Abstract
Hepatitis B virus (HBV) infections are a major worldwide health problem with chronic infections leading to cirrhosis and liver cancer. Viruses related to human HBV have been isolated from birds and rodents, but despite efforts to find hepadnaviruses that infect species intermediate in evolution between rodents and humans, none have been described. We recently isolated a hepadnavirus from a woolly monkey (Lagothrix lagotricha) that was suffering from fulminant hepatitis. Phylogenetic analysis of the nucleotide sequences of the core and surface genes indicated that the virus was distinct from the human HBV family, and because it is basal (ancestral) to the human monophyletic group, it probably represents a progenitor of the human viruses. This virus was designated woolly monkey hepatitis B virus (WMHBV). Analysis of woolly monkey colonies at five zoos indicated that WMHBV infections occurred in most of the animals at the Louisville zoo but not at four other zoos in the United States. The host range of WMHBV was examined by inoculation of one chimpanzee and two black-handed spider monkeys (Ateles geoffroyi), the closest nonendangered relative of the woolly monkey. The data suggest that spider monkeys are susceptible to infection with WMHBV and that minimal replication was observed in a chimpanzee. Thus, we have isolated a hepadnavirus with a host intermediate between humans and rodents and establishes a new animal model for evaluation of antiviral therapies for treating HBV chronic infections.
Hepatitis B virus (HBV) infections represent a worldwide health problem. The acute infection is usually self-limiting in adults but may be fatal in extreme cases. However, 5–10% of infected adults and up to 95% of infected newborns become chronic carriers. Chronic infections progress to cirrhosis and hepatocellular carcinoma in approximately 40% of those infected as newborns and 5% of infected adults (1). Worldwide there are an estimated 300 million chronic carriers with 1.5 million residing in the United States, making HBV infections the fourth leading cause of death due to infectious disease worldwide (2).
An efficacious vaccine has been developed, and it is currently recommended that it be included as one of the standard childhood vaccines. Although this will eventually lessen the incidence of this infectious disease, it will provide no benefit to the millions already infected. Current research is aimed at developing improved antiviral strategies., Several nonprimate animal models are available for in vivo testing of antivirals. Hepadnaviruses have been isolated from several species of birds and rodents including duck HBV (3), heron hepatitis virus (4), woodchuck hepatitis virus (5), ground squirrel hepatitis virus (6), and arctic ground squirrel hepatitis virus (7). Both ducks and woodchucks represent readily available small animal models, but in each case the representative viral genome is divergent from HBV and the metabolism of the model animal is very different from humans.
Despite extensive efforts to identify other members of this family (8), the only primate virus to be isolated is human HBV, with the possible exception of isolates from a chimpanzee (9) and a gibbon (10), both of which cluster genetically within the human HBV family and probably represent human viruses. The best animal model to date for infections with human HBV is the chimpanzee, but chimpanzees are highly intelligent and endangered animals. Thus, a critical need exists to develop small primate models with hepadnaviruses closely related to HBV. We have recently isolated a primate hepadnavirus from a New World monkey, and we propose this host–virus system as a significantly improved animal model for HBV research.
MATERIALS AND METHODS
Animals.
Serum samples from woolly monkeys housed at five zoos were examined. The samples were obtained from archived sources or as part of routine physical exams. The following zoos provided samples for analysis and agreed to be acknowledged for their contribution: Louisville Zoological Gardens, Louisville, KY; Woodlands Park Zoological Gardens, Seattle, WA; Pittsburgh Zoo, Pittsburgh, PA; and Columbus Zoological Garden, Columbus, OH. All of the samples from zoos other than Louisville were negative for woolly monkey hepatitis B virus (WMHBV) markers. The two black-handed spider monkeys and the two chimpanzees were housed at the Southwest Foundation for Biomedical Research. All procedures were approved by the Southwest Foundation for Biomedical Research Animal Research Committee.
PCR, Hybridization, and Sequence Analysis.
PCRs and Southern hybridization analyses were conducted exactly as described (11). Primers for PCR amplification were selected from a highly conserved domain overlapping the core gene. The forward primer 5′-CCTTGGGTGGCTTTGGGGCA-3′ spanned nucleotides 1884–1904, using the EcoRI site of the ayw3 sequence (12) as nucleotide 1, and the reverse primer 5′-GGGCATTTGGTGGTCTATA-3′, spanned nucleotides 2295–2274. HBV DNA was isolated from serum by digestion with proteinase K followed by phenol/chloroform extraction and ethanol precipitation as described (11). The DNA from 10 μl of serum or a dilution of serum was analyzed by PCR amplification with 3.6 min at 94°C for an initial denaturation step; followed by 35 cycles of 1 min at 94°C, 2 min at 42°C, and 3 min at 72°C; and a final step of 7 min at 72°C. Samples were analyzed by agarose gel electrophoresis and Southern blot hybridization with a 32P-labeled random-primed DNA probe. High-stringency hybridization conditions were in 50% formamide/7% SDS/0.25 M sodium phosphate, pH 7.2/0.25 M NaCl/1 mM EDTA at 42°C with washes in 0.1× SSC/0.1% SDS at 60°C. Low-stringency hybridization conditions were conducted in the same buffer at 37°C with washes in 0.2× SSC/0.1% SDS at 42°C.
The WMHBV genome was cloned as multiple overlapping PCR products with the pZErO-1 vector (Invitrogen), and sequence determinations were performed by automated dideoxynucleotide sequencing at the University of Texas Health Science Center in San Antonio sequencing core laboratory. The nucleotide sequences and encoded amino acids were aligned with the clustal program and percent similarities were calculated with the megalign program in the lasergene software package (DNAstar, Madison, WI).
Phylogenetic Analysis.
Phylogenetic analyses of nucleotide sequences were performed by using test version 4.0d57 of paup*, written by David Swofford (to be distributed by Sinauer Associates, Sunderland, MA). Maximum parsimony and maximum likelihood analyses were done by using unweighted characters, with gaps in sequences treated as missing characters. Results of exhaustive searches made statistical support for trees unnecessary (e.g., bootstrap). The GenBank accession numbers for the hepadnaviruses compared herein are as follows: woodchuck, J02442; chimpanzee, D00220; gibbon, U46935; human/adr/Japan, L08805; human/ayr/Japan, X04615; human/adw2/USA, X02763; human/adw2/Indonesia, M54923; human/ayw4/Senegal, X75664; human/ayw3/France, V01460; human/adw4/Colombia, X75663; and WMHBV, AF046996.
RESULTS AND DISCUSSION
Identification of a Hepadnavirus from a Woolly Monkey.
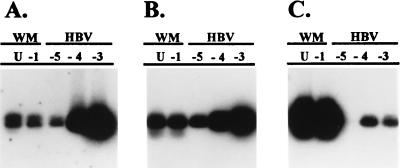
Recently, we had the opportunity to examine the serum from a woolly monkey (Lagothrix lagotricha) experiencing fulminant hepatitis at the Louisville Zoological Gardens. By using primers to the core gene that are conserved among all isolates of HBV, we obtained a PCR product of the correct size that was visible by ethidium bromide staining of the agarose gel; however, the product hybridized poorly with a HBV probe. With stringent hybridization conditions and the human HBV probe, greater hybridization was observed for the human HBV sample, even though much less DNA was present for the human sample (Fig. 1A). Use of lower-stringency hybridization conditions resulted in increased hybridization with the PCR product from the woolly monkey serum that was more proportional to the amount of DNA present (Fig. 1B). Use of the PCR fragment from the woolly monkey serum as a probe yielded very intense hybridization to the PCR products from the woolly monkey serum and greatly reduced hybridization to HBV DNA (Fig. 1C). Sequence analysis confirmed that the product amplified from the woolly monkey serum represented a hepadnavirus core gene that was considerably divergent from human HBV. These results suggested that we had identified a hepadnavirus from a New World primate, and the virus was provisionally designated woolly monkey hepatitis B virus (WMHBV).
Figure 1.
Southern blot hybridization of PCR products amplified from woolly monkey (lanes WM) serum or HBV containing serum. DNA was extracted from 1:10 dilutions of woolly monkey or HBV containing serum. U, undiluted; −1 to −5 = 10−1 to 10−5 dilutions. PCR was conducted with primers to the core gene. Hybridization of identical membranes was performed with an HBV DNA probe under high (A) or low (B) stringency conditions or with the isolated DNA fragment from amplification of the woolly monkey serum (C) under low-stringency conditions.
Demonstration of Chronic WMHBV Infections in Woolly Monkeys.
The initial isolate was obtained from a woolly monkey exhibiting fulminant hepatitis that resulted in death of the animal. The Louisville Zoo houses one of the most successful breeding programs of this endangered primate, so it was of immediate concern to determine the extent of the infection in the colony and in colonies housed at other zoos. Examination of sera from 16 animals from the Louisville colony revealed that 9 animals were positive by PCR for WMHBV DNA and 7 of these for which sufficient serum was available for testing were also positive by ELISA for HBV surface antigen (HBsAg). One animal was positive for antibody to HBsAg and was weakly positive (+/−) by PCR. Ten animals were positive for antibodies to HBV core antigen (HBcAg), 3 of which were negative for all other markers. Thus, of the animals tested from the Louisville Zoo colony, 81% (13 of 16) showed signs of ongoing or previous infections with WMHBV (Table 1 and Fig. 2). The animal with anti-HBsAg and the 3 animals for which anti-HBcAg was the only WMHBV marker had presumably convalesced from their infections, whereas the 9 PCR-positive animals had profiles consistent with either acute or chronic infections. PCR analysis of archived sera demonstrated that many of the infections were chronic and were present in the colony at least 9 years ago. In contrast, sera from 18 animals housed at four zoos with no relationship to the Louisville colony were tested and found to be negative for all WMHBV markers.
Table 1.
Virological status of woolly monkeys
| Name | PCR | HBsAg | Anti-HBsAg | Anti-HBcAg | All markers |
|---|---|---|---|---|---|
| Judy | − | ND | − | + | + |
| Cindy | + | + | ND | ND | + |
| Kizzy | ± | − | + | + | + |
| Zoey | + | + | ND | + | + |
| Phoebe | + | + | − | + | + |
| Mojo | + | + | − | − | + |
| Mr. B | − | − | − | − | − |
| Corey | + | + | − | + | + |
| Jethro | − | − | − | + | + |
| Sissi | − | − | − | − | − |
| Pancho | − | − | ND | + | + |
| Henry | − | − | − | − | − |
| Jay | + | ND | ND | + | + |
| Will | + | + | − | + | + |
| Brent | + | ND | ND | + | + |
| Lyle | + | + | − | − | + |
| Total positive | 9/15 | 7/13 | 1/10 | 10/15 | 13/16 |
| (60%) | (54%) | (10%) | (66%) | (81%) |
ND, not done (insufficient sample).
Figure 2.
Family tree of woolly monkeys from the Louisville Zoo. Shaded circles indicate animals from Scotland. The initials of two sires Herman (H) and Willie (W) are indicated to clarify parentage. ?, sire is unknown; open squares, male; open circles, female; *, index case; + or −, positive and negative for WMHBV markers. Serum samples for some animals were not available for analysis.
The Louisville colony is composed of animals from various sources. The family tree of animals currently housed at the Louisville zoo or pertinent to the establishment of the breeder colony is shown in Fig. 2. Animals were mostly donations from private owners until 1985 when six animals were obtained from a zoo in Scotland. Five of these animals are indicated by the shaded circles. Archived sera were available from four of these animals and all were positive for WMHBV markers. One of the samples dated back to 1989, suggesting that the source of the infections may trace back to Scotland. The zoo in Scotland is no longer in existence; thus, samples from this source were not available for testing. Of the four founding males in the colony, only two were available for testing and only one was positive. Samples from the founding dam were not available, but her three female offspring were positive, each of which was born in Scotland, and 80% of the offspring from these females were positive as well.
The pattern of WMHBV infections observed in the Louisville colony is consistent with vertical transmission of the virus. Approximately 90% of infants infected with HBV at birth will become long-term chronic carriers, but only 5–10% of exposed adults will develop a chronic infection (13). Chronic infections with hepadnaviruses are often asymptomatic for extended periods of time (decades in humans) especially when acquired at birth from the mother. Nonetheless, the chronic infections often progress to cirrhosis and liver cancer after several decades (1). To explore the possibility that some deaths of woolly monkeys in zoos in the United States might be related to chronic infection with WMHBV, the autopsy reports from 74 animals that died in captivity since 1974 were examined. Abnormal liver findings were cited in 28 of the reports with hepatitis and liver necrosis being the most common pathologies. Although cirrhosis and hepatocellular carcinoma were not reported, the high prevalence of liver-related disease is suggestive of a role for WMHBV in some deaths of captive woolly monkeys.
Analysis of the WMHBV Genome.
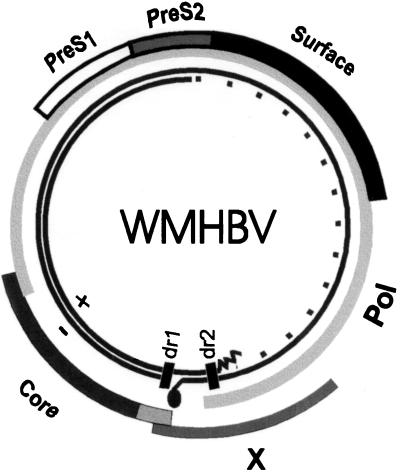
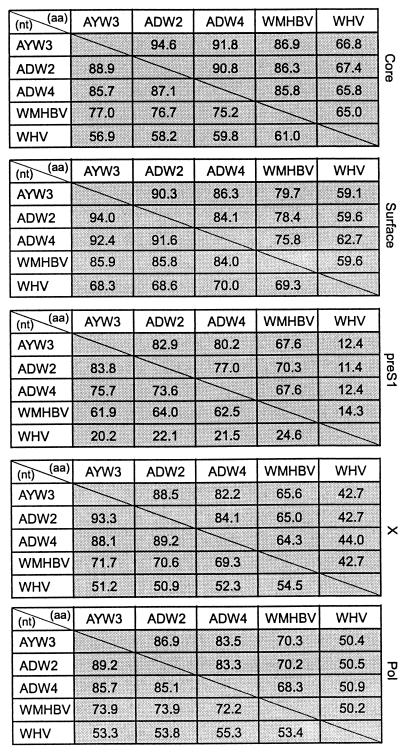
The WMHBV genome was cloned as multiple overlapping PCR fragments. The genome is 3,179 nucleotides long and has the same genetic organization as human HBV (Fig. 3). The different reading frames were compared for the percent similarity at both the nucleotide and amino acid level to three human HBV isolates from genotypes A, D, and F and a woodchuck HBV isolate (Fig. 4). Most of the genome of hepadnaviruses is evolutionarily constrained due to overlap with the polymerase ORFs (14). The pol ORF overlaps the entire surface gene and most of the X gene, but only the carboxyl-terminal 48 amino acids of core (Fig. 3). Despite this constraint, the WMHBV genome is considerably divergent from the human isolates, and the core gene is the most highly conserved, probably due to the constraint of forming an icosahedral nucleocapsid. The core gene of WMHBV exhibited 85.8–86.9% similarity to the human isolates at the amino acid level, and 75.2–77% similarity at the nucleotide level. The woodchuck core gene was 65 and 61% similar to WMHBV at the amino acid and nucleotide level, respectively. The X gene was the most divergent with 64.3–65.6% similarity between WMHBV and the HBV isolates at the amino acid level, whereas the human isolates exhibited 82.2–88.5% similarity among themselves. The pre-S1 region was nearly as divergent as the X gene, whereas the surface and polymerase genes were intermediate between core and X. The level of similarity between the polymerase genes of WMHBV and the human HBV isolates was unexpectedly low and was due in part to a high level of divergence in the spacer domain. The WMHBV amino acid sequence was approximately 66% divergent from the human isolates in this area.
Figure 3.
Genome organization of WMHBV. The WMHBV genome has the same genetic organization as human HBV. The outer bars show the various HBV ORFs. The inner circle depicts the viral DNA present in the virion that is partially double stranded with the polymerase covalently bound to the 5′ end of the minus strand of DNA and a viral RNA primer at the 5′ end of the plus strand of DNA. The small bars represent the DR1 and DR2 sequences involved in genomic replication.
Figure 4.
Comparison of amino acid and nucleotide similarity between hepadnaviruses. The sequences of WMHBV, three human HBV isolates (ayw3/France, genotype D; adw2/Indonesia, genotype B; and adw4/Colombia, genotype F), and woodchuck HBV are compared for percent similarity by using the megalign program of lasergene. Comparisons of the ORFs of the core, surface, pre-S1, X, and polymerase (pol) are shown. Numbers above and below the diagonal line are percent amino acid and nucleotide similarity, respectively.
Phylogenetic Analysis of the WMHBV Genome.
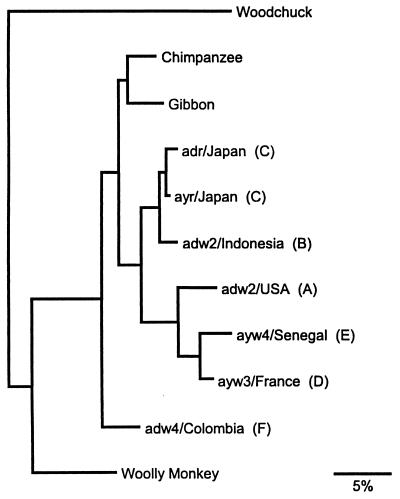
Phylogenetic analyses were performed to better determine the evolutionary relationship of WMHBV to the hepadnaviridae family. The nucleotide sequences of the core and surface genes of WMHBV were compared with seven human isolates representing genotypes A–F, a chimpanzee isolate, and a gibbon isolate, and woodchuck HBV was used as an outgroup to root the trees. Exhaustive searches for the most parsimonious trees yielded one tree when using core sequences and three trees when using surface sequences. The consensus (majority rule of the latter) of these is shown in Fig. 5. Maximum likelihood analyses yielded similar trees with only minor variations in the clustering of three isolates [adw2/Indonesia (type B), adw2/USA (type A), and adw4/Colombia (type F)]. The relationships between the human and other primate viruses were in general agreement with previous phylogenetic analyses, using complete genomic sequences (15, 16) or individual genes (14, 17). Contrary to earlier reports (10), our analyses suggest that the chimpanzee and gibbon viruses most likely originated from exposure to a human virus, because they were more closely related to human genotypes A–E than was the representative human genotype F virus (adw4/Colombia). The F genotype viruses are the most divergent of the human viruses and have been postulated to be the original New World strains (18). In this analysis, the closest relative to WMHBV was a human isolate from Colombia that represents the F genotype. The results demonstrate that WMHBV is distinct from the human HBV family, and because it is basal (ancestral) to the human monophyletic group, it probably represents a progenitor of the human viruses. Whether the woolly monkey virus adapted to infect humans in South and Central America remains unknown.
Figure 5.
Phylogenetic tree of hepadnaviruses. The consensus phylogenetic tree was based on nucleotide sequences from the core and surface genes of human, chimpanzee, gibbon, woolly monkey, and woodchuck HBV isolates. Distance along the horizontal axis among isolates is proportional to genetic divergence. The scale at the bottom represents 5% divergence. The genotypes (A–F) of the human HBV isolates are given in parentheses. The GenBank accession numbers for the various isolates are given in Materials and Methods.
Host Range of WMHBV and Establishment of a Small Primate Model for Antiviral Testing.
We have begun an analysis of the host range of WMHBV to establish a small primate model for HBV infections. Woolly monkeys are endangered and thus are not available for experimental purposes. Because hepadnaviruses typically have very restricted host ranges, we began our analyses with a close relative of the woolly monkey, the black-handed spider monkey (Ateles geoffroyi); both primates are within the Atelidae family and the Atelinae subfamily. Two animals were inoculated with 0.5 ml of woolly monkey serum containing approximately 109 genome equivalents per ml. The OD492 on the HBsAg ELISA was >2 within 1 week of inoculation and remained elevated for 10 weeks in one animal and 6 weeks in the other (Fig. 6 A and B). The animals were PCR-positive for WMHBV DNA for 6–4 weeks, with approximately 105 genome equivalent per ml. Antibodies to HBcAg were first detected on week 6. No antibodies to HBsAg were detected, but it is not certain that antibodies to WMHBV HBsAg react in the ELISA for human HBV anti-HBsAg, because only one of the four woolly monkeys that had apparently convalesced had anti-HBsAg reactivity in this assay (Table 1). No rise in alanine amino transferase was observed, and a liver biopsy taken from the animal shown in Fig. 5A on week 6 showed no abnormality. It appears that spider monkeys are susceptible to infection with WMHBV, but in these two animals, the infections were subclinical.
Figure 6.
WMHBV infections in spider monkeys and a chimpanzee. Two black-handed spider monkeys (A and B) and a chimpanzee (C) were inoculated with woolly monkey serum. The animals were monitored for HBsAg and antibodies to HBcAg by using diagnostic ELISAs from Abbott Laboratories and for WMHBV DNA by PCR. The anti-HBcAg assay is an inhibition assay in which lower OD492 values indicate positive responses.
We also have inoculated a chimpanzee with the same woolly monkey serum. The infection in this case appears minimal, but some replication appears to have occurred. The inoculum had an OD of 0.15 in the HBsAg ELISA, yet by week 1, the OD of the chimpanzee serum was >2 (Fig. 6C). This strongly argues against the reactivity being due to carry-over of the inoculum. The OD began to decline by week 3 but was considered positive in the assay until week 12. The level of replication was very low with approximately 103 genome equivalents per ml in the serum. Seroconversion for anti-HBcAg and anti-HBsAg was not observed. No rise in alanine amino transferase was observed and the histopathology was normal. These data suggest that the chimpanzee is minimally susceptible to infection with WMHBV. In a separate experiment, a chimpanzee that had convalesced from a previous infection with HBV was resistant to challenge with WMHBV, because no HBsAg or WMHBV DNA was detected in the serum over a prolonged time course. Although preliminary, these data suggest that cross-protective immunity between the two viruses may exist. Protection could be conferred by neutralizing antibodies to HBsAg or by a cytotoxic T lymphocyte response to any HBV antigen including the core antigen. Further studies are required to determine whether primates intermediate between the spider monkey and the chimpanzee are susceptible to infection with WMHBV and whether human HBV can infect spider monkeys.
A significant need exists for better animal models of HBV infections to test antiviral strategies for eliminating chronic liver disease. Currently, chimpanzees are the best model for HBV infections, but due to the large size, great expense, limited availability, and the highly intelligent nature of these animals, they are reserved for essential experiments. Considerable experimentation is performed with both the duck and woodchuck hepadnaviruses. However, due to the high level of divergence between HBV and these viruses and the considerable metabolic differences between their hosts and humans, the utility of these models is often limited. The isolation of WMHBV provides an opportunity to establish a new animal model to test antiviral strategies. The susceptibility of the black-handed spider monkey to infection with WMHBV suggests that development of this animal model is feasible. The reactivity of WMHBV surface antigen in the standard ELISA for HBsAg and the apparent cross-protection to WMHBV observed for the chimpanzee previously infected with HBV suggest that it may be possible to prevent future infections of woolly monkeys by use of the human vaccine. Alternatively, a vaccine specific to WMHBV can be produced, thus protecting this endangered primate.
Acknowledgments
This work was supported in part by National Institutes of Health Grants CA53246 and AI01124.
ABBREVIATIONS
- HBV
hepatitis B virus
- WMHBV
woolly monkey hepatitis B virus
- HBsAg
HBV surface antigen
- HBcAg
HBV core antigen
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF046996 for WMHBV).
References
- 1.Beasley R P, Hwang L-Y, Lin C-C, Chien C-S. Lancet. 1981;ii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 2.Young P. ASM News. 1996;62:450–451. [Google Scholar]
- 3.Mason W S, Seal G, Summers J. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprengel R, Kaleta E F, Will H. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Summers J, Smolec J M, Snyder R. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marion P L, Oshiro L S, Regnery D C, Scullard G H, Robinson W S. Proc Natl Acad Sci USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Testut P, Renard C-A, Terradillos O, Vitvitski-Trepo V, Tekaia F, Degott C, Blake J, Boyer B, Buendia M A. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deinhardt F. Adv Virus Res. 1976;20:113–157. doi: 10.1016/s0065-3527(08)60503-5. [DOI] [PubMed] [Google Scholar]
- 9.Vaudin M, Wolstenholme A J, Tsiquaye K N, Zuckerman A J, Harrison T J. J Gen Virol. 1988;69:1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]
- 10.Norder H, Ebert J W, Fields H A, Mushahwar I K, Magnius L O. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 11.Lanford R E, Michaels M G, Chavez D, Brasky K, Fung J, Starzl T E. J Med Virol. 1995;46:207–212. doi: 10.1002/jmv.1890460307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nature (London) 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 13.Ganem D. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 2703–2737. [Google Scholar]
- 14.Mizokami, M., Orito, E., Ohba, K., Ikeo, K., Lau, J. Y. N. & Gojobori, T. (1997) J. Mol. Evol. 44, Suppl. 1, S83–S90. [DOI] [PubMed]
- 15.Norder H, Couroucé A-M, Magnius L O. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R, Imai M, Miyakawa Y, Mayumi M. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 17.Orito E, Mizokami M, Ina Y, Moriyam E N, Kameshima N, Yamamoto M, Gojobori T. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arauz-Ruiz P, Norder H, Visona K A, Magnius L O. J Infect Dis. 1997;176:851–858. doi: 10.1086/516507. [DOI] [PubMed] [Google Scholar]