Abstract
Nucleotide pool sanitizing enzymes Dut (dUTPase), RdgB (dITPase), and MutT (8-oxo-dGTPase) of Escherichia coli hydrolyze noncanonical DNA precursors to prevent incorporation of base analogs into DNA. Previous studies reported dramatic AT→CG mutagenesis in mutT mutants, suggesting a considerable density of 8-oxo-G in DNA that should cause frequent excision and chromosomal fragmentation, irreparable in the absence of RecBCD-catalyzed repair and similar to the lethality of dut recBC and rdgB recBC double mutants. In contrast, we found mutT recBC double mutants viable with no signs of chromosomal fragmentation. Overproduction of the MutM and MutY DNA glycosylases, both acting on DNA containing 8-oxo-G, still yields no lethality in mutT recBC double mutants. Plasmid DNA, extracted from mutT mutM double mutant cells and treated with MutM in vitro, shows no increased relaxation, indicating no additional 8-oxo-G modifications. Our ΔmutT allele elevates the AT→CG transversion rate 27,000-fold, consistent with published reports. However, the rate of AT→CG transversions in our mutT+ progenitor strain is some two orders of magnitude lower than in previous studies, which lowers the absolute rate of mutagenesis in ΔmutT derivatives, translating into less than four 8-oxo-G modifications per genome equivalent, which is too low to cause the expected effects. Introduction of various additional mutations in the ΔmutT strain or treatment with oxidative agents failed to increase the mutagenesis even twofold. We conclude that, in contrast to the previous studies, there is not enough 8-oxo-G in the DNA of mutT mutants to cause elevated excision repair that would trigger chromosomal fragmentation.
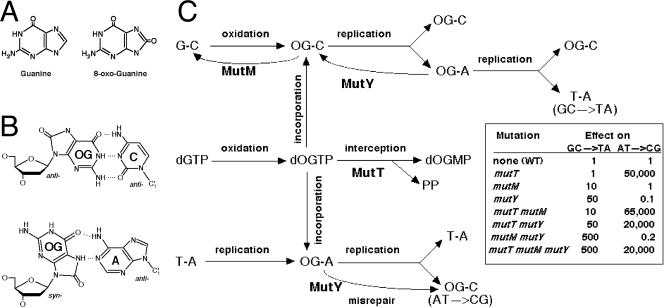
To avoid mutagenesis, it is important to remove base analogs not only from DNA itself but also from the DNA precursor pools, for example, by hydrolyzing modified deoxynucleoside triphosphates. MutT of Escherichia coli is an example of such a nucleoside triphosphatase that in vitro hydrolyzes an oxidized form of dGTP that contains 8-oxo-G (7,8-dihydro-8-oxoguanine) (54) (Fig. 1A). The proofreading-deficient variants of both DNA polymerase III (Pol III; the replicative polymerase) and DNA Pol I (the main repair polymerase) of E. coli are capable of inserting this modified guanine with equal, but low, efficiency across cytosine or adenine (54, 68), with other DNA polymerases showing preferences one way or the other (22). In the 8-oxo-G · C scenario, both C and 8-oxo-G bases are in the normal anti conformation, whereas in the 8-oxo-G · A scenario, the 8-oxo-guanine assumes the syn conformation to pair with adenine, another purine (Fig. 1B) (55). If 8-oxo-G in syn conformation is incorporated opposite the A residue in the template strand but during the subsequent replication round flips back to the normal anti conformation to pair with the C residue, this leads to the AT→CG transversion (Fig. 1C, the bottom pathway). The mutT mutant of E. coli is specifically elevated for the AT→CG transversions and is one of the strongest mutators known, with a 10,000- to 50,000-fold increase over the wild type (17, 27, 78) (Fig. 1C, inset table). The MutT enzyme is the founding member of the big class of enzymes, called Nudix hydrolases (of which there are 13 in E. coli alone) that are proposed to target abnormal nucleotides, thus cleansing the nucleotide pools of the cell (6, 28).
FIG. 1.
8-Oxo-guanine, its alternative pairing schemes, as well as the Mut proteins that counteract potential mutagenic consequences of 8-oxo-G (OG). (A) The structure of guanine compared to that of 8-oxo-guanine. (B) 8-Oxo-G · C pair (both in the anti conformation) versus 8-oxo-G · A pair (8-oxo-G is in the syn conformation). (C) The current understanding of the 8-oxo-G mutation-avoidance pathways (after references 27, 57, and 73). The top pathway leads toward the GC→TA transversion through A incorporation across template 8-oxo-G and is counteracted by MutM and MutY. Note that besides DNA-guanine oxidation, 8-oxo-guanine incorporation opposite the correct C residue without subsequent excision should also cause an increase in the GC→TA transversion down the line, as the template 8-oxo-G invites misincorporation of the A residue. The middle pathway shows 8-oxo-dGTP interception by MutT. The bottom pathway shows the AT→CG transversion through 8-oxo-G incorporation across the A residue in the template DNA and its enhancement by MutY. The table shows published data (27, 78) that were normalized to the wild type and rounded up to illustrate the logic of the scheme.
8-Oxo-G can form not only in the DNA precursor pools but also in DNA itself, causing the GC→TA transversions (Fig. 1C, top pathway). E. coli is proposed to employ the MutM and MutY glycosylases to prevent this mutagenesis due to 8-oxo-G formation in DNA (57). MutM is responsible for removing 8-oxo-G across from cytosine (9) while MutY removes a misincorporated adenine across from an existing 8-oxo-G in the template strand (3, 58) (Fig. 1C, top pathway). MutY also wrongly attacks DNA when 8-oxo-G misincorporates across template A, judging by the decreased AT→CG mutagenesis in the mutT mutY double mutants relative to the mutT single mutants (27, 78) (Fig. 1C, inset table). As MutY removes the correct adenine in this case, it leaves the newly incorporated 8-oxo-guanine in the opposite strand to be later paired with cytosine, causing the transversion (Fig. 1C, bottom pathway).
8-Oxo-G is not the only base analog whose incorporation or formation within DNA is actively countered by specialized enzymes. In an analogous situation, the product of the dut gene in E. coli (31), a dUTPase (5, 30), hydrolyzes another noncanonical DNA precursor, dUTP, to prevent uracil incorporation into the replicating DNA in place of thymine (79). On the other hand, if uracil forms within DNA as a result of cytosine deamination (52), uracil DNA glycosylase (UDG), with the help of exonuclease III, acts to remove this base (50, 75). Interestingly, UDG does not distinguish between the U · A base pairs (the products of uracil incorporation) and the U · G base pairs (the products of cytosine deamination), excising both uracils equally well (51, 63), which leads to the elevated frequency of excision repair intermediates (nicks) in the dut mutants of E. coli (1, 43, 76).
Similarly, RdgB is the E. coli dITPase (10, 14, 15), whose function is to intercept the noncanonical DNA precursor dITP before hypoxanthine incorporates into DNA in place of guanine. If hypoxanthine forms within DNA by adenine deamination (36), endonuclease V initiates its removal (84, 85). Analogous to the action of UDG on DNA-uracils, endonuclease V fails to distinguish between H · C base pairs (incorporated hypoxanthine) and H · T base pairs (deaminated adenine) (84, 86), and this is postulated to lead to an increased frequency of excision repair in the rdgB mutants (10).
The high level of mutagenesis in mutT mutants translates into a high density of 8-oxo-G in their DNA that should trigger elevated levels of excision repair. The process of excision repair of a base analog is generally considered harmless to the cell. However, in both dut and rdgB mutants, whose chromosomal DNA undergoes more frequent excision repair, chromosomal fragmentation occurs (44). The removal of the modification leaves a nick in the DNA backbone that is efficiently repaired most of the time but is postulated to turn into a double-strand break if run over by a replication fork (46). Consequently, dut and rdgB mutants are nonviable in combination with defects in recombinational repair (10, 42), the only repair pathway to mend double-strand DNA breaks in E. coli (47). We reasoned that, in mutT mutants, the expected increase in MutM- and MutY-catalyzed excision around the incorporated 8-oxo-G should similarly result in replication fork collapse (Fig. 2A) and, therefore, in recombination dependence. Here, we describe our experiments to test this prediction.
FIG. 2.
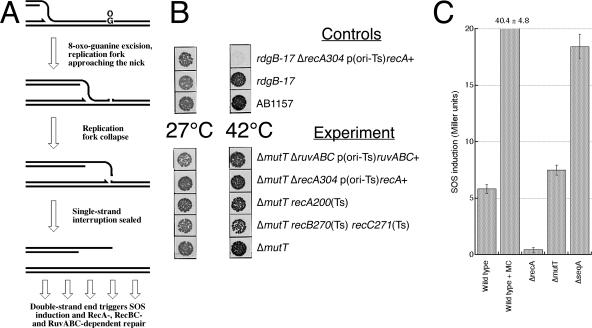
Do the mutT mutants depend on double-strand break repair and induce SOS? (A) A scheme of the assumed replication fork collapse as a result of 8-oxo-guanine (OG) excision in the template DNA in front of the fork. The same logic applies for the rdgB mutants, only 8-oxo-guanine is replaced with hypoxanthine. (B) Viability of mutT rec double and triple mutants at the 42°C temperature, nonpermissive for the rec alleles. A total of 10 μl of 10−4 dilutions of rapidly growing cultures was spotted on LB plates and incubated either at 27°C for 36 h or at 42°C for 20 h. The ΔrecA304 rdgB-17 p(ori-Ts) recA+ strain (44) is included as a positive control for the Rec dependence. The strains are as follows: AB1157, the wild-type strain; rdgB-17 ΔrecA304, EL002; rdgB-17, EL003; ΔmutT ΔruvABC, ER6; ΔmutT ΔrecA304, ER5; ΔmutT recA200(Ts), ER4; ΔmutT recBC(Ts), ER3; ΔmutT, ER2. (C) The level of SOS induction in ΔmutT mutant cultures. The seqA mutant is shown as an example of the level of SOS induction in RecA-dependent mutants (44). The strains are as follows: wild type, AK43; wild type+MC, AK43 grown in the presence of 100 ng/ml mitomycin C as a control for SOS induction (44); ΔrecA, ER65; ΔmutT, ER27; ΔseqA, ER26. The values are means ± standard error of the mean (for recA, n = 3; for others, n = 6 to 10). The SOS level in the mutT mutant is significantly different from the one in wild-type cells (t test, P0 = 0.012).
MATERIALS AND METHODS
Growth conditions.
Cells were grown in LB medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, 250 μl of 4 M NaOH per liter) or on LB agar (LB medium supplemented with 15 g of agar per liter). M9 minimal plates contained 1× M9 salts, 2 mM MgSO4, and 0.1 mM CaCl2 and were supplemented per liter with 10 mg of thiamine (B1), 15 g of agar, and 2 g of either glucose or lactose (61). Ampicillin (100 μg/ml), kanamycin (50 μg/ml), rifampin (25 or 100 μg/ml), IPTG (isopropyl-β-d-thiogalactopyranoside; 1 mM), and riboflavin (50 μg/ml) were added when needed (all concentrations are final). All cultures were grown at 30°C with reciprocal shaking, unless otherwise stated.
Reagents.
The Fpg (also named MutM) enzyme was purchased from New England Biolabs (NEB). Riboflavin, menadione, mitomycin C, and hydrogen peroxide were from Sigma. 8-Oxo-G was from Cayman Chemical. Methylene Blue was from Fisher Scientific. TE buffer contains 10 mM Tris-HCl, pH 8.0, and 1 mM EDTA.
Mutants.
All E. coli strains used in these experiments were K-12, and most of them were derivatives of AB1157 (Table 1). Alleles were moved between strains by P1 transduction (61, 62). Precise deletion-replacement alleles of selected genes were created by the method of Datsenko and Wanner (20) and confirmed by PCR and phenotypic tests (except for the Δorf135 and Δdgt mutants, for which no phenotypic tests are known). The ΔmutS and ΔmutT mutations were confirmed by increased formation of rifampin-resistant mutants (73). The ΔribA mutants were confirmed by their requirement for riboflavin (37). Upon P1 transduction, the ΔribA mutants formed large and small colonies; both required riboflavin and maintained their distinct size through subsequent restreaking. Since we did not know the reason for this difference, in subsequent tests we worked with only large colonies. The recA and recBCD mutants were confirmed by their characteristic sensitivity to UV irradiation (80). Overproduction of MutM and MutY was confirmed by restoration of low titers of rifampin-resistant mutants in a mutY background (56).
TABLE 1.
Bacterial strains
| Strain source and name | Relevant genotype or descriptiona | Reference or derivation |
|---|---|---|
| Strains from previous studies | ||
| AB1157b | Rec+ | 4 |
| AK43 | sfiA::lacZ (MuΔX cat) | 44 |
| AK105 | dut-1 | 42 |
| CC101c | lacZ E461stop | 18 |
| CC104c | lacZ E461A | 18 |
| CM1319d | mutM103::mini-Tn10 | CGSC 7738; 11 |
| JC9941 | recA200(Ts) | A. J. Clark |
| JC10287 | Δ(srlR-recA)304 | 19 |
| JB1 | ΔrecBCD3::kan | 62 |
| JJC754 | ΔruvABC::cat | 60 |
| L-41e | DH5α recA+ ΔmltB::cat | 42 |
| SK129 | recB270(Ts) recC271(Ts) | 45 |
| Strains from the present study | ||
| BB034 | Δdgt-10::cat | Deletion-replacement |
| EL003 | rdgB-17 | KanpRL27 insertion |
| EL002 | ΔrecA304 rdgB-17 | JC10287 × P1 EL003 |
| ER2 | ΔmutT9::kan | Deletion-replacement |
| ER3 | ΔmutT9::kan recB270(Ts) recC271(Ts) | SK129 × P1 ER2 |
| ER4 | ΔmutT9::kan recA200(Ts) | JC9941 × P1 ER2 |
| ER5 | ΔmutT9::kan ΔrecA304 | JC10287f × P1 ER2 |
| ER6 | ΔmutT9::kan ΔruvABC | JJC754 × P1 ER2 |
| ER8 | ΔrecBCD4 | JB1; kan removed by pCP20 |
| ER11 | DH5α recA+mutM103::mini-Tn10 | L-41 × P1 CM1319 |
| ER12 | mutM103::mini-Tn10 | AB1157 × P1 ER11 |
| ER13 | ΔmutT9::kan mutM103::mini-Tn10 | ER2 × P1 ER12 |
| ER14 | ΔmutT9::kan ΔrecBCD4 | ER8g × P1 ER2 |
| ER15 | ΔseqA20::kan | Deletion-replacement |
| ER16 | ΔseqA21 | ER15; kan removed by pCP20 |
| ER26 | sfiA::lacZ ΔseqA20::kan | AK43 × P1 ER15 |
| ER27 | sfiA::lacZ ΔmutT9::kan | AK43 × P1 ER2 |
| ER46 | ΔseqA20::kan recBC(Ts) | SK129 × P1 ER15 |
| ER48 | ΔmutT10 | ER9, kan removed by pCP20 |
| ER49 | ΔribA::cat | Deletion-replacement |
| ER50 | ΔmutT9::kan ΔribA::cat | ER2 × P1 ER49 |
| ER51 | ΔmutT9::kan ΔribA::cat recA(Ts) | ER4 × P1 ER49 |
| ER52 | Δorf135::kan | Deletion-replacement |
| ER53 | ΔmutT10 Δorf135::kan | ER48 × P1 ER52 |
| ER54 | ΔmutT10 recA(Ts) | ER4; kan removed by pCP20 |
| ER55 | ΔmutT10 Δorf135::kan recA(Ts) | ER54 × P1 ER52 |
| ER56 | ΔmutT10 ΔribA::cat Δorf135::kan recA(Ts) | ER54 × P1 ER49 |
| ER57c | CC101 ΔmutT9:: kan | CC101 × P1 ER2 |
| ER58c | CC104 ΔmutT9::kan | CC104 × P1 ER2 |
| ER59c | CC101 ΔmutT10 | ER57; kan removed by pCP20 |
| ER60c | CC101 ΔmutT10 Δorf135::kan | ER59 × P1 ER52 |
| ER61c | CC101 ΔribA::cat | CC101 × P1 ER49 |
| ER62c | CC101 ΔmutT9::kan ΔribA::cat | ER57 × P1 ER49 |
| ER63 | ΔmutT10 ΔribA::cat Δorf135::kan | ER53 × P1 ER49 |
| ER64c | CC101 Δorf135::kan | CC101 × P1 ER52 |
| ER65 | sfiA::lacZ ΔrecA304 | JC10287f × P1 AK43 |
| ER66c | CC101 Δdgt-10::cat | CC101 × P1 BB034 |
| ER67c | CC101 ΔmutS234::cat | CC101 × P1 LA24 |
| ER68c | CC101 ΔmutT9::kan Δdgt-10::cat | ER57 × P1 BB034 |
| ER69c | CC101 ΔmutT9::kan ΔmutS234::cat | ER57 × P1 LA24 |
| LA24 | ΔmutS234::cat | Deletion-replacement |
All strains have the AB1157 background unless indicated otherwise (see below).
The complete genotype includes the following: F− λ− rac- thi-1 hisG4 Δ(gpt-proA)62 argE3 thr-1 leuB6 kdgK51 rfbD1 araC14 lacY1 galK2 xylA5 mtl-1 tsx-33 glnV44 rpsL31.
The complete genotype included in addition the following: ara Δ(lac proB)XIII F lacI-Z-proB+.
This is a B/r (not K-12) strain. Other mutations include the following: F− sulA1 trpE65(Oc).
A complete genotype includes in addition the following: F′ endA1 hsdR17(r− m+) glnV44 thi-1 recA1 relA1 Δ(lacIZYA-argF)U169 deoR(φ80-ΔlacZM15) gyrA(NalR).
The recA defect was complemented with the pEAK2 plasmid for the purpose of P1 transduction. The plasmid was cured at 37°C once the construct was confirmed.
The recBC defect was complemented with the pK134 plasmid for the purpose of P1 transduction. The plasmid was cured at 37°C once the construct was confirmed.
Plasmids.
The general features of the plasmids used are described in Table 2. pER5 is pMTL21 (13) into which a PCR-amplified fragment containing the mutY gene with its native promoter was cloned into the BamHI site codirectional with the bla gene. pER6 is pMTL21 into which a PCR-amplified fragment containing the mutM gene with its native promoter was cloned into the PstI site codirectional with the bla gene. The pmutM plasmid (82) is pTrc99A containing the mutM gene under the strong IPTG-inducible trc promoter. Plasmids pEAK2, pJSB2, pK96, pK134, pTrc99A, pX25A8L, and RP4 were described previously (Table 2).
TABLE 2.
Plasmids
| Plasmid source and name | Description (ori, drug resistance, other genes) | Reference or derivation |
|---|---|---|
| Previous studies | ||
| pEAK2 | pSC101(Ts), bla, recA | 42 |
| pJSB2 | pSC101(Ts), bla, ruvABC | 10 |
| pK96 | pSC101, aad, recC ptr recB recD | 49 |
| pK134 | pSC101(Ts), bla, recC ptr recB | 62 |
| pMTL21 | pMTL, bla | 13 |
| pMutM | pBR322, bla, mutM | 82 |
| pTrc99A | pBR322, bla | 2 |
| RP4 | IncPα, bla tet aph | 67 |
| pX25A8L | pBR322, bla, Neurospora crassa DNA | 41 |
| This study | ||
| pER5 | pMTL, bla, mutY | pMTL21::mutY |
| pER6 | pMTL, bla, mutM | pMTL21::mutM |
Quantitative mutagenesis.
To verify the mutator phenotypes, saturated cultures were serially diluted (10-fold at each step), and dilutions were spotted on LB plates (for titer) and on LB plates supplemented with 100 μg/ml rifampin (for mutants). The titer of resistant colonies was scored after 1 day of incubation at 37°C. To quantify lac reversion in the CC101 background, strains were propagated on M9-glucose minimal medium to maintain the episome, and 2- to 7-day-old colonies were inoculated in LB broth. For CC101, 10 ml of a saturated culture was concentrated and spread on M9-lactose minimal medium (12), while the ΔmutT mutants had 10 μl of each dilution spotted on the plate. Since the small amount of glucose present in lactose preparations allowed formation of Lac− microcolonies, cell titer was determined by counting these microcolonies on M9-lactose plates using a stereomicroscope. The counts of total cells obtained this way were identical to counts obtained by regular plating on M9-glucose plates. To determine the effect of mutagenic treatments on the reversion frequency, colonies of CC101 and CC101 ΔmutT were inoculated into LB medium and grown to an optical density at 600 nm (OD600) of ∼0.01, and the culture was split, with one-half receiving treatment while the other served as a control. Both halves were then grown for another 15 to 17 h before being plated on M9-lactose. Lac+ colonies of CC101 were counted after 48 h, while Lac+ CC101 ΔmutT colonies were counted after 24 h under the microscope because the strain exhibited adaptive-like mutagenesis (26) at later times.
Spot test for synthetic lethality.
Growth at the nonpermissive temperature was first assayed by diluting an overnight culture 100-fold, growing it to 5 × 108 cells/ml, diluting 0.2 μl in 5 ml of 1% NaCl, and spotting by 10-μl amounts. Plates were incubated at either 27 to 30°C [permissive temperature for rec(Ts) alleles] or 42°C (the nonpermissive temperature). Colonies were given approximately 24 h to grow at both 42°C and at 30°C. Since preliminary results indicated that the viability of saturated cultures was similar to that of rapidly growing cultures, in subsequent assays we spotted 10 μl of a 10−6 dilution of a saturated culture.
SOS induction.
To determine the level of SOS induction in the cell, ΔmutT::kan was P1 transduced into AK43 (44), a strain containing a MuΔX::cat-derived construct with the lacZ gene fused under the sfiA promoter (66). When the cells are under SOS-induced stress, the promoter is expressed, and the level of β-galactosidase can be quantitatively measured by the modified protocol of Miller (61), using 200 μl of culture (44). As a positive control, wild-type cells containing the sfiA::lacZ fusion were treated with 100 ng/ml mitomycin C, a cross-linking agent. As a control for no SOS induction we used ΔrecA cells.
Pulsed-field gel electrophoresis.
The pulsed-field gel electrophoresis protocol was adapted from reference 44. Overnight LB cultures were diluted to an OD600 of 0.02 in LB medium and grown in the presence of 2.5 to 10 μCi of [32P]orthophosphoric acid to an OD600 of 0.35 (approximately 4 to 5 h) at 30°C for ΔrecBCD strains or for 1 h at 22° and 3 h at 37° (to OD600 values of 0.6 to 0.9) for recBC(Ts) strains. All cultures were then brought to an OD600 of 0.35. Cells from 0.5- to 1-ml aliquots were spun down, washed in 1 ml of TE buffer, and resuspended in 60 μl of TE buffer. Five microliters of proteinase K (10 mg/ml) and 65 μl of 1.2% agarose in lysis buffer (see below) were added and mixed by pipetting. A total of 110 μl of the mixture was then pipetted into the plug mold and allowed to solidify. The plugs were incubated overnight at 60°C in the lysis buffer (1% sarcosine, 50 mM Tris-HCl, and 25 mM EDTA). Samples were loaded into a 1.0% agarose gel in 0.5× Tris-borate-EDTA buffer and run at 6.5 V/cm with a pulse time of 90 s for 7 h, 105 s for 8 h, and 125 s for 8 h in a Gene Navigator (Pharmacia) instrument. The gel was vacuum dried onto a piece of chromatography paper (Fisher) for 2 h at 80°C and then exposed to a PhosphorImager screen until signals from the wells reached between 300,000 and 900,000 counts.
Quantification of in vivo nicking.
Cultures of AB1157, ΔmutT, and dut-1 strains, containing plasmid pX25A8L, pK96, or RP4, respectively, were grown to an OD600 of 0.6 to 0.7. A total of 450 μl of the culture was mixed with 50 μl of 10% sodium dodecyl sulfate, and the total DNA was extracted with 500 μl of phenol, then with 500 μl of phenol:chloroform, and finally with 500 μl of chloroform, followed by two ethanol precipitations (48). Then, 250 ng (pK96), or 500 ng (pX25A8L), or 1 μg (RP4) of the DNA preparation was run on a 1.1% gel at 2.5 V/cm for 20 h. The DNA was transferred to a hybridization membrane (Amersham) by capillary transfer and probed with 32P-labeled plasmid DNA (random hexamer labeling; NEB kit).
Treatment with Fpg (MutM) in vitro.
Plasmid DNA from 40 ml of saturated culture and 120 ml of log phase culture (OD600 of 0.8) of strains harboring plasmid RP4 was prepared by the alkaline lysis procedure (8). A total of 150 ng of this DNA was treated with 0.08 U of the MutM DNA glycosylase (the Fpg protein; NEB) in a 100-μl reaction mixture using 1× NEB buffer 1 (10 mM bis-Tris propane-HCl, 10 mM MgCl2, 1 mM dithiothreitol [pH 7.0 at 25°C]) at 37°C for 5 to 10 min. Negative controls were treated with the buffer only. The reaction was stopped with 10 μl of 10% sodium dodecyl sulfate, and DNA was ethanol precipitated prior to loading on a 1.1% gel, which was run for 20 h at 2.5 V/cm. The DNA was transferred to Hybond N+ membrane (Amersham) by capillary transfer and probed with 32P-labeled plasmid. To induce 8-oxo-G in DNA in vitro, DNA solutions were supplemented with 10 to 100 μM methylene blue and illuminated by a 60-W bulb for 15 min at a distance of 15 cm (25). Two consecutive ethanol precipitations removed the bulk of methylene blue before the gel electrophoresis.
RESULTS
The mutT and recA mutations do not show synthetic interactions.
MutT was the first recognized example of a DNA precursor pool sanitizer (7), the prototype for the class of functions to which Dut and RdgB also belong (28). Since both dut and rdgB mutants show dependence on recombinational repair for viability (10, 42), we tested mutT mutants for a similar requirement. The mutT gene was deleted and replaced with a kanamycin resistance cassette. The allele was P1 transduced into strains carrying conditional alleles of recA, recBC, or ruvABC, either temperature-sensitive alleles or complete deletions complemented with functional genes on ori temperature-sensitive plasmids. In these strains, a mutant dependent on recombinational repair is able to grow at 30°C but is nonviable or severely weakened at 42°C (10, 42). We found that, in contrast to dut or rdgB mutations, the mutT mutation did not confer synthetic lethality or even inhibition at 42°C in recombinational repair-deficient strains (Fig. 2B). This suggests that, in contrast to the dut or rdgB mutants, the level of DNA modifications and/or excision repair in mutT mutants is not high enough to cause such a dramatic phenotype as dependence on recombinational repair. We decided to detect the elevated level of DNA modification or excision repair by more sensitive assays, measuring SOS induction, chromosomal fragmentation, or accumulation of MutM-recognized DNA modifications.
The mutT mutant is slightly induced for the SOS response.
Recombinational repair mends chromosomal damage—a unique class of DNA damage that disables the whole chromosome. Whenever cells deploy recombinational repair, they also initiate the SOS response, boosting the cell's capacity to repair and tolerate DNA damage, while at the same time inhibiting cell division to allow more time for repair. The SOS response increases the expression of some 30 genes and decreases the expression of approximately 20 more genes (16, 24). The RecA protein senses the damage or disruption in replication and stimulates self-cleavage of the LexA repressor, thereby allowing transcription of LexA-repressed genes (53). The level of SOS induction can be measured by placing a reporter gene downstream of a LexA-inducible promoter. If the level of SOS induction in recA mutants is taken for the background of the procedure, there is a measurable SOS induction in wild-type cells, reflecting the ongoing repair of continuously generated chromosomal lesions (Fig. 2C). All known RecA-dependent mutants show various degrees of additional SOS over the wild-type level (44), consistent with the detectable additional chromosomal damage. Even such RecA-independent mutants as rnhA, rep, and uvrD also induce SOS (38, 66, 77), so mutT mutants, even though shown to be Rec independent, might still induce some SOS. Indeed, when we introduced the ΔmutT mutation into the SOS indicator strain, we detected a small statistically significant SOS induction over the wild-type level (Fig. 2C), suggesting a possibility of additional chromosomal damage in mutT mutants.
The mutT mutation does not increase chromosomal fragmentation.
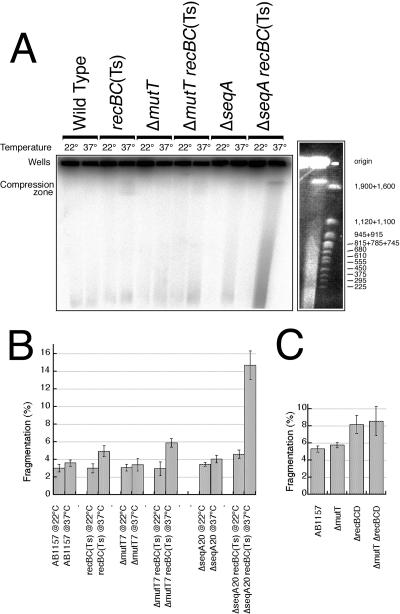
An even more sensitive assay for elevated levels of excision in chromosomal DNA is the direct detection of chromosomal fragmentation in pulsed-field gels. Chromosomal fragmentation reflects formation of double-strand DNA breaks as a result of excision repair in replicating DNA (43). The recA and recBC mutants, deficient in recombinational repair, cannot mend these double-strand breaks and, consequently, accumulate subchromosomal fragments. In addition, recBC mutants are also deficient in the major linear DNA degradation activity (exonuclease V) (47), the absence of which makes linear chromosomal DNA stable in these mutants. These DNA pieces of several hundred kilobase pairs in length can be separated from the intact chromosome by pulsed-field gel electrophoresis. During pulsed-field gel electrophoresis, intact (circular) chromosomes remain in the wells, while linear subchromosomal pieces migrate into the lane (59). The fraction of the total DNA in the lanes is measured, and a greater percentage indicates a higher level of fragmentation. Single mutants of dut and rdgB do not show excessive chromosomal fragmentation, but the double mutants dut recBC(Ts) and rdgB recBC(Ts) accumulate 20 to 25% of the total DNA in subchromosomal fragments after 4 h of incubation at 37°C, the nonpermissive temperature for the recBC(Ts) allele (44). In contrast, a ΔmutT recBC(Ts) double mutant does not show an increase in fragmentation over a recBC single mutant at 37°C (Fig. 3A and B), and the double mutant ΔmutT ΔrecBCD does not show an increase over the level of a ΔrecBCD single mutant (Fig. 3C). The absence of chromosomal fragmentation in the mutT recBC double mutants is consistent with the findings that the mutT rec double mutants are not synthetically lethal, and mutT single mutants are only slightly induced for the SOS response.
FIG. 3.
The level of chromosomal fragmentation in the mutT mutants. The seqA mutants are shown as an example of a significant chromosomal fragmentation (44). (A) A representative pulsed-field gel of the chromosomal DNA isolated in agarose plugs from the indicated strains, grown at the indicated temperatures. To the right, the last two lanes after ethidium bromide staining show their relation to molecular weight markers (yeast chromosomes; size indicated in kbp). Strains are the following: wild type, AB1157; recBC(Ts), SK129; ΔmutT, ER2; ΔmutT recBC(Ts), ER3; ΔseqA, ER15; ΔseqA recBC(Ts), ER46. (B) The level of chromosomal fragmentation, averaged from three independent experiments run on different days. The strains are the same as in panel A. The data are means ± standard error of the mean (n = 3). The level of fragmentation in ΔmutT recBC(Ts) at 37°C is not significantly different from that in recBC(Ts) at 37°C (t test, P0 = 0.12). (C) The level of chromosomal fragmentation in the ΔmutT ΔrecBCD mutant, averaged from seven independent experiments run on different days. The data are means ± standard error of the mean. The strains are the following: wild type, AB1157; ΔmutT, ER2; ΔrecBCD, ER8; ΔmutT ΔrecBCD, ER14.
The mutT mutation does not increase the level of excision.
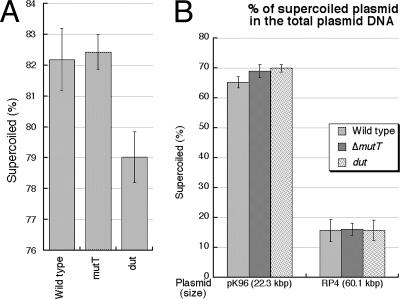
Next, we considered the possibility that base analogs are incorporated into the DNA of mutT mutants and are then excised, but this excision is somehow different from the excision in dut or rdgB mutants in that it does not cause chromosomal fragmentation. In dut mutant strains, the high level of uracil misincorporation combined with the efficient excision repair elevates the steady-state level of nicks in DNA (1, 43, 76). Nicks can be detected as relaxation of supercoiled plasmids. Total DNA, extracted from wild-type and mutant cells, contains both supercoiled and relaxed plasmid forms. A mutant with a high level of nicks will show more relaxed plasmid species compared to the wild-type cells. To detect possible plasmid relaxation due to 8-oxo-G excision in mutT mutants, we used a 30-kbp plasmid that we had successfully used before with dut mutants (43). We found no significant difference in the levels of relaxed plasmid between mutT mutants and wild-type cells (Fig. 4), indicating no increase in excision repair due to the mutT defect. The level of detection in this assay with this number of repetitions (Fig. 4A) is six additional relaxation events per genome equivalent (the statistically significant difference in supercoiling of 3.8% [(82/79 − 1) × 100] translates into 1 relaxation event per 1.58 × 106 nucleotides [60,000 nucleotides in the plasmid divided by 0.038], which, with almost 9.3 × 106 nucleotides in the E. coli genome equivalent, gives ∼6).
FIG. 4.
The level of in vivo plasmid relaxation in a mutT mutant. (A) Total plasmid DNA of pX25A8L (30.0 kbp) was extracted from wild-type (AB1157), ΔmutT mutant (ER2), and dut-1 mutant (AK105) strains as described previously (43) and run on a 1.1% agarose, and the fraction of supercoiled plasmid species in the total plasmid DNA was determined after blot hybridization with plasmid-specific probes. The data points are means ± standard error of the mean (n = 11). The dut value is significantly different from the wild-type value (t test, P0 = 0.025). (B) The procedure described in panel A was performed with plasmids pK96 and RP4. The data points are means ± standard error of the mean (4 ≤ n ≤ 10). The plasmid size is shown in parentheses. The differences in the levels of supercoiling likely reflect the different replicons.
When we carried out the same assay in two other big plasmids, 22.3 kbp and 60.1 kbp long, driven by different replicons, we again found no difference between the wild type and the mutT mutant strain, but this time the dut mutant was also no different from the wild type (Fig. 4B), suggesting some replicon-dependent variations in the sensitivity of the assay. The lack of increased relaxation in mutT mutants can be due either to the low level of 8-oxo-G incorporation or perhaps to an inefficient action of MutM and MutY glycosylases in vivo. For example, endonuclease V, which initiates excision of hypoxanthine from DNA, is known to act slowly and to be swamped by increased hypoxanthine incorporation in rdgB mutants (10).
The mutT and recA mutations do not show synthetic lethality even when MutM and MutY proteins are overproduced.
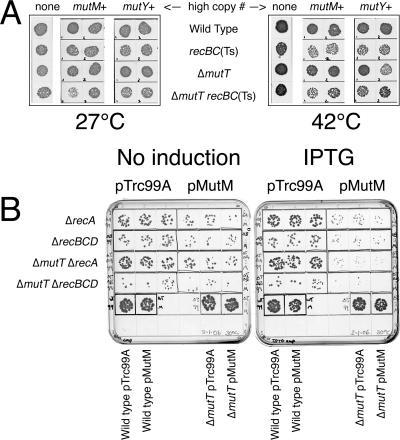
To test the possibility that MutM and MutY reactions in vivo are slow, we increased the copy number of either MutM or MutY proteins in mutT mutant cells by placing the functional mutM and mutY genes on a high-copy-number vector. The mutT rec strains, transformed with the resulting plasmids, were still viable at 42°C, although all strains were slightly weakened by the high copy number of mutY+ or mutM+ genes (Fig. 5A). The recBC mutants were affected more than rec+ cells or recA mutants (Fig. 5A; also data not shown). Even though the constructed plasmids complemented the mutator phenotype of the mutY mutant, there was a possibility that the mutM and mutY genes are downregulated under their native promoters, negating our attempt to overproduce them. However, a plasmid expressing the mutM gene under the control of a strong inducible promoter (82) gave essentially the same results: all recA and recBCD mutants showed inhibition in response to mutM induction, regardless of the presence or absence of the mutT deletion (Fig. 5B).
FIG. 5.
High copy number of mutM+ and mutY+ does not kill mutT rec mutants. (A) The effect of high-copy-number plasmids carrying the mutM+ or mutY+ genes on the viability of mutT and recBC mutant strains. A total of 10 μl of 10−4 dilutions of rapidly growing cultures was spotted on LB plates and incubated either at 27°C for 36 h or at 42°C for 20 h. Strains are as follows: wild type, AB1157; recBC(Ts), SK129; ΔmutT, ER2; ΔmutT recBC(Ts), ER3. Plasmids are the following: mutM+, pER6; mutY+, pER5. (B) No mutT rec synthetic lethality even when MutM is overexpressed. A total of 10 μl of 10−6 dilutions of saturated cultures was spotted on LB plates either supplemented with IPTG to induce mutM+ expression or left untreated (no induction) and incubated at 30°C for 24 h. The left three spots on each plate are three independent cultures carrying the pTrc99A vector plasmid; the right three spots are three independent cultures carrying the overexpression pMutM plasmid. The strains are the following: wild type, AB1157; ΔmutT, ER2; ΔrecA, JC10287; ΔrecBCD, ER8; ΔmutT ΔrecA, ER5; ΔmutT ΔrecBCD, ER14.
Upon closer examination, we noticed that the original ΔmutT recBC(Ts) transformants with the mutM+ multicopy plasmid pER6 were heterogeneous, giving rise to stable clones that showed various degrees and patterns of temperature sensitivity (not shown). The four major patterns were (i) no sensitivity, (ii) slower growth and no loss of titer, (iii) normal growth and loss of titer, and (iv) slow growth and loss of titer. Some heterogeneity was also noticed for ΔmutT single mutants (Fig. 5A). Since these patterns were stable and all transformants were confirmed for all three expected phenotypes—mutT (increased mutagenesis), recBC (UV sensitivity), and MutM overproduction (decreased mutagenesis in mutY mutants)—at face value this result suggests that MutM overproduction is detrimental in mutT recBC conditions, but some additional unknown factors are at play. However, our general conclusion is that overproduction of DNA glycosylases acting on 8-oxo-G-containing DNA does not create a problem for the mutT mutant cells, suggesting little additional 8-oxo-G in the DNA of mutT mutants.
No additional MutM-recognized DNA modifications in DNA from mutT mutM double mutants.
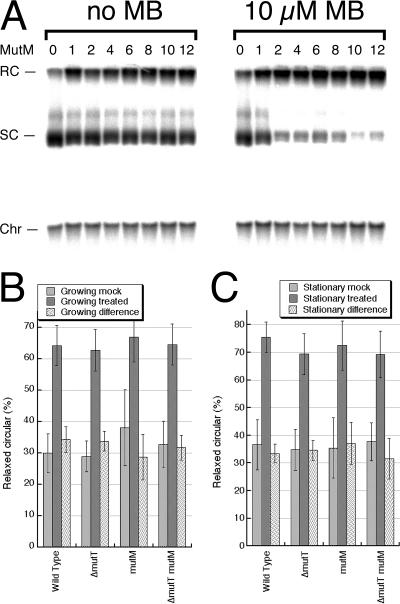
To assess the level of MutM-recognizable modifications in the DNA of mutT mutants directly, we measured the in vitro relaxation of the supercoiled plasmid by purified MutM (Fpg). The MutM glycosylase has an associated DNA lyase activity that, after removal of the modified base, also nicks the DNA backbone at the generated abasic site (64). Thus, removal of a single base modification by MutM converts a supercoiled plasmid into its relaxed form. As a control for the MutM activity and specificity, we treated pure supercoiled plasmid DNA in vitro with 10 μM methylene blue in the presence of light to specifically generate 8-oxo-G in this DNA (25, 72) and observed an almost complete plasmid relaxation upon MutM treatment (Fig. 6A). It should be pointed out that the treatment combining methylene blue and light by itself breaks DNA strands (72), which can be seen as a slight increase of relaxed species without the enzyme treatment (Fig. 6A).
FIG. 6.
The level of MutM-recognized DNA modifications in plasmid DNA. (A) RP4 DNA (positive control), treated in vitro with methylene blue (MB) and light, can be subsequently nicked in vitro with the MutM (Fpg) enzyme, indicating the presence of 8-oxo-guanine residues. Amount of MutM is expressed in 102 units, so that, for example, 8 corresponds to 0.08 units. RC, relaxed circular plasmid DNA; SC, supercoiled plasmid DNA; Chr, chromosomal DNA. (B) The level of relaxation caused by MutM-recognized DNA modifications in plasmid DNA isolated from growing cultures of wild-type (AB1157), ΔmutT (ER2), mutM (ER12), and ΔmutT mutM (ER13) strains. Mock, plasmid incubated in the buffer only; treated, plasmid incubated in the complete reaction mixture; difference, the result of the subtraction of the mock value from the treated value. The data points are means of five independent measurements done on different days ± standard error of the mean. (C) The level of relaxation caused by MutM-recognized DNA modifications in plasmid DNA isolated from stationary cultures of the same strains.
The in vitro MutM-nicking assay for 8-oxo-G was performed once before, using a 4.4-kbp plasmid isolated from mutT, mutT mutM, and mutT mutY mutant cells, and no MutM-recognized DNA modifications were found (81). To maximize the sensitivity of the assay, we used the 60.1-kbp plasmid RP4. When isolated by the alkaline lysis procedure, approximately 60% of the plasmid is in the supercoiled form, while after the treatment with MutM, only 30% of the plasmid still remains supercoiled (Fig. 6B and C). Plasmid DNA was extracted from wild-type cells and mutT and mutM single mutants, as well as from mutT mutM double mutants, from both stationary and exponentially growing cells (Fig. 6B and C). Stationary cells should have time to remove lesions before the DNA extraction, but the log-phase cells would contain unexcised 8-oxo-G. Plasmid preparations were treated with MutM or mock treated with buffer alone and run in agarose gels for subsequent blot hybridization. We expected to see an increase in relaxed species for the mutT mutM double mutant, which should incorporate elevated levels of 8-oxo-G but would be unable to remove them.
We found that although the plasmid from both stationary and growing cells was significantly relaxed by the enzyme treatment, the degree of relaxation was independent of the mutT or mutM status of the strains from which the plasmid DNA was purified (Fig. 6B and C). The lowest standard error in these experiments was ∼10% of the mean, allowing us to reliably detect a ∼30% difference in the degree of relaxation between the two sets of data (which gives a zero class calculated as 1 − 0.3 = 0.7). Since the frequency of zero class permits calculation of the average number of events per molecule using the Poisson distribution, −ln(0.7) is 0.36. Dividing the 122,000 nucleotides of RP4 by this frequency of events per molecule gives one event per ∼3.4 × 105 nucleotides, or fewer than 27 MutM-recognizable DNA modifications per E. coli genome equivalent (∼9.3 × 106 nucleotides); this is the sensitivity limit of our current measurements with this number of repetitions. On the basis of this in vitro MutM relaxation study, we conclude that, compared with the wild-type cells, there are fewer than 27 additional MutM-recognized DNA modifications per genome of mutT mutants.
The expected high AT→CG transversion mutagenesis in our ΔmutT mutants.
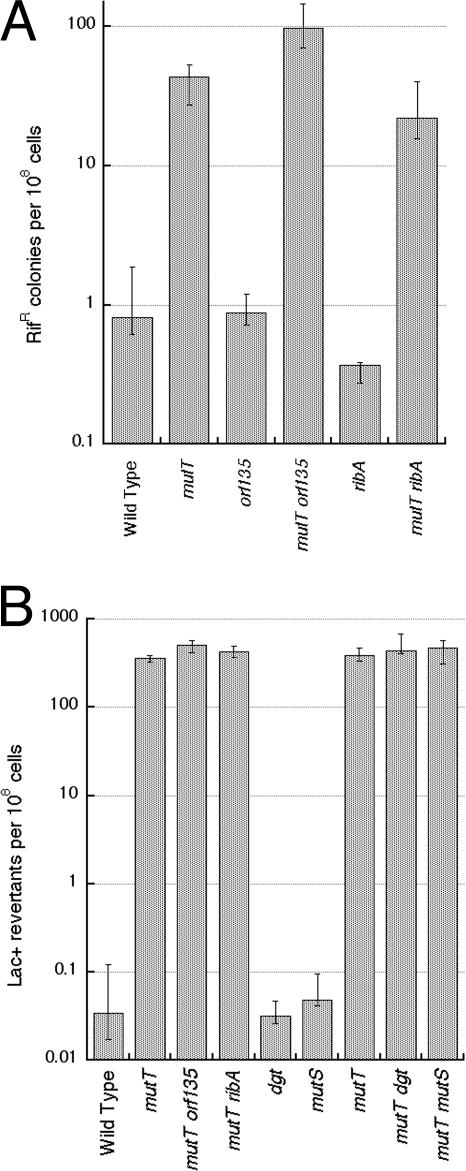
At this point we had to address the possibility that our ΔmutT allele is an atypical one in that it is not highly mutagenic. Although a qualitative rifampin test confirmed the mutator phenotype of our ΔmutT mutants, the increase was modest in the quantitative rifampin test (Fig. 7A), as expected for the rpoB mutagenic target that detects all types of base substitutions (29). To quantify the highly specific mutator phenotype of the ΔmutT allele, we introduced it into CC101 and CC104 indicator strains (18). The E. coli CC101 strain is incapable of growing on lactose as the sole carbon source due to a point mutation in the lacZ gene, but reverts to Lac+ via AT→CG transversion, the type of mutation specifically induced in mutT mutants (83). We found that our ΔmutT allele elevates the spontaneous Lac+ reversion frequency in the CC101 strain more than 104-fold (Fig. 7B), which is consistent with the published values (27, 78). The mutagenesis is highly specific, because the ΔmutT allele does not increase reversion to Lac+ in CC104, an otherwise isogenic strain that detects GC→TA transversions (not shown). These findings indicate that our ΔmutT allele induces a high level of DNA modification, which causes the expected high level of mutagenesis yet somehow does not translate into chromosomal breakage.
FIG. 7.
Mutagenesis tests of mutT mutants and their derivatives. (A) Rifampin test. (B) Lac+ reversion in CC101 (a specific test for AT→CG transversion). Values are medians ± first and third quartiles (n = 4 to 19).
The density of 8-oxo-G is still too low for chromosomal fragmentation.
The high level of specific mutagenesis in the mutT mutant suggests a considerable density of the modified guanine base in the DNA of mutT mutants. Remarkably, since 8-oxo-G is so mutagenic, pairing equally well with C and with A, the actual density of 8-oxo-G in DNA can be assessed from the mutation rates per cell per generation. Such calculations using the values reported for the same CC101 mutT construct in previous studies (27, 78) give from 54 to 560 8-oxo-G modifications per genome equivalent (Table 3). Excision repair of this level of DNA modification should cause a considerable chromosomal fragmentation. However, our values for the rate of AT→CG transversions in the CC101 mutT strain translate into only 3.0 to 4.4 8-oxo-G modifications per genome equivalent. The main reason is that while the mutagenicity of our mutT allele is the same as the normalized mutation rates used in the previous studies (Table 3), the mutation rate in the CC101 strain is 22 to 125 times lower. It should be noted that a similarly low level of mutagenesis was already reported for this CC101 strain in an independent study from this laboratory (12).
TABLE 3.
Mutation rates in CC101 versus CC101 mutT strains for the AT→CG transversions and the calculated densities of 8-oxo-G in the chromosomal DNA of CC101 mutT strains
| Studya | Mutation rate per cell generationb
|
Density of 8-oxo-G (no. of modifications)c | ||
|---|---|---|---|---|
| μ of CC101 (10−11) | μ of CC101 mutT (10−6) | Relative increase (n-fold) | ||
| 1 | 134 | 23 | ∼17,200 | 54 |
| 2 | 750 | 240 | 32,000 | 560 |
| This study | 4.9-6.9 | 1.3-1.9 | ∼27,000 | 3.0-4.4 |
To calculate mutation rates for studies 1 and 2, the frequencies that these studies report were treated as medians (21). Mutation rates (μ values) for all three studies were calculated by the method of Lea and Coulson, as explained by Rosche and Foster (70). In addition, mutation rates for this study were also calculated by the method of Drake or by the P0 method, again as explained by Rosche and Foster (70). Therefore, a range is reported instead of a single value.
The density of 8-oxo-G per genome equivalent in mutT DNA was calculated by multiplying the obtained mutation rates per cell per generation at this single site by the size of the E. coli genome (4.64 × 106), dividing by 4 (since only A residues, which are 25% of all bases in the E. coli DNA, can mutate into C residues) and multiplying by 2 (since 8-oxo-G pairs equally well with C [no mutation] and with A [the detected transversion].
The calculated level of three to four 8-oxo-G modifications in the DNA of mutT mutants is 10 times lower than the steady-state level of endonuclease V-recognized modifications in the DNA of rdgB mutants (about 42 per genome equivalent) (10) and is even lower than the steady-state level of uracil excision events in dut mutants (16 per genome equivalent) (43) (Table 4). We would not be able to detect this level of modification in mutT mutants using our most sensitive assay that measures plasmid relaxation by Fpg treatment in vitro because it is below the level of its sensitivity (about 27 modifications per genome equivalent) (Table 4). It is also unlikely that excision of these infrequent modifications can cause detectable chromosomal fragmentation.
TABLE 4.
Comparison of the density of DNA modifications and the steady-state level of relaxation events in mutants affecting either interception of noncanonical DNA precursors or excision of noncanonical DNA bases or both
| System namea | Mutant | Density of modifications (no.)b | Steady-state level of relaxation events (no.)c |
|---|---|---|---|
| Dut+Ung | dut | <22* | 16.3 |
| ung | 286 | NA | |
| dut ung | 174,000 | NA | |
| RdgB+Nfi | rdgB | 42 | ND |
| nfi | <2* | NA | |
| rdgB nfi | 42 | NA | |
| MutT+MutM | mutT | <27* | <6 |
| mutM | <27* | NA | |
| mutT mutM | <27* | NA |
For Dut+Ung, all data are from Kouzminova and Kuziminov (43), except for data for dut ung, which are averaged from Warner et al. (79). For RdgB+Nfi, all data are from Bradshaw and Kuzminov (10). Data for MutT+MutM are from the present study.
All values are densities per genome equivalent. Density of modifications was measured by following relaxation of a big plasmid DNA in vitro with a DNA repair enzyme specific for this modification (as shown, for example, in Fig. 6). Values marked with an asterisk are equivalent to wild-type levels.
Steady-state level of relaxation events per genome equivalent was measured by assaying for plasmid relaxation in vivo (as shown, for example, in Fig. 4). NA, not applicable; ND, not done.
Attempts to further increase 8-oxo-G incorporation in mutT mutants. (i) H2O2, menadione and 8-oxo-G.
To increase the level of 8-oxo-G incorporation into DNA artificially, we used the mutT derivative of CC101 to seek conditions that would further increase the level of AT→CG transversions. Since 8-oxo-G is the product of guanine oxidation and the two most common reactive oxygen species are hydrogen peroxide and superoxide (32), we treated CC strains with subinhibitory concentrations of hydrogen peroxide (300 μm) and the superoxide-generating chemical menadione (400 μm) but did not find any increased mutagenesis (not shown). We also sought to increase mutagenesis in the CC101 ΔmutT strain by growing it in the presence of 10 mM concentrations of exogenous 8-oxo-G, but this did not increase the number of AT→CG transversions either (not shown).
(ii) ribA.
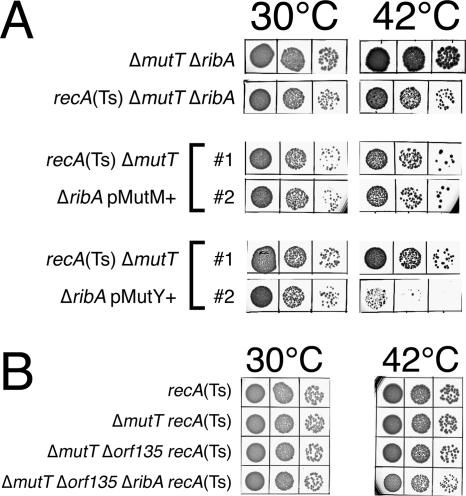
Although the AT→CG transversion mutagenesis is dramatically increased in mutT mutants (Fig. 7B), our failure to further increase it with various treatments can be rationalized by the existence of other activities in the cell specifically preventing 8-oxo-G incorporation into DNA. One of the proposed back-up enzymes for 8-oxo-dGTP interception is RibA, encoding GTP-cyclohydrolase II, the enzyme catalyzing the first step in riboflavin biosynthesis that converts dGTP into pyrophosphate, formate, and 2,5-diamino-6-(ribosylamino)-4-(3H)-pyrimidinone 5′-phosphate (37). The proposed intermediate of this reaction resembles 8-oxo-G, and the RibA enzyme was shown to have an 8-oxo-dGTPase activity in vitro, while the mutT ribA double mutant was claimed to exhibit an increased level of Rifr mutagenesis (37), implying that the RibA protein can partially fulfill MutT's role in cleansing the nucleotide pools of 8-oxo-dGTP. We deleted the ribA gene from the chromosome but found that the mutT ribA double mutant had the same level of Rifr mutagenesis as the mutT single mutant (Fig. 7A), whereas the mutT ribA recA triple mutant was still viable (Fig. 8A). When the MutM and MutY proteins were overexpressed, the triple mutant mutT ribA recA cells were viable but lost titer (Fig. 8A), suggesting that the ribA defect does increase slightly the level of MutM- and MutY-recognized DNA modifications.
FIG. 8.
High copy numbers of mutM+ and mutY+, as well as other potential defects in 8-oxo-dGTP interception, do not kill mutT rec mutants. (A) The effect of high-copy-number plasmids carrying the mutM+ or mutY+ genes on the viability of the recA(Ts) mutT ribA triple mutant (ER51). A total of 10 μl of serial dilutions of saturated cultures was spotted on LB plates and incubated at either 30°C or 42°C for 24 h. pMutM+, pER6 plasmid; pMutY+, pER5 plasmid. (B) The recA200(Ts) mutT ribA orf135 quadruple mutant is viable. Ten microliters of serial dilutions of saturated cultures was spotted on LB plates and incubated at either 30°C or 42°C for 24 h. Strains are the following: recA(Ts), JC9941; ΔmutT recA(Ts), ER4; ΔmutT Δorf135 recA(Ts), ER54; ΔmutT Δorf135 ΔribA recA(Ts), ER56.
(iii) orf135.
orf135 (also termed YnjG and NudG) is another Nudix hydrolase with reported activity against 8-oxo-dGTP (35). Although the oxidized dGTP does not seem to be its primary substrate (65), the level of Rifr mutagenesis of an orf135 mutant is claimed to be elevated twofold (34), suggesting possible overlap of function between the MutT and Orf135 enzymes. We deleted orf135 from the chromosome to generate a ΔmutT Δorf135 double mutant and a ΔmutT ΔribA Δorf135 triple mutant. We tested the mutation frequency (either to Lac+ or Rifr phenotypes) of the single and double deletion mutants in CC101 and found no increase in the specific AT→CG transversions (Fig. 7B) but a threefold increase in Rifr mutagenesis in the mutT orf135 double mutant (Fig. 7A). We also tested for recA dependence of the ΔmutT ΔribA Δorf135 mutant and found that the quadruple mutant mutT ribA orf135 recA grew at 42°C almost as well as the double and the triple mutants (Fig. 8B), indicating the absence of a significant increase in excision of modified bases.
(iv) dgt.
One more possible backup enzyme is dGTP-triphosphohydrolase (the product of the dgt gene) (69), the enzyme identified by Kornberg and colleagues as a dGTP-hydrolyzing contamination in early preparations of DNA Pol I (40). Since the only known substrate of the enzyme in vitro is dGTP, there is a suspicion that the real substrate in vivo is a modified dGTP, with 8-oxo-dGTP being a possible candidate. We inactivated the dgt gene by constructing a precise deletion in the chromosome but found that the Δdgt mutation increases the level of AT→CG transversions in neither wild-type nor mutT mutant backgrounds (Fig. 7B).
(v) mutS.
It can be calculated from the published lacI spontaneous mutagenesis data that the mismatch repair defect in E. coli (due to the mutH or mutL or mutS mutation) significantly increases the frequency of both GC→TA transversions (87 times) and AT→CG transversions (18 times) (71). Although data from one locus may not be representative, they are suggestive nonetheless, so we introduced ΔmutS mutation into both CC101 and CC101 ΔmutT strains. However, we found that the original AT→CG transversion mutagenesis was not significantly affected in either case (Fig. 7B).
DISCUSSION
We tested whether a ΔmutT mutant showed dependence on double-strand break repair due to a postulated incorporation and excision of 8-oxo-G. The extremely high reported rates of AT→CG transversions in mutT mutants suggested significant 8-oxo-dGTP incorporation, resulting in a high steady-state level of 8-oxo-G in their DNA and elevated excision. Similar situations in dut and rdgB mutants lead to chromosomal fragmentation and dependence on recombinational repair (10, 42, 44). To our surprise, we found no synthetic interactions between the mutT defect, on the one hand, and recA, recBCD, or ruvABC mutations on the other. It should be noted that the mutT1 recA mutants were reported to be viable (17); however, this is the first time that the viability of mutT recBC mutants was tested. For example, the rep and rnhA mutants are synthetically lethal with the recBC defect but viable with the recA defect (33, 77). On the other hand, the synthetically lethal dut recA mutants were for a long time considered viable (23, 39), apparently due to a rapid accumulation of suppressors (42). Finally, our study was the first one to employ a complete mutT deletion allele, which could have behaved differently from the mutT1 allele. Three additional assays corroborated the observed independence of the mutT mutants of recombinational repair. In the first, we used pulsed-field gel electrophoresis to detect chromosomal fragmentation, and by that test, the mutT mutant did not exhibit a level significantly above the wild type. In the second test, we measured SOS induction, a sensitive indicator of DNA damage. All RecA-dependent mutants show SOS induction (44), and even some RecA-independent mutants do (38, 66, 77). The mutT mutant showed little SOS induction, in agreement with the lack of chromosomal fragmentation. In the third assay, we measured the level of relaxed plasmid DNA in vivo due to MutM and MutY DNA-glycosylase activity. A large plasmid extracted from the mutT mutant showed the same ratio of supercoiled-to-relaxed DNA as the plasmids from wild-type cells. We conclude that the absence of MutT does not cause increased excision repair and, therefore, does not translate into elevated chromosomal fragmentation.
Since we assumed that our mutT mutants have the reported high absolute levels of mutagenesis (27, 78), we next considered the possibility that the 8-oxo-G-triggered excision by MutM and MutY is inefficient. If so, overproduction of the MutM and MutY glycosylases should elevate the excision and may cause chromosomal fragmentation requiring the recombinational repair system. However, while recA and recBCD single mutants did show some inhibition with the extra glycosylases, the result was largely independent of the mutT status of the strains. Moreover, when we extracted plasmid DNA from mutT and mutM mutants and treated it in vitro with a pure MutM enzyme, we found that the level of MutM-recognizable lesions was the same in DNA from all backgrounds, indicating that the mutT defect does not detectably increase the level of MutM-recognized DNA modifications. At this point we measured the rate of AT→CG transversions in our wild-type and mutT mutant strains to compare them with the reported data. We found that, whereas the increase in mutagenesis due to the mutT defect matches the values reported in the previous studies, the absolute level of AT→CG transversions is up to two orders of magnitude lower because of the much lower basal level in our wild-type strain (which is confirmed in an independent study [12]). We have noticed an adaptive-like mutagenesis (26) in the CC101 strain, about a twofold increase in the number of Lac+ colonies between day 2 and day 8 (Fig. 9), but its magnitude is not enough to account for the difference in results from different laboratories. Although we still do not know the reason for the disparate levels of mutation rates, the reduced absolute level of AT→CG transversions in our mutT mutants translates into only three to four 8-oxo-G modifications per genome equivalent, which is not enough to cause chromosomal fragmentation of the type observed in the dut and rdgB mutants (10, 42, 44). These results generally agree with the earlier studies that were unable to detect either 8-oxo-dGTP in extracts of E. coli cells (74) or 8-oxo-G in the DNA from mutT mutM and mutT mutY double mutants (81).
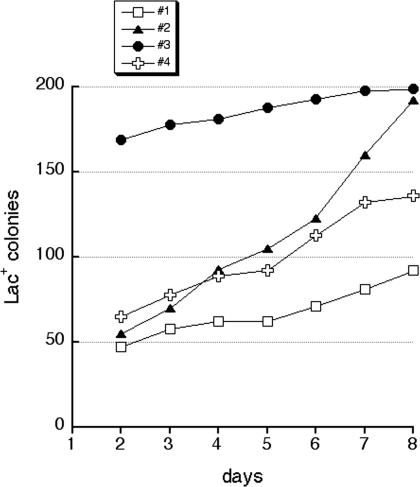
FIG. 9.
Adaptive mutagenesis in CC101 strain. Four independent 12-ml cultures were grown in LB medium from fresh single colonies to saturation and plated on M9 plates supplemented with Lac and incubated for 8 days at 37°C. Each day, the total number of Lac+ colonies on these plates was counted, and the results are shown in this graph. Culture 3 had an initial jackpot but then showed a similar rate of appearance of additional Lac+ colonies as cultures 1 and 4, while culture 2 originally had a normal, low level of Lac+ colonies but then showed an increased adaptive mutation rate relative to the other three cultures.
Using the CC101 indicator strain, we tried to find conditions that would further increase the AT→CG transversions in mutT mutants. In particular, we removed RibA and Orf135, two proposed back-up enzymes for MutT, as well as the Dgt dGTPase, a suspected noncanonical nucleotide interceptor. In our hands, these additional mutations failed to increase specific mutagenesis in the mutT-deficient background. Moreover, the mutT orf135 and mutT ribA double mutants were both viable in combination with the recA defect, as was the mutT ribA orf135 recA quadruple mutant. Inactivation of the methyl-directed mismatch repair system with a mutS deletion also failed to increase the level of AT→CG transversions in mutT mutants. We also tried oxidizing conditions and direct 8-oxo-guanine supplementation but without success. We conclude that 8-oxo-G incorporation in the DNA of mutT mutants is at the maximal level but does not translate into chromosomal fragmentation because the absolute levels of 8-oxo-G in DNA still remain low. Therefore, the mutT case turned out to be an exception that proves the rule that the high density of DNA modifications leads, via more frequent excision, to chromosomal fragmentation. On the other hand, we cannot exclude the possibility that there are conditions for increased 8-oxo-G incorporation into DNA in mutT mutants that would cause MutM/Fpg-dependent chromosomal fragmentation.
Acknowledgments
We are grateful to Luciana Amado for the ΔmutS allele and to Brian Budke for the Δdgt allele. We also thank the unknown referees and Elena Kouzminova for really helpful comments on the manuscript.
This work was supported by grant RSG-05-135-01-GMC from the American Cancer Society.
Footnotes
Published ahead of print on 6 July 2007.
REFERENCES
- 1.Amado, L., and A. Kuzminov. 2006. The replication intermediates in Escherichia coli are not the product of DNA processing or uracil excision. J. Biol. Chem. 281:22635-22646. [DOI] [PubMed] [Google Scholar]
- 2.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 3.Au, K. G., S. Clark, J. H. Miller, and P. Modrich. 1989. Escherichia coli mutY gene encodes an adenine glycosylase active on G-A mispairs. Proc. Natl. Acad. Sci. USA 86:8877-8881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bachmann, B. J. 1987. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 1190-1219. In F. C. Neidhardt, L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology. American Society for Microbiology, Washington, DC.
- 5.Bertani, L. E., A. Haggmark, and P. Reichard. 1963. Enzymatic synthesis of deoxyribonucleotides. II. Formation and interconversion of deoxyuridine phosphates. J. Biol. Chem. 238:3407-3413. [PubMed] [Google Scholar]
- 6.Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271:25059-25062. [DOI] [PubMed] [Google Scholar]
- 7.Bhatnagar, S. K., L. C. Bullions, and M. J. Bessman. 1991. Characterization of the MutT nucleoside triphosphatase of Escherichia coli. J. Biol. Chem. 266:9050-9054. [PubMed] [Google Scholar]
- 8.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 9.Boiteux, S., E. Gajewski, J. Laval, and M. Dizdaroglu. 1992. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry 31:106-110. [DOI] [PubMed] [Google Scholar]
- 10.Bradshaw, J. S., and A. Kuzminov. 2003. RdgB acts to avoid chromosome fragmentation in Escherichia coli. Mol. Microbiol. 48:1711-1725. [DOI] [PubMed] [Google Scholar]
- 11.Bridges, B. A., M. Sekiguchi, and T. Tajiri. 1996. Effect of mutY and mutM/fpg-1 mutations on starvation-associated mutation in Escherichia coli: implications for the role of 7,8-dihydro-8-oxoguanine. Mol. Gen. Genet. 251:352-357. [DOI] [PubMed] [Google Scholar]
- 12.Budke, B., and A. Kuzminov. 2006. Hypoxanthine incorporation is nonmutagenic in Escherichia coli. J. Bacteriol. 188:6553-6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers, S. P., S. E. Prior, D. A. Barstow, and N. P. Minton. 1988. The pMTL nic− cloning vectors. I. Improved pUC polylinker regions to facilitate the use of sonicated DNA for nucleotide sequencing. Gene 68:139-149. [DOI] [PubMed] [Google Scholar]
- 14.Chung, J. H., J. H. Back, Y. I. Park, and Y. S. Han. 2001. Biochemical characterization of a novel hypoxanthine/xanthine dNTP pyrophosphatase from Methanococcus jannaschii. Nucleic Acids Res. 29:3099-3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung, J. H., H. Y. Park, J. H. Lee, and Y. Jang. 2002. Identification of the dITP- and XTP-hydrolyzing protein from Escherichia coli. J. Biochem. Mol. Biol. 35:403-408. [DOI] [PubMed] [Google Scholar]
- 16.Courcelle, J., A. Khodursky, B. Peter, P. O. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox, E. C., and C. Yanofsky. 1969. Mutator gene studies in Escherichia coli. J. Bacteriol. 100:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cupples, C. G., and J. H. Miller. 1989. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 86:5345-5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Czonka, L. N., and A. J. Clark. 1979. Deletions generated by the transposon Tn10 in the srl-recA region of the Escherichia coli K-12 chromosome. Genetics 93:321-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drake, J. W. 1991. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl. Acad. Sci. USA 88:7160-7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Einolf, H. J., N. Schnetz-Boutaud, and F. P. Guengerich. 1998. Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dehydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry 37:13300-13312. [DOI] [PubMed] [Google Scholar]
- 23.El-Hajj, H. H., L. Wang, and B. Weiss. 1992. Multiple mutant of Escherichia coli synthesizing virtually thymineless DNA during limited growth. J. Bacteriol. 174:4450-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernández de Henestrosa, A. R., T. Ogi, S. Aoyagi, D. Chafin, J. J. Hayes, H. Ohmori, and R. Woodgate. 2000. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 35:1560-1572. [DOI] [PubMed] [Google Scholar]
- 25.Floyd, R. A., M. S. West, K. L. Eneff, and J. E. Schneider. 1989. Methylene blue plus light mediates 8-hydroxyguanine formation in DNA. Arch. Biochem. Biophys. 273:106-111. [DOI] [PubMed] [Google Scholar]
- 26.Foster, P. L. 1999. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu. Rev. Genet. 33:57-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fowler, R. G., S. J. White, C. Koyama, S. C. Moore, R. L. Dunn, and R. M. Schaaper. 2003. Interactions among the Escherichia coli mutT, mutM, and mutY damage prevention pathways. DNA Repair 2:159-173. [DOI] [PubMed] [Google Scholar]
- 28.Galperin, M. Y., O. V. Moroz, K. S. Wilson, and A. G. Murzin. 2006. House cleaning, a part of good housekeeping. Mol. Microbiol. 59:5-19. [DOI] [PubMed] [Google Scholar]
- 29.Garibyan, L., T. Huang, M. Kim, E. Wolff, A. Nguyen, T. Nguyen, A. Diep, K. Hu, A. Iverson, H. Yang, and J. H. Miller. 2003. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair 2:593-608. [DOI] [PubMed] [Google Scholar]
- 30.Greenberg, G. R., and R. L. Somerville. 1962. Deoxyuridylate kinase activity and deoxyuridine triphosphatase in Escherichia coli. Proc. Natl. Acad. Sci. USA 48:247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochhauser, S. J., and B. Weiss. 1978. Escherichia coli mutants deficient in deoxyuridine triphosphatase. J. Bacteriol. 134:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 33.Itaya, M., and R. J. Crouch. 1991. A combination of RNase H (rnh) and recBCD or sbcB mutations in Escherichia coli K12 adversely affects growth. Mol. Gen. Genet. 277:424-432. [DOI] [PubMed] [Google Scholar]
- 34.Kamiya, H., E. Iida, N. Murata-Kamiya, Y. Yamamoto, T. Miki, and H. Harashima. 2003. Suppression of spontaneous and hydrogen peroxide-induced mutations by a MutT-type nucleotide pool sanitization enzyme, the Escherichia coli Orf135 protein. Genes Cells 8:941-950. [DOI] [PubMed] [Google Scholar]
- 35.Kamiya, H., N. Murata-Kamiya, E. Iida, and H. Harashima. 2001. Hydrolysis of oxidized nucleotides by the Escherichia coli Orf135 protein. Biochem. Biophys. Res. Commun. 288:499-502. [DOI] [PubMed] [Google Scholar]
- 36.Karran, P., and T. Lindahl. 1980. Hypoxanthine in deoxyribonucleic acid: generation by heat-induced hydrolysis of adenine residues and release in free form by a deoxyribonucleic acid glycosylase from calf thymus. Biochemistry 19:6005-6011. [DOI] [PubMed] [Google Scholar]
- 37.Kobayashi, M., Y. Ohara-Nemoto, M. Kaneko, H. Hayakawa, M. Sekiguchi, and K. Yamamoto. 1998. Potential of Escherichia coli GTP cyclohydrolase II for hydrolyzing 8-oxo-dGTP, a mutagenic substrate for DNA synthesis. J. Biol. Chem. 273:26394-26399. [DOI] [PubMed] [Google Scholar]
- 38.Kogoma, T., X. Hong, G. W. Cadwell, K. G. Barnard, and T. Asai. 1993. Requirement of homologous recombination functions for viability of the Escherichia coli cells that lack RNase HI and exonuclease V activities. Biochimie 75:89-99. [DOI] [PubMed] [Google Scholar]
- 39.Konrad, E. B., and I. R. Lehman. 1975. Novel mutants of Escherichia coli that accumulate very small DNA replicative intermediates. Proc. Natl. Acad. Sci. USA 72:2150-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornberg, S. R., I. R. Lehman, M. J. Bessman, E. S. Simms, and A. Kornberg. 1958. Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J. Biol. Chem. 233:159-162. [PubMed] [Google Scholar]
- 41.Kouzminova, E., and E. U. Selker. 2001. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 20:4309-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kouzminova, E. A., and A. Kuzminov. 2004. Chromosomal fragmentation in dUTPase-deficient mutants of Escherichia coli and its recombinational repair. Mol. Microbiol. 51:1279-1295. [DOI] [PubMed] [Google Scholar]
- 43.Kouzminova, E. A., and A. Kuzminov. 2006. Fragmentation of replicating chromosomes triggered by uracil in DNA. J. Mol. Biol. 355:20-33. [DOI] [PubMed] [Google Scholar]
- 44.Kouzminova, E. A., E. Rotman, L. Macomber, J. Zhang, and A. Kuzminov. 2004. RecA-dependent mutants in E. coli reveal strategies to avoid replication fork failure. Proc. Natl. Acad. Sci. USA 101:16262-16267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kushner, S. R. 1974. In vivo studies of temperature-sensitive recB and recC mutants. J. Bacteriol. 120:1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuzminov, A. 1995. Collapse and repair of replication forks in Escherichia coli. Mol. Microbiol. 16:373-384. [DOI] [PubMed] [Google Scholar]
- 47.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuzminov, A., E. Schabtach, and F. W. Stahl. 1994. χ-sites in combination with RecA protein increase the survival of linear DNA in E. coli by inactivating ExoV activity of RecBCD nuclease. EMBO J. 13:2764-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuzminov, A., and F. W. Stahl. 1997. Stability of linear DNA in recA mutant Escherichia coli cells reflects ongoing chromosomal DNA degradation. J. Bacteriol. 179:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindahl, T. 1974. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. USA 71:3649-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindahl, T., S. Ljungquist, W. Siegert, B. Nyberg, and B. Sperens. 1977. DNA N-glycosidases. Properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 252:3286-3294. [PubMed] [Google Scholar]
- 52.Lindahl, T., and B. Nyberg. 1974. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry 13:3405-3410. [DOI] [PubMed] [Google Scholar]
- 53.Little, J. W. 1991. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 73:411-421. [DOI] [PubMed] [Google Scholar]
- 54.Maki, H., and M. Sekiguchi. 1992. MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis. Nature 355:273-275. [DOI] [PubMed] [Google Scholar]
- 55.McAuley-Hecht, K. E., G. A. Leonard, N. J. Gibson, J. B. Thomson, W. P. Watson, W. N. Hunter, and T. Brown. 1994. Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry 33:10266-10270. [DOI] [PubMed] [Google Scholar]
- 56.Michaels, M. L., C. Cruz, A. P. Grollman, and J. H. Miller. 1992. Evidence that MutY and MutM combine to prevent mutations by an oxidatively damaged form of guanine in DNA. Proc. Natl. Acad. Sci. USA 89:7022-7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Michaels, M. L., J. Tchou, A. P. Grollman, and J. H. Miller. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964-10968. [DOI] [PubMed] [Google Scholar]
- 59.Michel, B., S. D. Ehrlich, and M. Uzest. 1997. DNA double-strand breaks caused by replication arrest. EMBO J. 16:430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Michel, B., G. D. Recchia, M. Penel-Colin, S. D. Ehrlich, and D. J. Sherratt. 2000. Resolution of Holliday junctions by RuvABC prevents dimer formation in rep mutants and UV-irradiated cells. Mol. Microbiol. 37:180-191. [DOI] [PubMed] [Google Scholar]
- 61.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 62.Miranda, A., and A. Kuzminov. 2003. Chromosomal lesion suppression and removal in Escherichia coli via linear DNA degradation. Genetics 163:1255-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nilsen, H., S. P. Yazdankhah, I. Eftedal, and H. E. Krokan. 1995. Sequence specificity for removal of uracil from U · A pairs and U · G mismatches by uracil-DNA glycosylase from Escherichia coli, and correlation with mutational hotspots. FEBS Lett. 362:205-209. [DOI] [PubMed] [Google Scholar]
- 64.O'Connor, T. R., and J. Laval. 1989. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc. Natl. Acad. Sci. USA 86:5222-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O'Handley, S. F., C. A. Dunn, and M. J. Bessman. 2001. Orf135 from Escherichia coli is a Nudix hydrolase specific for CTP, dCTP and 5-methyl-dCTP. J. Biol. Chem. 276:5421-5426. [DOI] [PubMed] [Google Scholar]
- 66.Ossanna, N., and D. W. Mount. 1989. Mutations in uvrD induce the SOS response in Escherichia coli. J. Bacteriol. 171:303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pansegrau, W., E. Lanka, P. T. Barth, D. H. Figurski, D. G. Guiney, D. Haas, D. R. Helinski, H. Schwab, V. A. Stanisich, and C. M. Thomas. 1994. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J. Mol. Biol. 239:623-663. [DOI] [PubMed] [Google Scholar]
- 68.Purmal, A. A., Y. W. Kow, and S. S. Wallace. 1994. 5-Hydroxypyrimidine deoxynucleoside triphosphates are more efficiently incorporated into DNA by exonuclease-free Klenow fragment than 8-oxopurine deoxynucleoside triphosphates. Nucleic Acids Res. 22:3930-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quirk, S., S. K. Bhatnagar, and M. J. Bessman. 1990. Primary structure of the deoxyguanosine triphosphate triphosphohydrolase-encoding gene (dgt) of Escherichia coli. Gene 89:13-18. [DOI] [PubMed] [Google Scholar]
- 70.Rosche, W. A., and P. L. Foster. 2000. Determining mutation rates in bacterial populations. Methods 20:4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schaaper, R. M., and R. L. Dunn. 1991. Spontaneous mutation in the Escherichia coli lacI gene. Genetics 129:317-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schneider, J. E., S. Price, L. Maidt, J. M. Gutteridge, and R. A. Floyd. 1990. Methylene blue plus light mediates 8-hydroxy 2′-deoxyguanosine formation in DNA preferentially over strand breakage. Nucleic Acids Res. 18:631-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tajiri, T., H. Maki, and M. Sekiguchi. 1995. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat. Res. 336:257-267. [DOI] [PubMed] [Google Scholar]
- 74.Tassotto, M. L., and C. K. Mathews. 2002. Assessing the metabolic function of the MutT 8-oxodeoxyguanosine triphosphatase in Escherichia coli by nucleotide pool analysis. J. Biol. Chem. 277:15807-15812. [DOI] [PubMed] [Google Scholar]
- 75.Taylor, A. F., and B. Weiss. 1982. Role of exonuclease III in the base excision repair of uracil-containing DNA. J. Bacteriol. 151:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tye, B.-K., and I. R. Lehman. 1977. Excision repair of uracil incorporated in DNA as a result of a defect in dUTPase. J. Mol. Biol. 117:293-306. [DOI] [PubMed] [Google Scholar]
- 77.Uzest, M., S. D. Ehrlich, and B. Michel. 1995. Lethality of rep recB and rep recC double mutants of Escherichia coli. Mol. Microbiol. 17:1177-1188. [DOI] [PubMed] [Google Scholar]
- 78.Vidmar, J. J., and C. G. Cupples. 1993. MutY repair is mutagenic in mutT—strains of Escherichia coli. Can. J. Microbiol. 39:892-894. [DOI] [PubMed] [Google Scholar]
- 79.Warner, H. R., B. K. Duncan, C. Garrett, and J. Neuhard. 1981. Synthesis and metabolism of uracil-containing deoxyribonucleic acid in Escherichia coli. J. Bacteriol. 145:687-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Willetts, N. S., and A. J. Clark. 1969. Characteristics of some multiply recombination-deficient strains of Escherichia coli. J. Bacteriol. 100:231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wójcik, A., E. Grzesiuk, B. Tudek, and C. Janion. 1996. Conformation of plasmid DNA from Escherichia coli deficient in the repair systems protecting DNA from 8-oxyguanine lesions. Biochimie 78:85-89. [DOI] [PubMed] [Google Scholar]
- 82.Wyrzykowski, J., and M. R. Volkert. 2003. The Escherichia coli methyl-directed mismatch repair system repairs base pairs containing oxidative lesions. J. Bacteriol. 185:1701-1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yanofsky, C., E. C. Cox, and V. Horn. 1966. The unusual mutagenic specificity of an E. coli mutator gene. Proc. Natl. Acad. Sci. USA 55:274-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yao, M., Z. Hatahet, R. J. Melamede, and Y. W. Kow. 1994. Purification and characterization of a novel deoxyinosine-specific enzyme, deoxyinosine 3′ endonuclease, from Escherichia coli. J. Biol. Chem. 269:16260-16268. [PubMed] [Google Scholar]
- 85.Yao, M., and Y. W. Kow. 1997. Further characterization of Escherichia coli endonuclease V. Mechanism of recognition for deoxyinosine, deoxyuridine, and base mismatches in DNA. J. Biol. Chem. 272:30774-30779. [DOI] [PubMed] [Google Scholar]
- 86.Yao, M., and Y. W. Kow. 1995. Interaction of deoxyinosine 3′-endonuclease from Escherichia coli with DNA containing deoxyinosine. J. Biol. Chem. 270:28609-28616. [DOI] [PubMed] [Google Scholar]