observed in sulfur-free medium or with pig mucin, demonstrating that the human mucin acts as a specific growth substrate. The other cystic fibrosis P. aeruginosa isolates tested grew at rates similar to that of strain E601 with human mucin as a sulfur source but were unable to grow with pig mucin. The ability to grow slowly with LS174T mucin as a sulfur source therefore seems to be typical for these clinical isolates. By contrast, strain PAO1 was unable to desulfurize either human mucin or pig mucin, though it grew well with sulfate, cyclamate, or the combination of cyclamate and human mucin (Table 1).
TABLE 1.
Growth rates of P. aeruginosa with different sulfur sourcesa (h−1)
| Sulfur source | Growth ratea (h−1) (mean ± SD) of:
|
|
|---|---|---|
| P. aeruginosa E601 | P. aeruginosa PA01 | |
| Cyclamate | 0.105 ± 0.008 | 0.135 ± 0.022 |
| Sulfate | 0.100 ± 0.004 | 0.313 ± 0.010 |
| Human mucin | 0.0047 ± 0.0001 | NG |
| Pig mucin | NG | NG |
| Cyclamate + human mucin | 0.108 ± 0.005 | 0.141 ± 0.002 |
Maximum growth rates were determined in the exponential phase of growth in a vigorously shaken microplate by measuring the A600 every 10 min. Values were determined in three independent experiments, with eightfold replication in each experiment. NG, no growth.
Genome comparison of P. aeruginosa strains PA01 and E601.
Pseudomonas aeruginosa E601 is a recent clinical isolate and was therefore expected to be genetically different from strain PAO1, which has been in laboratory culture for several decades. Differences between the genomes of different isolates of P. aeruginosa and strain PAO1 have been reported to vary, with 10 to 17% of genes being present in other isolates but not in P. aeruginosa PAO1 (43). The genomic differences between strains PAO1 and E601 were investigated by hybridization of the total genomic DNA from each strain to the Affymetrix P. aeruginosa PAO1 GeneChip. The P. aeruginosa E601 DNA hybridized to 5,198 of 5,570 annotated open reading frames (ORFs) of the P. aeruginosa PAO1 GeneChip and to six intergenic regions and one non-PAO1 gene that did not give a signal with genomic DNA from P. aeruginosa PAO1. Overall, the genome cross-hybridization showed 97% coverage for the detection of transcripts between the two species, thus allowing the reasonable usage of the P. aeruginosa PAO1 chip with P. aeruginosa E601. No further attempt was made at this stage to identify additional genes that were present in the genome of strain E601 but not in strain PAO1.
P. aeruginosa sulfate starvation response—upregulation of gene families.
Expression levels of specific P. aeruginosa genes are expected to change when mucin rather than sulfate is supplied as the sole sulfur source, primarily because many genes are upregulated in the absence of sulfate as part of the sulfate starvation response (34). However, changes were also expected to include specific mucin-induced genes and changes in gene expression due to growth rate effects, since growth with mucin is considerably slower than with sulfate (Table 1). To avoid these growth rate effects, we compared gene expression in sulfate-grown cells with that in cells grown with cyclamate (N-cyclohexylsulfamate) as an alternative sulfur source. Cyclamate allows a growth rate comparable to that observed with sulfate (Table 1) and acts as a synthetic, chemically defined surrogate for N-linked mucin. In addition, the use of cyclamate-grown cells facilitates a comparison with previous proteomic studies of P. aeruginosa PAO1 (34). The sulfate starvation responses of P. aeruginosa strains PAO1 and E601 were examined by isolating the total RNA of each strain from exponential-phase batch cultures grown with sulfur supplied as cyclamate, sulfate, or cyclamate and sulfate together, followed by GeneChip analysis. Cells were harvested at the same optical density in order to minimize growth-phase-dependent effects.
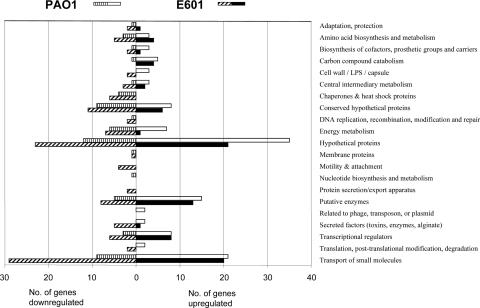
Growth with cyclamate in the absence of added sulfate led to the differential upregulation (greater than twofold; P < 0.05) of 132 genes in both of the two strains studied. It is important to note that under these conditions, cells are not starved for sulfur, but the absence of inorganic sulfate leads to the change in gene expression (34). Transcriptomic responses can vary dramatically between different strains of the same species (38), and comparison of these gene lists showed that 155 and 134 genes were upregulated in strain PAO1 and strain E601, respectively, while 67 and 273 genes were downregulated more than twofold in the two strains, respectively (Fig. 1; see Table S3 in the supplemental material). Of the 132 upregulated genes, 43 were strongly upregulated (>10-fold) (Table 2). When differentially regulated genes were classified by metabolic category (Fig. 1), broadly similar patterns were observed for the two strains. The most strongly upregulated groups of genes correspond to hypothetical and conserved hypothetical proteins, but a large group of genes related to transport functions was also upregulated. Interestingly, we detected significant downregulation of genes related to energy metabolism (Fig. 1). Comparison of sulfate-grown cells with those provided with mixed cyclamate and sulfate revealed that only two cyclamate-upregulated genes were not repressed by cyclamate-sulfate (PA2298 and PA2299), suggesting that these may be specifically cyclamate induced.
FIG. 1.
Functional classification of P. aeruginosa genes that displayed greater-than-twofold altered expression (P < 0.05) in the absence of sulfate compared to growth with sulfate as the sulfur source. The responses of P. aeruginosa PAO1 and P. aeruginosa E601 are shown separately. LPS, lipopolysaccharide.
TABLE 2.
Genes whose expression was strongly upregulated (>10-fold) in P. aeruginosa in the absence of sulfate relative to growth with inorganic sulfatea
| Gene name | Fold change in expression | % of S-amino acidsb | COGc | Protein description |
|---|---|---|---|---|
| PA0183 (atsA) | 32.95 | 1.87 | 3119 | Arylsulfatase |
| PA0184 (atsC) | 41.30 | 1.43 | 1116 | Probable ATP-binding component of ABC transporter |
| PA0185 (atsB) | 16.18 | 1.86 | 600 | Probable permease of ABC transporter |
| PA0186 (atsR) | 24.03 | 0.00 | 715 | Probable binding protein component of ABC transporter |
| PA0192 | 25.42 | 0.51 | 1629 | Probable TonB-dependent receptor |
| PA0193 (atsK) | 42.40 | 0.00 | 2175 | Alkylsulfatase |
| PA0194 | 26.92 | 0.00 | 2175 | Hypothetical protein |
| PA0197 (tonB2) | 38.60 | 0.74 | 810 | Hypothetical protein |
| PA0198 (exbB1) | 55.97 | 0.42 | 811 | Transport protein ExbB |
| PA0199 (exbD1) | 35.72 | 1.50 | 848 | Transport protein ExbD |
| PA2085 | 11.08 | 0.59 | 5517 | Probable ring-hydroxylating dioxygenase small subunit |
| PA2086 | 13.53 | 2.00 | 596 | Probable epoxide hydrolase |
| PA2087 | 12.16 | 2.75 | None | Hypothetical protein |
| PA2088 | 13.07 | 2.13 | 4106 | Hypothetical protein |
| PA2092 | 12.03 | 1.76 | 2814 | Probable MFS transporter |
| PA2292 | 13.41 | 4.30 | 3536 | Hypothetical protein |
| PA2293 | 20.70 | 1.25 | 1413 | Hypothetical protein |
| PA2294 | 22.32 | 3.87 | 1116 | Probable ATP-binding component of ABC transporter |
| PA2295 | 35.52 | 3.40 | 600 | Probable permease of ABC transporter |
| PA2296 | 44.80 | 0.85 | 715 | Hypothetical protein |
| PA2297 | 18.52 | 13.58 | 2221 | Probable ferredoxin |
| PA2298 | 14.83 | 4.70 | 1053 | Probable oxidoreductase |
| PA2299 | 19.46 | 2.41 | 2188 | Probable transcriptional regulator |
| PA2307 | 15.12 | 0.69 | 600 | Probable permease of ABC transporter |
| PA2308 | 15.43 | 2.13 | 1116 | Probable ATP-binding component of ABC transporter |
| PA2309 | 11.83 | 0.00 | 715 | Hypothetical protein |
| PA2310 | 10.81 | 1.02 | 2175 | Hypothetical protein |
| PA2324 | 26.10 | 0.48 | 1960 | Hypothetical protein |
| PA2325 | 14.64 | 0.00 | 1960 | Hypothetical protein |
| PA2326 | 11.14 | 1.07 | 2141 | Hypothetical protein |
| PA2346 | 24.30 | 0.73 | 1960 | Conserved hypothetical protein |
| PA2347 | 34.83 | 0.49 | 1960 | Hypothetical protein |
| PA2348 | 46.96 | 1.52 | 2141 | Conserved hypothetical protein |
| PA2349 | 36.85 | 0.00 | 1464 | Conserved hypothetical protein |
| PA2350 | 19.24 | 1.90 | 1135 | Probable ATP-binding component of ABC transporter |
| PA2351 | 10.54 | 4.61 | 2011 | Probable permease of ABC transporter |
| PA2354 | 16.04 | 1.06 | 3829 | Probable transcriptional regulator |
| PA2355 (msuC) | 22.58 | 1.78 | 1960 | Probable FMNH2-dependent monooxygenase |
| PA2356 (msuD) | 18.44 | 1.05 | 2141 | Methanesulfonate sulfonatase |
| PA2357 (msuE) | 17.80 | 0.00 | 431 | NADH-dependent FMN reductase |
| PA3447 | 13.47 | 2.41 | 1116 | Probable ATP-binding component of ABC transporter |
| PA3448 | 14.59 | 0.73 | 600 | Probable permease of ABC transporter |
| PA3954 | 20.21 | 0.27 | 2141 | Hypothetical protein |
Other sulfur sources were supplied for growth of P. aeruginosa. For a complete list of P. aeruginosa genes upregulated and downregulated (greater than twofold) under the conditions tested, see Table S3 in the supplemental material. Analysis was performed using Genespring GX 7.3, with a Benjamini-Hochberg multiple-testing correction and a false detection rate of 5%. Significance of the called genes was also assessed using SAM (49), giving q values of <0.05.
Percentages of sulfur-amino acids were calculated as the proportion of cysteine/methionine residues in the encoded protein, not including the initiating formylmethionine.
COG to which the gene has been assigned (46).
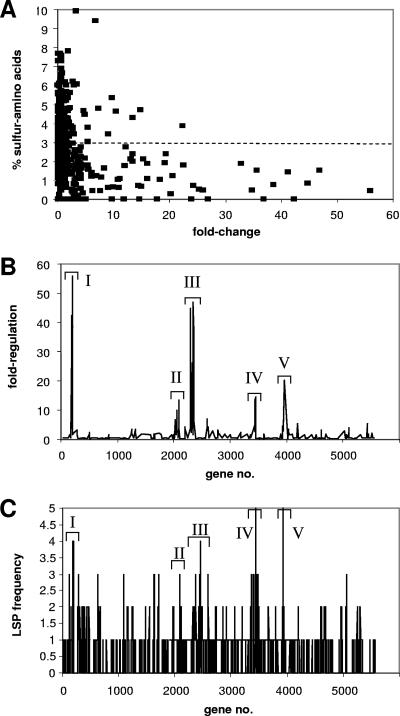
Although cells grown with cyclamate as a sulfur source grow at a rate comparable to that for cells grown with sulfate, it has been suggested that part of the sulfate starvation response may be the synthesis of proteins with reduced levels of cysteine and methionine (24). The predicted amino acid composition of the proteins encoded by the sulfate starvation-induced (SSI) genes was calculated (Fig. 2A) and provided startling confirmation of this, with 77% of the SSI genes encoding proteins with below-average content of S-amino acids (Fig. 2A) (the S-amino acid content was calculated as the number of encoded Cys and Met residues per ORF, not including fMet, relative to the overall ORF length) and >90% of them having a below-average cysteine content. The regulated genes also fell into distinct regions on the genome (Fig. 2B), forming five islands where most of the sulfate-starvation-induced genes were located (PA183 to PA199, PA2050 to PA2094, PA2289 to PA2359, PA3441 to PA3450, and PA3931 to PA3954) and within which >50% of the genes were upregulated greater than twofold during sulfate limitation. These islands also contained enhanced levels of genes encoding low-sulfur proteins (LSPs) (defined as ORFs in which the percentage of encoded cysteine and methionine residues [not including fMet] per ORF was <0.64%, i.e., more than 1.5 standard deviations below the average value for all the ORFs on the genome of 2.87%.) (Fig. 2C), though it should be noted that there were also many LSP-encoding genes that were not strongly regulated by sulfur supply. The clustering of genes encoding proteins with reduced sulfur content has been reported before for E. coli (37) and Pseudomonas putida (41), but a correlation with changes in gene expression was not reported.
FIG. 2.
(A) Sulfur content of sulfate starvation-regulated genes (see Table S2 in the supplemental material) in P. aeruginosa PAO1. (B) Distribution of SSI genes (see Table S3 in the supplemental material) on the P. aeruginosa genome, showing the presence of five islands that contain enhanced levels of sulfate starvation-regulated genes (sulfur islands I to V). (C) Distribution of genes encoding LSPs in the P. aeruginosa genome. The frequency is given as the number of LSP-encoding genes in a 10-gene window. LSPs were defined as proteins for which the percentage of encoded cysteine and methionine residues (not including fMet) was more than 1.5 standard deviations below the average value for all ORFs in the genome of 2.87%. The islands of SSI genes (I to V) (as described for panel B) are shown for comparison.
Although only putative function can be assigned to the upregulated group of hypothetical proteins, orthologue analysis (clusters of orthologous groups [COGs]) (47) yields valuable additional information. Forty-nine of the 132 genes upregulated during sulfate limitation in both strain PAO1 and E601 belong to only nine COGs. The dominant function in these clusters is transport: eight genes appear to encode proteins related to substrate-binding proteins of sulfonate-dependent ABC transporters (COG715) while the cognate permease (COG600; six genes) and ATPase (COG1116; five genes) components of these transporters are also detected as being upregulated under conditions of sulfur limitation. A small group of HisM-type amino acid permeases is also upregulated (COG765; four genes). The other dominant COG families are classes of enzymes with representatives acting in sulfonate degradation of organosulfonates, including FMNH2-dependent monooxygenases such as SsuD and MsuD (COG2141; eight genes) and α-ketoglutarate-dependent dioxygenases such as TauD and AtsK (COG2175; five genes). Upregulation of multiple uncharacterized members of these five COGs under sulfate-limited conditions suggests that the encoded proteins catalyze the uptake and desulfurization of a variety of environmentally relevant sulfonates and/or sulfate esters.
Interestingly, the cysteine biosynthetic pathway was only weakly upregulated by sulfate limitation in P. aeruginosa, with only the sulfate transporter genes sbp and cysTWA being derepressed more than twofold during growth with cyclamate. This contrasts strongly with the changes observed in E. coli during growth in the absence of sulfate (13), where all the genes of the cys regulon are upregulated under sulfate starvation conditions and the cells do not upregulate multiple members of particular COG families. Sulfate limitation was linked to an oxidative stress response, as also observed in E. coli, with upregulation of ahpC (alkylhydroperoxide reductase), katB (catalase), ohr (organic hydroperoxide resistance protein), and lsfA (thiol-specific antioxidant). Multiple members of two additional COGs potentially related to sulfur metabolism were also upregulated. The most interesting of these was COG1960 (six genes), which includes proteins with homology to acylCoA dehydrogenases, some of which are known to be involved in desulfurization of dibenzothiophene and methanesulfonate in this and other species (DszC and MsuC, respectively) (11, 25). With the exception of DszC (which acts as an FMNH2-dependent monooxygenase in Rhodococcus [11]), the function of these proteins has yet to be clarified. A second group is made up of TonB-dependent receptor proteins (COG1629; four genes). Glutathione S-transferases (COG625) represent another large protein family connected to sulfur metabolism, with 18 members of this group on the P. aeruginosa genome, mostly of unknown function (53). No glutathione S-transferase genes, and indeed no other glutathione-related genes, were significantly regulated by shifts in the sulfur source, providing evidence that changes in the sulfur source for growth do not affect glutathione utilization or synthesis.
Role of the TonB complex in the sulfate starvation response.
Three of the most strongly upregulated genes in the absence of sulfate were tonB2 (PA0197) and the two genes encoding TonB accessory proteins, exbB and exbD. The TonB-ExbB-ExbD complex is essential in the uptake of siderophores into the cell and also plays a role in the transport of vitamin B12 (8). Its potential importance in sulfur metabolism is underlined by the fact that several putative TonB-dependent receptors and transducers are upregulated during growth in the absence of sulfate. Recent work has shown that these outer membrane proteins act either as transporters or as transducers of external signals, depending on their domain structure (26), and are energized by the TonB-ExbB-ExbD complex in the cytoplasmic membrane and periplasm. The strong upregulation of the tonB2 gene under sulfate-limited conditions suggested that it might be required for uptake of alternative sulfur sources and contrasts strongly with the expression levels of tonB1 (PA5531) and tonB3 (17), which were unchanged under sulfate limitation. We cultivated a tonB2 mutant strain of P. aeruginosa (strain 1407 [58]) with a variety of different sulfur sources but found that it grew normally compared to the wild-type strain (data not shown). This suggested that the tonB1 gene (PA5531) might be able to complement the deficiency in tonB2. In contrast, TonB2 cannot fully compensate for a deficiency in TonB1, since growth of a tonB1 mutant strain (strain K1440M) required iron supplementation during growth with either sulfate or cyclamate (i.e., even when tonB2 expression was induced by sulfate limitation). We conclude that the sulfur-regulated TonB2 complex is important for uptake of an unknown sulfur-containing substrate. It is located on the genome in sulfur island I (Fig. 2B), in close proximity to a putative TonB-dependent receptor (PA0192) and a number of genes that are involved in sulfate ester metabolism (22), so it may be involved in utilization of an unidentified sulfate ester.
Sulfate starvation and the influence of mucin.
Cells of P. aeruginosa E601 were grown in batch cultures containing human mucin as the sole source of sulfur, and RNA extracted from these cultures was used for GeneChip analysis. Compared to growth results with sulfate, provision of mucin as the sole sulfur source led to upregulation of a slightly larger group of genes than that observed during growth with cyclamate (see Table S4 in the supplemental material). This group included almost all the sulfate starvation-induced genes that were upregulated during growth with cyclamate, confirming that the observed sulfate limitation response is not specific to cyclamate as the sulfur source and is seen during both rapid and slow growth. Several additional genes were also upregulated under these slow-growth conditions, including exoenzyme S, components of the type III secretion apparatus, and multidrug efflux systems. However, most of these were also upregulated during rapid growth with mucin and cyclamate together, relative to growth with cyclamate alone, and they therefore constitute the cells’ response to mucin independent of the overall sulfate starvation response.
DISCUSSION
Pseudomonas aeruginosa is known to attach to mucin produced by the lung epithelium of patients suffering from cystic fibrosis (6, 9), and it has been suggested that P. aeruginosa uses mucin as a carbon source (1). In this report we show that P. aeruginosa E601 and other cystic fibrosis isolates are also able to utilize a model human mucin as a sole sulfur source for growth. Strain E601 responds to these growth conditions by upregulation of a general sulfate starvation response (Table 2) and a specific response to the presence of mucin that includes genes that encode a number of virulence factors.
The homeostatic response of P. aeruginosa to sulfate limitation involved the upregulation of some 132 genes (>2-fold, P < 0.05) (see Table S2 in the supplemental material), with 44 of these showing >10-fold regulation (Table 2). Regulation of these genes by sulfur supply was observed in two different strains of P. aeruginosa, the cystic fibrosis isolate E601 and the laboratory strain PAO1. Some additional genes were upregulated in each strain specifically, but these were considered less relevant for the present study. Importantly, the same set of SSI genes was observed during growth with cyclamate and with mucin for both strains investigated, and this included the genes encoding all the SSI proteins identified in a previous proteomic study (34). Compared with that of other bacteria, the P. aeruginosa SSI response is quite extensive. E. coli, for example, responds to sulfate starvation (growth with taurine or glutathione) with changed regulation of 64 genes (13), while in Bacillus subtilis some 56 genes were similarly regulated (growth with methionine) (2). The most-expected response to sulfate limitation is increased transcription of the cysteine and methionine biosynthetic pathways, and this is indeed observed in E. coli, with all the genes of the cys regulon and several methionine biosynthesis genes upregulated under sulfate starvation conditions (13). In P. aeruginosa, however, this was not observed, and although expression of sulfate transport genes was increased, the rest of the cysteine and methionine biosynthesis pathways were not significantly affected. In B. subtilis, there was upregulation of cysteine biosynthesis, but since methionine was supplied as the derepressing growth substrate, no conclusions can be drawn concerning methionine biosynthesis (2).
The P. aeruginosa genome is characterized by the presence of larger numbers of regulatory proteins and uptake systems than in most other bacteria whose genomes have been sequenced (27). This allows a flexibility of response to different environmental conditions and helps explain the ability of this species to colonize a wide variety of habitats. The P. aeruginosa SSI response observed here reflects this genomic flexibility, since its main feature is the upregulation of families of related genes. Many of these are of unknown function, and they are likely to catalyze uptake and desulfurization of unidentified sulfur-containing substrates. B. subtilis also increases transcription of a range of transport systems under sulfate limitation, but E. coli appears to be limited to the two characterized sulfonate utilization operons, ssu and tau (50, 51). All three species respond to sulfate starvation with the synthesis of proteins that help to counter oxidative stress, such as catalases and peroxidases. It has been suggested that this may be linked to peroxide production by autoxidation of enzymes containing flavin adenine dinucleotide, such as sulfite oxidase (13). However, all three species studied respond to sulfate limitation by strongly upregulating the expression of FMNH2-dependent monooxygenases (mainly sulfonatases such as SsuD or MsuD; in P. aeruginosa, no fewer than eight genes of this family were upregulated) and the corresponding flavin mononucleotide (FMN) reductases. The resulting increase in FMN reductase activity will lead to enhanced levels of reduced FMN in the cell, and if the appropriate sulfonatase substrates are not present, rapid autoxidation will release active oxygen species, potentially causing the observed oxidative stress response.
Comparison of the sulfate starvation responses observed in E. coli during growth with taurine (rapid growth) and glutathione (slow growth), and during various shifts in sulfur supply, led to the conclusion that a set of responses that occurred both for N and S limitation were in fact consequences of slow growth and were controlled to a great extent by the RpoS protein (12, 13). We were therefore surprised to observe that in P. aeruginosa, the SSI response seen with cyclamate (rapid growth) was very similar to that observed during slow growth with mucin as the sulfur source. Of all the sulfate starvation-regulated genes (see Table S3 in the supplemental material), only six corresponded with P. aeruginosa genes found to be RpoS controlled in a previous study (40), suggesting that RpoS does not play a large role in the homeostatic sulfate starvation response in this species. However, other sigma factors are also important in the SSI response of Pseudomonas species. In P. putida, for example, the RpoN sigma factor is known to play a role in sulfur metabolism, controlling expression of the sfn genes that are required for dimethylsulfone metabolism (10). Very few of the P. aeruginosa SSI genes carried a conserved RpoN-binding site (46), but these included the P. aeruginosa sfn orthologues PA2346 and PA2347 and the nearby msuD and msuE genes, which are involved in methanesulfonate metabolism (25). Apart from these, only the porin gene oprB (PA3186) appeared to be RpoN controlled. In P. putida, expression of the sfn genes also required the CysB protein, which is a LysR family regulator of sulfur metabolism in a number of gram-negative species. In P. aeruginosa, CysB has been shown to be required for the utilization of sulfate esters and to be involved in sulfonate utilization (19, 52), but the extent to which it controls the other SSI genes reported here has not yet been investigated. A consensus CysB-binding sequence has been reported in Salmonella enterica serovar Typhimurium (16), but it is not well conserved, and a search for conserved sequences upstream of the P. aeruginosa SSI genes (Table 2) was unsuccessful.
The largest group of sigma factors in P. aeruginosa is that associated with extracytoplasmic function (the ECF group), of which the best characterized are AlgU and PvdS (15, 21). The 19 ECF sigma factors found in P. aeruginosa reflect the versatility of this species in adapting to a variety of different habitats and responding to environmental signals (33). Under sulfate-limited conditions, P. aeruginosa upregulated transcription of one of these ECF sigma factors, PA2093, and this transcriptional activation was further increased when mucin was present in the growth medium. This gene was located within a larger cluster of sulfate-regulated genes (PA2087 to PA2094) within sulfur island II and was associated with a TonB-dependent transducer system and a putative sulfonatase gene, suggesting that it is involved in sensing extracellular sulfate limitation and especially in responding to the presence of mucin as a sulfur source in the environment. Cells of P. aeruginosa also responded to the presence of mucin with increased transcription of several genes involved in the synthesis of the type III secretion system and exoenzyme S. The upregulation of the type III secretion system genes under the conditions tested here suggests that the cells recognize mucin not only as a source of sulfur but also as a component of the host environment and induce the type III secretion system accordingly. Previous studies on the response of P. aeruginosa to mucus exposure have identified the migA and npt20 genes (PA0705 and PA5499, respectively) as mucin induced (54). These genes were not induced in the present study, possibly because the concentrations of mucin applied here were much lower.
Supplementary Material
Acknowledgments
We are grateful to Gail Preston for assistance with the RpoN analysis, to Daniel Jameson, Andrew Hayes, and Leo Zeef for advice on analyzing the microarray data, and to Carola Tralau for technical assistance. We thank the University of Washington for transposon mutants of P. aeruginosa.
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council to M.K. and by a CNRS ATIP grant to S.V. We are grateful for support from Cystic Fibrosis Foundation Therapeutics.
Footnotes
Published ahead of print on 3 August 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aristoteli, L. P., and M. D. Willcox. 2003. Mucin degradation mechanisms by distinct Pseudomonas aeruginosa isolates in vitro. Infect. Immun. 71:5565-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beil, S., M. A. Kertesz, T. Leisinger, and A. M. Cook. 1996. The assimilation of sulfur from multiple sources and its correlation with expression of the sulfate-starvation-induced stimulon in Pseudomonas putida S-313. Microbiology 142:1989-1995. [DOI] [PubMed] [Google Scholar]
- 4.Bykowski, T., J. R. van der Ploeg, R. Iwanicka-Nowicka, and M. M. Hryniewicz. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43:1347-1358. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, B. J., G. E. Rowe, K. Leiper, and J. M. Rhodes. 2001. Increasing the intra-Golgi pH of cultured LS174T goblet-differentiated cells mimics the decreased mucin sulfation and increased Thomsen-Friedenreich antigen (Gal beta1-3GalNac alpha-) expression seen in colon cancer. Glycobiology 11:385-393. [DOI] [PubMed] [Google Scholar]
- 6.Carnoy, C., A. Scharfman, E. Vanbrussel, G. Lamblin, R. Ramphal, and P. Roussel. 1994. Pseudomonas aeruginosa outer membrane adhesins for human respiratory mucus glycoproteins. Infect. Immun. 62:1896-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chace, K. V., D. S. Leahy, R. Martin, R. Carubelli, M. Flux, and G. P. Sachdev. 1983. Respiratory mucous secretions in patients with cystic fibrosis—relationship between levels of highly sulfated mucin component and severity of the disease. Clin. Chim. Acta 132:143-155. [DOI] [PubMed] [Google Scholar]
- 8.Cornelis, P., and S. Matthijs. 2002. Diversity of siderophore-mediated iron uptake systems in fluorescent pseudomonads: not only pyoverdines. Environ. Microbiol. 4:787-798. [DOI] [PubMed] [Google Scholar]
- 9.Devaraj, N., M. Sheykhnazari, W. S. Warren, and V. P. Bhavanandan. 1994. Differential binding of Pseudomonas aeruginosa to normal and cystic-fibrosis tracheobronchial mucins. Glycobiology 4:307-316. [DOI] [PubMed] [Google Scholar]
- 10.Endoh, T., H. Habe, H. Nojiri, H. Yamane, and T. Omori. 2005. The sigma-54 dependent transcriptional activator SfnR regulates the expression of the Pseudomonas putida sfnFG operon responsible for dimethyl sulphone utilization. Mol. Microbiol. 55:897-911. [DOI] [PubMed] [Google Scholar]
- 11.Gray, K. A., O. S. Pogrebinsky, G. T. Mrachko, L. Xi, D. J. Monticello, and C. H. Squires. 1996. Molecular mechanisms of biocatalytic desulfurization of fossil fuels. Nat. Biotechnol. 14:1705-1709. [DOI] [PubMed] [Google Scholar]
- 12.Gyaneshwar, P., O. Paliy, J. McAuliffe, A. Jones, M. I. Jordan, and S. Kustu. 2005. Lessons from Escherichia coli genes similarly regulated in response to nitrogen and sulfur limitation. Proc. Natl. Acad. Sci. USA 102:3453-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gyaneshwar, P., O. Paliy, J. McAuliffe, D. L. Popham, M. I. Jordan, and S. Kustu. 2005. Sulfur and nitrogen limitation in Escherichia coli K-12: specific homeostatic responses. J. Bacteriol. 187:1074-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagelueken, G., T. M. Adams, L. Wiehlmann, L. Widow, H. Kolmar, B. Tümmler, D. W. Heinz, and W. D. Schubert. 2006. The crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, defines a third class of sulfatases. Proc. Natl. Acad. Sci. USA 103:7631-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmann, J. D. 2002. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 46:47-110. [DOI] [PubMed] [Google Scholar]
- 16.Hryniewicz, M. M., and N. M. Kredich. 1995. Hydroxyl radical footprints and half-site arrangements of binding sites for the CysB transcriptional activator of Salmonella typhimurium. J. Bacteriol. 177:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, B. X., K. Ru, Z. Yuan, C. B. Whitchurch, and J. S. Mattick. 2004. tonB3 is required for normal twitching motility and extracellular assembly of type IV pili. J. Bacteriol. 186:4387-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hummerjohann, J., E. Kuttel, M. Quadroni, J. Ragaller, T. Leisinger, and M. A. Kertesz. 1998. Regulation of the sulfate starvation response in Pseudomonas aeruginosa: role of cysteine biosynthetic intermediates. Microbiology 144:1375-1386. [DOI] [PubMed] [Google Scholar]
- 19.Hummerjohann, J., S. Laudenbach, J. Rétey, T. Leisinger, and M. A. Kertesz. 2000. The sulfur-regulated arylsulfatase gene cluster of Pseudomonas aeruginosa, a new member of the cys regulon. J. Bacteriol. 182:2055-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jansen, H. J., C. A. Hart, J. M. Rhodes, J. R. Saunders, and J. W. Smalley. 1999. A novel mucin-sulphatase activity found in Burkholderia cepacia and Pseudomonas aeruginosa. J. Med. Microbiol. 48:551-557. [DOI] [PubMed] [Google Scholar]
- 21.Jensen, L. J., M. Skovgaard, T. Sicheritz-Ponten, N. T. Hansen, H. Johannson, M. K. Joergensen, K. Kiil, P. F. Hallin, and D. W. Ussery. 2004. Comparative genomics of four Pseudomonas species, p. 139-164. In J.-L. Ramos (ed.), Pseudomonas, vol. 1. Genomics, lifestyle and molecular architecture. Kluwer Academic, New York, NY. [Google Scholar]
- 22.Kertesz, M. 2004. Metabolism of sulphur-containing organic compounds, p. 323-357. In J.-L. Ramos (ed.), Pseudomonas, vol. 3. Biosynthesis of macromolecules and molecular metabolism. Kluwer Academic/Plenum, New York, NY. [Google Scholar]
- 23.Kertesz, M. A. 2000. Riding the sulfur cycle—metabolism of sulfonates and sulfate esters in Gram-negative bacteria. FEMS Microbiol. Rev. 24:135-175. [DOI] [PubMed] [Google Scholar]
- 24.Kertesz, M. A., T. Leisinger, and A. M. Cook. 1993. Proteins induced by sulfate limitation in Escherichia coli, Pseudomonas putida, or Staphylococcus aureus. J. Bacteriol. 175:1187-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kertesz, M. A., K. Schmidt-Larbig, and T. Wüest. 1999. A novel reduced flavin mononucleotide-dependent methanesulfonate sulfonatase encoded by the sulfur-regulated msu operon of Pseudomonas aeruginosa. J. Bacteriol. 181:1464-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koebnik, R. 2005. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 13:343-347. [DOI] [PubMed] [Google Scholar]
- 27.Kulasekara, B. R., and S. Lory. 2004. The genome of Pseudomonas aeruginosa, p. 47-75. In J.-L. Ramos (ed.), Pseudomonas, vol. 1. Genomics, lifestyle and molecular architecture. Kluwer Academic, New York, NY. [Google Scholar]
- 28.Lam, J., R. Chan, K. Lam, and J. W. Costerton. 1980. Production of mucoid microcolonies by Pseudomonas aeruginosa within infected lungs in cystic fibrosis. Infect. Immun. 28:546-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamblin, G., S. Degroote, J. M. Perini, P. Delmotte, A. E. Scharfman, M. Davril, J. M. Lo-Guidice, N. Houdret, V. Dumur, A. Klein, and P. Roussel. 2001. Human airway mucin glycosylation: a combinatory of carbohydrate determinants which vary in cystic fibrosis. Glycoconjugate J. 18:661-684. [DOI] [PubMed] [Google Scholar]
- 30.Li, C., and W. H. Wong. 2003. DNA-Chip analyzer (dChip), p. 120-141. In G. Parmigiani, E. S. Garett, R. Irizarry, and S. L. Zeger (ed.), The analysis of gene expression data: methods and software. Springer, New York, NY.
- 31.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 32.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oguiza, J. A., K. Kiil, and D. W. Ussery. 2005. Extracytoplasmic function sigma factors in Pseudomonas syringae. Trends Microbiol. 13:565-568. [DOI] [PubMed] [Google Scholar]
- 34.Quadroni, M., P. James, P. Dainese-Hatt, and M. A. Kertesz. 1999. Proteome mapping, mass spectrometric sequencing and reverse transcriptase-PCR for characterisation of the sulfate starvation-induced response in Pseudomonas aeruginosa PAO1. Eur. J. Biochem. 266:986-996. [DOI] [PubMed] [Google Scholar]
- 35.Quadroni, M., W. Staudenmann, M. Kertesz, and P. James. 1996. Analysis of global responses by protein and peptide fingerprinting of proteins isolated by two-dimensional gel electrophoresis: application to the sulfate-starvation response of Escherichia coli. Eur. J. Biochem. 239:773-781. [DOI] [PubMed] [Google Scholar]
- 36.Robertson, A. M., and D. P. Wright. 1997. Bacterial glycosulphatases and sulphomucin degradation. Can. J. Gastroenterol. 11:361-366. [DOI] [PubMed] [Google Scholar]
- 37.Rocha, E. P. C., A. Sekowska, and A. Danchin. 2000. Sulphur islands in the Escherichia coli genome: markers of the cell's architecture? FEBS Lett. 476:8-11. [DOI] [PubMed] [Google Scholar]
- 38.Salunkhe, P., T. Topfer, J. Buer, and B. Tummler. 2005. Genome-wide transcriptional profiling of the steady-state response of Pseudomonas aeruginosa to hydrogen peroxide. J. Bacteriol. 187:2565-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 40.Schuster, M., A. C. Hawkins, C. S. Harwood, and E. P. Greenberg. 2004. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51:973-985. [DOI] [PubMed] [Google Scholar]
- 41.Scott, C., M. E. Hilton, C. W. Coppin, R. J. Russell, J. G. Oakeshott, and T. D. Sutherland. 2007. A global response to sulfur starvation in Pseudomonas putida and its relationship to the expression of low-sulfur-content proteins. FEMS Microbiol. Lett. 267:184-193. [DOI] [PubMed] [Google Scholar]
- 42.Smart, C. H. M., M. J. Walshaw, C. A. Hart, and C. Winstanley. 2006. Use of suppression subtractive hybridization to examine the accessory genome of the Liverpool cystic fibrosis epidemic strain of Pseudomonas aeruginosa. J. Med. Microbiol. 55:677-688. [DOI] [PubMed] [Google Scholar]
- 43.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sriramulu, D. D., H. Lunsdorf, J. S. Lam, and U. Römling. 2005. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J. Med. Microbiol. 54:667-676. [DOI] [PubMed] [Google Scholar]
- 45.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 46.Studholme, D. J., and R. Dixon. 2003. Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185:1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai, H. H., C. A. Hart, and J. M. Rhodes. 1991. Production of mucin degrading sulphatases and glycosidases by Bacteroides thetaiotaomicron. Lett. Appl. Microbiol. 13:97-101. [Google Scholar]
- 49.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Ploeg, J. R., R. Iwanicka-Nowicka, T. Bykowski, M. Hryniewicz, and T. Leisinger. 1999. The Cbl-regulated ssuEADCB gene cluster is required for aliphatic sulfonate-sulfur utilization in Escherichia coli. J. Biol. Chem. 174:29358-29365. [DOI] [PubMed] [Google Scholar]
- 51.van der Ploeg, J. R., R. Iwanicka-Nowicka, M. A. Kertesz, T. Leisinger, and M. M. Hryniewicz. 1997. Involvement of CysB and Cbl regulatory proteins in expression of the tauABCD operon and other sulfate starvation-inducible genes in Escherichia coli. J. Bacteriol. 179:7671-7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermeij, P., C. Wietek, A. Kahnert, T. Wüest, and M. A. Kertesz. 1999. Genetic organization of sulfur-controlled aryl desulfonation in Pseudomonas putida S-313. Mol. Microbiol. 32:913-926. [DOI] [PubMed] [Google Scholar]
- 53.Vuilleumier, S., and M. Pagni. 2002. The elusive roles of bacterial glutathione S-transferases: new lessons from genomes. Appl. Microbiol. Biotechnol. 58:138-146. [DOI] [PubMed] [Google Scholar]
- 54.Wang, J. Y., S. Lory, R. Ramphal, and S. G. Jin. 1996. Isolation and characterization of Pseudomonas aeruginosa genes inducible by respiratory mucus derived from cystic fibrosis patients. Mol. Microbiol. 22:1005-1012. [DOI] [PubMed] [Google Scholar]
- 55.Wolfgang, M. C., B. R. Kulasekara, X. Y. Liang, D. Boyd, K. Wu, Q. Yang, C. G. Miyada, and S. Lory. 2003. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:8484-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, Z. J., R. A. Irizarry, R. Gentleman, F. Martinez-Murillo, and F. Spencer. 2004. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 99:909-917. [Google Scholar]
- 57.Xia, B. Y., J. A. Royall, G. Damera, G. P. Sachdev, and R. D. Cummings. 2005. Altered O-glycosylation and sulfation of airway mucins associated with cystic fibrosis. Glycobiology 15:747-775. [DOI] [PubMed] [Google Scholar]
- 58.Zhao, Q. X., and K. Poole. 2000. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol. Lett. 184:127-132. [DOI] [PubMed] [Google Scholar]
- 59.Zhong, S., C. Li, and W. H. Wong. 2003. Chip Info: software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res. 3:3483-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.