Abstract
The human pathogen Mycoplasma genitalium is known to mediate cell adhesion to target cells by the attachment organelle, a complex structure also implicated in gliding motility. The gliding mechanism of M. genitalium cells is completely unknown, but recent studies have begun to elucidate the components of the gliding machinery. We report the study of MG312, a cytadherence-related protein containing in the N terminus a box enriched in aromatic and glycine residues (EAGR), which is also exclusively found in MG200 and MG386 gliding motility proteins. Characterization of an MG_312 deletion mutant obtained by homologous recombination has revealed that the MG312 protein is required for the assembly of the M. genitalium terminal organelle. This finding is consistent with the intermediate-cytadherence phenotype and the complete absence of gliding motility exhibited by this mutant. Reintroduction of several MG_312 deletion derivatives into the MG_312 null mutant allowed us to identify two separate functional domains: an N-terminal domain implicated in gliding motility and a C-terminal domain involved in cytadherence and terminal organelle assembly functions. In addition, our results also provide evidence that the EAGR box has a specific contribution to mycoplasma cell motion. Finally, the presence of a conserved ATP binding site known as a Walker A box in the MG312 N-terminal region suggests that this structural protein could also play an active function in the gliding mechanism.
Mycoplasma genitalium is an important sexually transmitted pathogen capable of colonizing the human genitourinary tract. It is considered the principal causative agent of nonchlamydial, nongonococcal urethritis in men (18), and it has also been associated with genital tract diseases in women, such as endometritis (3), pelvic inflammatory disease, and cervicitis (7, 23). Furthermore, M. genitalium has been related to other extragenitourinary pathologies (40), including pneumonia, chronic fatigue, and arthritis. In addition to its clinical significance, M. genitalium has the smallest known genome among the organisms able to grow in axenic culture. In this sense, with just 525 genes (8, 9), this cell wall-less prokaryote is considered one of the most suitable models to understand the cellular biology of a self-replicating cell (30). Nonetheless, behind this apparent simplicity, a remarkable complexity exists, exemplified by the presence of a complex and differentiated polar structure, known as the attachment or terminal organelle. M. genitalium and several mycoplasma species accomplish surface adhesion by using this structure, which has been traditionally studied in Mycoplasma pneumoniae (21). The attachment organelle of M. pneumoniae is defined by the presence of an electron-dense core (15, 16, 36), and it is composed of a filamentous network of cytoskeletal proteins, including HMW1, HMW2, HMW3, B, C, P65, and the major cytadhesin P1 (29).
M. genitalium cells have also the ability to locomote across solid surfaces by a novel and poorly characterized mechanism known as gliding motility, which is believed to be involved in the colonization and dissemination processes. It is noteworthy that the terminal organelle is also responsible for this movement (13), reinforcing the importance of this polar structure in mycoplasma pathogenesis. Many gliding motility proteins identified in M. pneumoniae (P1 and P30) and Mycoplasma mobile (Gli349, Gli521, and Gli123) appear also to be implicated in glass binding and hemadsorption (HA) (12, 33, 35, 42, 43), supporting the close relationship between the gliding motility and cytadherence machineries. However, proteins other than cytadhesins and cytadherence-associated components of the attachment organelle contribute to mycoplasma motion (14). Recently, we identified two proteins, MG200 and MG386, involved in M. genitalium gliding motility but not in cytadherence (25). Interestingly, both proteins possess sequence motifs enriched in aromatic and glycine residues (EAGR boxes). This feature is exclusively shared with the cytadherence-accessory MG312 protein (homologous to M. pneumoniae HMW1), suggesting the involvement of this protein in gliding motility. HMW1 is a huge peripheral membrane protein (1) located in the attachment organelle (34), and it has a modular structure characterized by separate N-terminal and C-terminal domains (6), which are well conserved in the homologous M. genitalium MG312 protein. These two domains are joined by a less conserved central domain characterized by repeating acidic and proline-rich motifs. Characterization of an M. pneumoniae cytadherence-negative mutant lacking HMW1 and with defects in the P30 adhesin gene (22) showed that HMW1 is required for normal cell morphology and cytadherence (10). More-detailed analyses revealed that the HMW1 C-terminal domain is needed for HMW2 stabilization, proper cell morphology, P1 trafficking to the tip structure (45), and HMW1 turnover in the absence of HMW2 (27).
In the present study we have examined the contribution of the MG312 protein to the cytadherence and gliding motility of M. genitalium. Our results demonstrate that, in the absence of the MG312 protein, the M. genitalium cells adhere poorly to surfaces and they are nonmotile. In addition, we have found that the MG312 protein has two separate functional domains: a C-terminal domain related to cytadherence and cell morphology, as previously described for M. pneumoniae (45), and an N-terminal domain not directly related to surface attachment and terminal organelle development but clearly involved in M. genitalium cell locomotion. The presence in this region of a conserved ATP binding site, known as the P-loop or Walker A box (44), suggests that this structural protein could also play an active function in the gliding mechanism.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Mycoplasma genitalium strain G37 and the derivative mutants obtained in this study were grown in SP-4 broth (41) at 37°C under 5% CO2 in tissue culture flasks (TPP). Tetracycline (2 μg ml−1) or gentamicin (100 μg ml−1) was added to SP-4 medium for isolation and culture of transformants. For gliding motility assessment based on colony morphology, the obtained mutant strains were grown in agar overlays as previously described (25).
Escherichia coli strain XL1-Blue was used for plasmid amplification and grown as previously described (26).
Construction of pΔMG_312 and MG_312 deletion derivatives.
General DNA manipulations were performed by following standard procedures (32). Primers used in this work are summarized in Table 1. Genomic DNA of the wild-type (WT) strain, isolated as previously described (26), was used as a template for PCRs. To obtain the pΔMG_312 plasmid, the 1-kb PCR fragment encompassing the upstream region and the first ≈100 bp of the 5′ end of the MG_312 gene was amplified using primers 5′BEKOMG312 and 3′BEKOMG312, which incorporate, respectively, at their ends Acc65I and EcoRI restriction sites. A second 1-kb fragment containing the last ≈100 bp of the MG_312 gene and a downstream region was also amplified using primers 5′BDKOMG312 and 3′BDKOMG312. These primers contained at their ends BamHI and XbaI restriction sites, respectively. Both PCR fragments were cloned into EcoRV-digested pBE (26), excised with the corresponding restriction enzymes, and ligated together with a 2-kb fragment containing the tetM438 selectable marker (26) and Acc65I/XbaI-digested pBSKII+ (Invitrogen). The tetM438 selectable marker was released from the pMTnTetM438 plasmid (26) by digestion with EcoRI and BamHI.
TABLE 1.
Primers
| Primer | Sequence (5′-3′)a |
|---|---|
| 5′BEKOMG312 | GGTACCCCTGCTTTGTGAATCCATGG |
| 3′BEKOMG312 | GAATTCAGTAATGGATTCAATGAGTGC |
| 5′BDKOMG312 | GGATCCAGGGTCTTCTTCAACAGTGG |
| 3′BDKOMG312 | TCTAGATAGTGTTTTATTAGCACTAACC |
| 5′MG312 | GTCGACTAGTATTTAGAATTAATAAAGTATGGCTAAAAACAAGCAATCG |
| 3′MG312 | CTGCAGTTAATAATCAAGGCTAAAATCAC |
| 5′MG312ΔN-t | GTCGACTAGTATTTAGAATTAATAAAGTATGGAATCTCAACCAACTGATG |
| 3′MG312ΔC-t | CTGCAGTTAAAGATCTGATTTTTTATTGAAATTATC |
| 3′MG312ΔCD | AGATCTGATTTAGTGGAGATCCACTGGTC |
| 5′MG312ΔEAGR | AAGCAAGAAGAGAATGATTCCACTAAAGAATCTCAAC |
| 3′MG312ΔEAGR | GTTGAGATTCTTTAGTGGAATCATTCTCTTCTTGCTT |
| 3′MG312EcoRI | GAATTCAGAATTTAATGAATCATTG |
The restriction sites introduced in the 5′ ends of the primers are underlined; the 22 nucleotides from the putative MG_438 gene promoter region (26) are in italics; the ATG and stop codons introduced into two primers are in boldface.
Construction of MG_312 deletion derivatives was as follows. The WT MG_312 allele was amplified using primers 5′MG312 and 3′MG312, while the MG_312ΔN-t deletion derivative was amplified using primers 5′MG312ΔN-t and 3′MG312. Primers 5′MG312 and 5′MG312ΔN-t include at their ends SalI restriction sites and the 22-bp promoter region of the MG_438 open reading frame, previously described (26), while the 3′MG312 primer includes a PstI restriction site. Primer 5′MG312ΔN-t incorporates in addition one ATG codon after the promoter region. The MG_312ΔC-t derivative was amplified using primers 5′MG312 and 3′MG312ΔC-t. The 3′MG312ΔC-t primer includes one stop codon followed by a PstI restriction site. Finally, the three PCR fragments were then cloned separately into the SalI/PstI-digested pMTnGm minitransposon (26), creating the pMTnGmMG_312, pMTnGmMG_312ΔN-t, and pMTnGmMG_312ΔC-t plasmids. Alternatively, the MG_312ΔRep derivative was obtained by SacI digestion and subsequent religation of the pMTnGmMG_312 plasmid. For the construction of the MG_312ΔCD deletion derivative, a 381-bp fragment corresponding to the 5′ region of MG_312 was amplified by PCR using primers 5′MG312 and 3′MG312ΔCD. The 3′MG312ΔCD primer includes the bases AG in the 5′ end of the BglII restriction site to keep the reading frame of the MG_312 fragment encoding the C terminus. This PCR fragment was cloned into a SalI/BglII-digested pMTnGmMG_312 plasmid, creating the pMTnGmMG_312ΔCD plasmid. Finally, the MG_312ΔEAGR deletion derivative was obtained by PCR mutagenesis. The two PCR products obtained using the primers 5′MG312 and 3′MG312ΔEAGR and the primers 5′MG312ΔEAGR and 3′MG312EcoRI were used as templates for a third PCR using the primers 5′MG312 and 3′MG312EcoRI. The recombinant PCR product was cloned into a pBE plasmid (26) and sequenced to confirm the deletion of the sequence coding for the EAGR box. A positive clone was digested with SalI/EcoRI, and the resulting 1,148-bp fragment was cloned into the SalI/EcoRI-digested pMTnGmMG_312 plasmid, creating the pMTnGmMG_312ΔEAGR plasmid.
Electroporation and transformation.
Transformation of M. genitalium strain G37 with suicide plasmid pΔMG_312 was performed by electroporation as previously described (26) using 30 μg of DNA. For transformation of ΔMG_312 mutants, cells were grown to mid-log phase, centrifuged, and passed five times through a 25-gauge needle. Cells were then separately electroporated with 5 μg of plasmids containing the different MG_312 deletion derivatives. Transformants were filter cloned (0.45-μm filter) by three consecutive passages before analysis (25).
Southern blotting.
Genomic DNAs from pΔMG_312 transformants were digested with HindIII and electrophoresed on 0.8% agarose gels. DNA fragments were then transferred to a nylon membrane (Roche) and probed as previously described (26). The MG_312 flanking regions previously amplified for pΔMG_312 construction were used as a probe.
Preparation of monoclonal antibodies.
A panel of monoclonal antibodies against M. genitalium proteins was obtained as follows. Antigen was prepared by solubilizing 109 M. genitalium cells in 3 ml of 0.1% Triton X-100 in sterile phosphate-buffered saline (PBS). After 30 min at room temperature, aliquots of 150 μl were stored at −80°C until needed. No viable mycoplasma cells were detected after plating several aliquots in SP-4 medium. BALB/c mice were then immunized by four adjuvant-free intraperitoneal injections by following standard procedures. Splenocyte fusion, hybridoma testing, and monoclonal antibody production were performed as described previously (11).
SDS-PAGE and Western immunoblotting.
Total mycoplasma cell proteins were prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoresed on 8% polyacrylamide gels by following standard procedures. Gels were stained with Coomassie blue or transferred electrophoretically to nitrocellulose membranes and probed as previously described (32) with monoclonal antibody anti-MG217 at a 1:500 dilution or anti-P140 (24) at a 1:1,000 dilution, anti-HMW1 serum (37) at a 1:8,000 dilution, and anti-HMW3 serum (38) at a 1:5,000 dilution.
Scanning electron microscopy (SEM).
The WT strain and ΔMG_312 derivative transformants were grown in cell culture Permanox chamber slides (Nunc) for 16 h, while the ΔMG_312 mutants were cultured on poly-l-lysine-coated glass coverslips as previously described (2). Samples were then processed as described previously (2) before being examined in a Hitachi (Tokyo, Japan) S-570 scanning electron microscope.
Microcinematographies.
Characterization and quantitative analysis of the gliding motility of the WT strain and ΔMG_312 and ΔMG_312 derivative mutants were performed by microcinematography as previously described (25). Particular movements of ≈350 individual cells from each strain were analyzed to determine the gliding frequency. Gliding speed was monitored by measuring the tracks described by 50 individual cells from eight different fields for 2 min. To obtain comparable mean gliding speeds for each strain studied, all samples contained similar cell densities.
Quantitative plastic binding assay.
Frozen stocks of the WT strain and ΔMG_312 and ΔMG_312 derivative mutants were diluted in 1 ml of SP-4 medium to give a final concentration of approximately 107 CFU ml−1. The binding assay was performed by dispensing 800-μl aliquots of the different mycoplasma suspensions into cell culture dishes 35 mm in diameter (Corning), which were then incubated for 3 h at 37°C. The 200 μl remaining was used to precisely determine the total number of mycoplasmas in each binding experiment. After the incubation period, mycoplasma cultures were gently shaken to remove nonadhered mycoplasmas, which were recovered by pipetting the culture medium. Determination of ATP is considered a proper method to estimate mycoplasma cell mass (31). To determine the total numbers of mycoplasmas and nonattached mycoplasmas, the respective ATP contents were measured by dispensing triplicate 25-μl aliquots into a 96-well sample plate (B&W Isoplate; Wallac) and processed using the ATP bioluminescence assay kit HS II (Roche) as specified by the manufacturer. A Victor3V 1420 multilabel counter was used for bioluminescence measurements. To calculate the number of mycoplasmas attached to the plastic surface, the number of unattached mycoplasmas was subtracted from the total number of mycoplasmas in each binding experiment. Experiments were repeated three times per strain.
Surface proteolysis of M. genitalium.
To detect possible surface-exposed regions of the MG312 protein, intact cells from the WT strain were digested with proteinase K. Two mid-log-phase cultures grown in 75-cm2 tissue culture flasks were washed three times with PBS and resuspended in 4 ml of the same buffer. The cell suspension was divided into eight 500-μl aliquots. Six aliquots were subjected to proteolysis by adding proteinase K (80 μg ml−1) and incubating for 5, 10, 15, 30, 45, and 60 min at room temperature. The digestions were stopped by adding phenylmethylsulfonyl fluoride (PMSF; 1 mM final concentration), and cells were then centrifuged, washed with 1 mM PMSF in PBS, and analyzed by Western blotting using anti-P140 and anti-HMW1 antibodies. For control purposes, an aliquot of 500 μl of cells was processed as described above but no proteinase K or PMSF was added to the cell suspension. An additional control was performed by adding PMSF prior to the proteinase K treatment and then processing the sample as described above.
RESULTS
Obtaining MG_312 null mutants by homologous recombination.
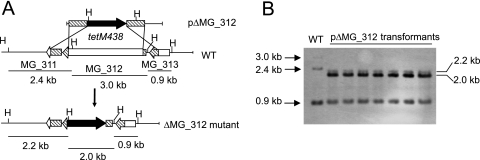
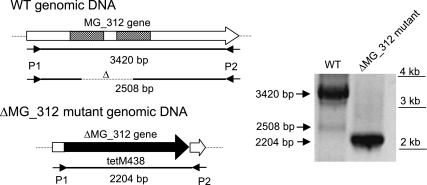
Since gene replacement by homologous recombination has been reported in M. genitalium (2, 4, 5), this approach was used to obtain MG_312 null mutants. For this purpose we constructed the suicidal plasmid pΔMG_312, which contains the tetM438 selectable marker (26) enclosed by the flanking regions of the MG_312 gene. A recombination event involving double crossover between plasmid pΔMG_312 and the M. genitalium genome results in the deletion of bases 111 to 3281 of the MG_312 gene (92.6% of the coding sequence). Deleted sequences were replaced by the tetM438 selectable marker, which confers tetracycline (Tc) resistance to transformed cells (Fig. 1A). After electroporation of M. genitalium WT cells with the pΔMG_312 plasmid, we obtained 125 resistant colonies, representing a transformation efficiency of 2.75 × 10−7 per viable cell. This transformation efficiency is comparable to those obtained for the recently reported ΔMG_191 and ΔMG_192 deletion mutants (2). Among the Tc-resistant colonies arising from the transformation assay with pΔMG_312, several colonies were picked and propagated in SP-4 medium containing Tc. All pΔMG_312 transformants isolated showed severe plastic attachment deficiencies and most of their cells grew aggregated in cell suspension. To further confirm the absence of the deleted sequence, genomic DNAs from the WT strain and pΔMG_312 transformants were subjected to Southern blot analyses. Genomic DNAs were digested with HindIII and probed with complementary sequences corresponding to the flanking regions of the MG_312 gene. As can be seen in Fig. 1B, all transformants (designated ΔMG_312 mutants) exhibited a hybridization pattern compatible with a double-crossover event, demonstrating replacement of the MG_312 gene by the tetM438 selectable marker.
FIG. 1.
Homologous recombination at the MG_312 gene. (A) Schematic representation of a double-crossover recombination between pΔmg312 and the MG_312 gene and gene replacement by the tetM438 selectable marker. Hatched boxes represent flanking regions of the MG_312 gene in suicide plasmid pΔmg312. Black, tetM438 selectable marker. Sizes of fragments resulting from the HindIII (H) digestion of DNAs from the WT strain and several pΔmg312 transformants are also indicated. (B) Southern blot hybridization profiles of genomic DNA from the WT strain and pΔmg312 transformants using the MG_312 flanking regions as a probe.
Protein profile, cytadherence, and cell morphology characterization of ΔMG_312 mutants.
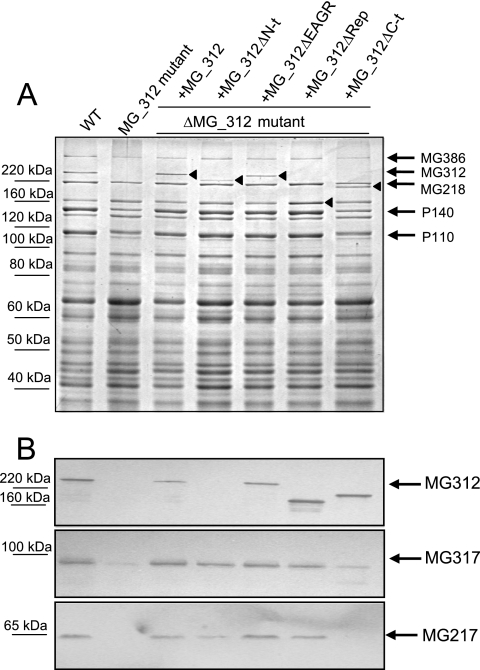
Protein profiles of the ΔMG_312 mutants and the WT strain were examined by SDS-PAGE and Western blotting. As expected, the loss of a ca. 220-kDa band, which was previously identified by mass spectrometry as an MG312 protein (39), was evident in the ΔMG_312 mutant protein profile, as shown by SDS-PAGE (Fig. 2A) and Western blotting using anti-HMW1 antibodies (Fig. 2B). Loss of the MG312 protein was also correlated with significantly reduced levels of the major cytadhesins P140 and P110 and a slight reduction in the MG386 and MG218 protein levels (Fig. 2A). Western blot analyses using an anti-MG217 monoclonal antibody and anti-HMW3 antibodies also revealed a remarkable reduction in the MG217 (homologue to M. pneumoniae P65) and MG317 (homologue to M. pneumoniae HMW3) protein levels (Fig. 2B). These results indicate that loss of the MG312 protein affects the stability of many components of the attachment organelle, suggesting a role for MG312 as a structural protein maintaining the integrity of the M. genitalium terminal organelle.
FIG. 2.
Protein profile of the WT strain and ΔMG_312 and ΔMG_312 derivative mutants. The protein profile was analyzed by 8% SDS-PAGE (A) and Western immunoblotting using anti-HMW1, anti-HMW3, and anti-MG217 antibodies (B). Arrowheads show expression of the different ΔMG_312 derivatives.
Cytadherence properties of the ΔMG_312 mutants were tested by qualitative HA assessment, revealing intermediate HA activity (Table 2). Cell surface adsorption of ΔMG_312 mutants was also tested by quantitative plastic binding assays, indicating as well a limited cell adsorption phenotype (29.2%) (Table 3) compared to that of the WT. These results are in agreement with the involvement of the MG312 protein in M. genitalium cytadherence.
TABLE 2.
Summary of phenotypes of the ΔMG_312 and ΔMG_312 derivative mutants
| Strain genotype | Protein levela
|
HA phenotypec | Cell morphology | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MG312b | MG386 | MG218 | P140 | P110 | MG317 | MG217 | |||
| WT | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | Flask shape |
| ΔMG_312 | − | ++ | ++ | + | + | ± | ± | + | Rounded/pleomorphic |
| ΔMG_312 + MG_312 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | Near WT |
| ΔMG_312 + MG_312ΔRep | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | Near WT |
| ΔMG_312 + MG_312ΔEAGR | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | Near WT |
| ΔMG_312 + MG_312ΔN-t | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | Near WT/elongated |
Relative amounts of protein: −, none; ±, very reduced; +, significantly reduced; ++, slightly reduced; +++, WT level.
For the MG_312 derivative mutants, symbols indicate the stability of the MG312 derivative form.
Qualitative HA activity: +, intermediate HA activity; +++, WT HA activity.
TABLE 3.
Characterization of cell gliding and plastic binding capabilities of the WT strain and ΔMG_312 and ΔMG_312 derivative mutantsa
| Strain genotype | Mean gliding speed (%) | Mean gliding frequency (%) | Plastic binding activity (%) |
|---|---|---|---|
| WT | 100 ± 13.1 | 100 | 100 ± 3.9 |
| ΔMG_312 | 0 | 0 | 29.2 ± 5.0 |
| ΔMG_312 + MG_312 | 98.4 ± 18.2 | 84.8 | 84.7 ± 7.9 |
| ΔMG_312 + MG_312ΔRep | 85.7 ± 18.9 | 73.1 | 109.9 ± 6.9 |
| ΔMG_312 + MG_312ΔEAGR | 59.0 ± 21.8 | 72.7 | 90.3 ± 2.5 |
| ΔMG_312 + MG_312ΔN-t | 40.1 ± 17.5 | 55.4 | 82.5 ± 6.9 |
Values (± standard deviations) are normalized to WT levels.
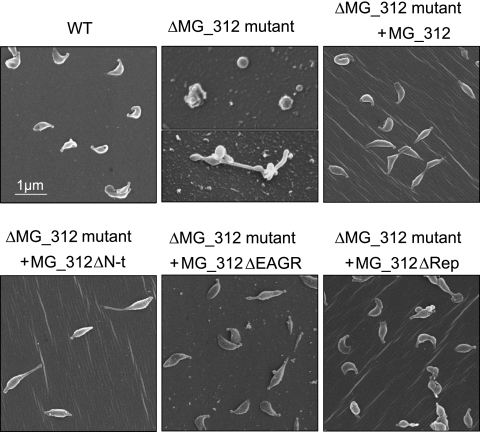
Cells from ΔMG_312 mutants also showed altered cell morphology, as demonstrated by SEM. Because ΔMG_312 mutant cells adhere poorly to plastic surfaces, poly-l-lysine-coated coverslips were used to promote cell attachment. In contrast to WT M. genitalium with its characteristic flask shape appearance, showing a well-defined terminal structure (2), the ΔMG_312 mutants were characterized by the loss of the attachment organelle (Fig. 3). In particular, the ΔMG_312 mutants exhibited rounded and pleomorphic cells reminiscent of those observed for the ΔMG_191 and ΔMG_192 mutants (2).
FIG. 3.
SEM analyses of the WT strain and ΔMG_312 and ΔMG_312 derivative mutants. All pictures are shown at the same magnification.
Gliding motility of ΔMG_312 mutants.
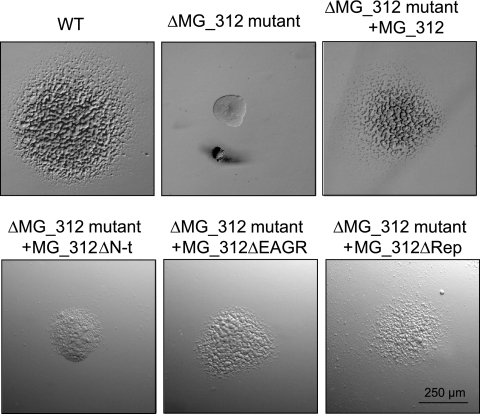
We have previously shown that the colony morphology of M. genitalium cells, attached to cell culture dishes and covered with SP-4 medium containing 0.5% low-melting-point agarose, reflects the gliding status of the studied strain (25). Under these culture conditions, WT cells developed noncompact colonies with satellite microcolonies, a feature that characterizes gliding-proficient strains (Fig. 4). In contrast, cells from the ΔMG_312 mutants developed colonies showing smooth surfaces without satellite formation, indicating the existence of severe gliding deficiencies in this mutant (Fig. 4). Gliding motility of the ΔMG_312 mutants was further characterized by cinematography (Table 3). Among the cells attached in the preparation, most were aggregated and frequently detached from plastic and moved vigorously, exhibiting a Brownian movement. However, the few individual cells that remained attached during the examination period were completely nonmotile, demonstrating in this way the involvement of the MG312 protein in gliding motility.
FIG. 4.
Colony morphology of the WT and ΔMG_312 and ΔMG_312 derivative mutants. Cells from the WT strain and ΔMG_312 and ΔMG_312 derivative mutants were attached to cell culture dishes and covered with SP-4 medium containing 0.5% low-melting-point agarose. All pictures are shown at the same magnification.
Construction of MG_312 deletion derivatives.
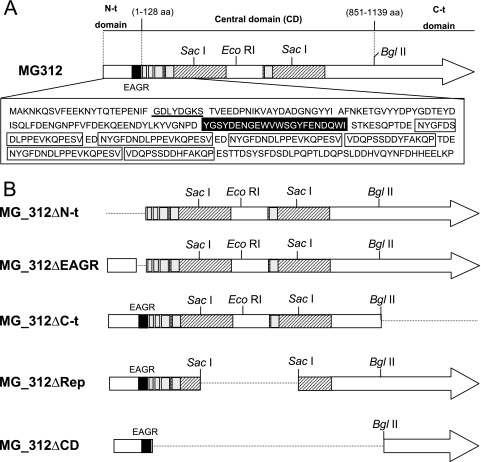
The involvement of MG312 in gliding motility seems evident, as we demonstrated above. However, it remains unclear whether the contribution of MG312 to gliding motility is a direct consequence of the loss of the terminal organelle and/or whether MG312 may also play a specific and independent role in M. genitalium cell locomotion. The C terminus of HMW1 has been previously linked with functions related to cytadherence and cell morphology (45). We have addressed the question of whether the functions related to adhesion and locomotion may be located in separate MG312 protein domains, as suggested by the presence of an EAGR box in the MG312 N terminus (Fig. 5A). To answer this question, we engineered several deletion derivatives of the MG_312 gene (Fig. 5B). These constructions were designed according to the modular structure previously described for the M. pneumoniae HMW1 protein (6), which remains well conserved in the M. genitalium MG312 protein. From their deduced amino acid sequences, both homologue proteins consist of three distinct domains (6). In particular, the MG312 N-terminal domain (Met1 to Glu128), which includes the EAGR box, and the C-terminal domain (Leu851 to Tyr1139) are especially well conserved in both proteins (Fig. 5A). The MG312 central domain, which is not conserved in HMW1, comprises two large and nonconsecutive repeats of 196 amino acids each. These repeats are identical in their amino acid sequences as well as their DNA coding sequences, excepting nucleotide 934, which for the first repeat is an A and for the second repeat is a G, changing the amino acid Lys to a Glu. In addition, several smaller repeating motifs of 18 or 32 amino acids are found in the first half of this central domain (Fig. 5A).
FIG. 5.
Schematic representation of the modular structure of the MG312 protein (A) and the construction designed to obtain the MG_312 deletion derivatives (B). The EAGR box is indicated in black. The Walker A box is underlined. Hatched boxes represent the two large identical repeats found in the central domain of MG312 protein; the small repeating motifs are indicated by gray boxes. Amino acid sequences corresponding to the EAGR box and the small repeating motifs are also indicated. Regions deleted in MG_312 deletion derivatives are represented by discontinuous lines.
In the PCRs performed to obtain the full-length MG_312 and MG_312ΔN-t derivatives, it was noticeable that some PCR products were shorter than expected. When primers 5′BEKOMG312 and 5′BDKOMG312 were used, we also detected, in addition to the expected 3.4-kb band, corresponding to the MG_312 gene product, a 2.5-kb product, suggesting the presence within the WT population of specific deletion mutants (Fig. 6). These mutants can arise by double recombination between the two large repeats found in the MG_312 gene. To ensure PCR specificity, a control was performed using DNA from the ΔMG_312 mutant. In this mutant, no bands other than a single 2.2-kb fragment are expected because the MG_312 gene is replaced by the tetM438 marker (Fig. 6). The recombinant origin of the 2.5-kb PCR band was further confirmed by sequencing the corresponding DNA fragment, demonstrating the existence of deletion mutants analogous to the MG_312ΔRep derivative in natural populations of M. genitalium.
FIG. 6.
PCR amplification from DNA of the M. genitalium WT strain and the ΔMG_312 mutant. The primers 5′BEKOMG312 (P1) and 5′BDKOMG312 (P2) were used in both PCR amplifications. In addition to the expected 3.4-kb band, corresponding to the MG_312 gene product, we also detected a 2.5-kb product, suggesting the presence within the WT population of specific MG_312 deletion mutants. As a control, a single 2.2-kb product was amplified from ΔMG_312 mutant DNA.
Analyses of the ΔMG_312 derivatives.
The different MG_312 deletion derivatives were reintroduced separately into the ΔMG_312 mutant by transposon delivery (Fig. 5B). Several transformants for each ΔMG_312 derivative were isolated, cloned, and further analyzed. Since we aimed to identify putative domains of the MG312 protein related to gliding motility but not to adherence, those constructions that did not restore plastic adherence were not further analyzed. This was the case for the MG_312ΔC-t and MG_312ΔCD derivatives, which failed to restore a plastic adherence proficiency phenotype as well as a WT protein profile, even when multiple clones were screened (Fig. 2 and data not shown). It is noteworthy that the MG_312ΔC-t derivative, which lacks the C-terminal domain, was expressed in a stable form in the ΔMG_312 mutant, as demonstrated by SDS-PAGE and Western blotting (Fig. 2A and B), ruling out transcriptional or translational defects in the construction. Hence, the C-terminal domain of MG312 seems particularly important for MG312 function in adherence and proper terminal organelle assembly. On the other hand, no detectable levels of the MG_312ΔCD derivative product could be distinguished by SDS-PAGE or Western blotting (data not shown), indicating that the whole central domain is probably necessary for maintaining MG312 stability.
The reintroduction of the full MG_312 gene into the ΔMG_312 mutant restored the WT protein profile as expected (Fig. 2). In addition, steady-state levels of recombinant MG312 protein were similar to those of the resident MG312 protein in the WT strain (Fig. 2). Restoration of the WT protein profile was also correlated with the reestablishment of normal cell morphology, as demonstrated by SEM, in addition to a plastic adherence proficiency phenotype when cells were grown in tissue culture flasks (Fig. 3 and data not shown). The same was true for the MG_312ΔEAGR and MG_312ΔRep derivatives (Fig. 2 and 3 and Table 3), indicating that the EAGR box is not required to restore the adherence properties and proper cell morphology and that these functions can be fulfilled by the presence of a single repeat of the central domain.
In the case of the MG_312ΔN-t derivative, a slight instability of the recombinant MG_312ΔN-t protein was observed by SDS-PAGE (Fig. 2A) among the multiple transformants screened. The failure of detection by Western blotting of the MG_312ΔN-t deletion derivative using the anti-HMW1 serum (Fig. 2B) indicates that this serum reacts only against the N-terminal region of the MG312 protein. The observed instability in the MG_312ΔN-t deletion derivative may indicate that the N-terminal domain is required for MG312 stability. However, restoration of surface adherence, a WT protein profile, and terminal organelle formation was also achieved (Table 3; Fig. 2 and 3), demonstrating that the first 128 amino acids are not essential for the attachment functions and terminal organelle assembly. Nevertheless, SEM analyses also showed that, in contrast to the WT strain, some cells harboring the recombinant MG_312ΔN-t derivative exhibited a more elongated shape, leaving a long filament at the end (Fig. 3). In Table 2 are summarized the phenotypes of the ΔMG_312 and ΔMG_312 derivative mutants.
Gliding motility of the ΔMG_312 derivatives.
As stated above, neither the large repeats of the central domain nor the first 128 amino acids of the MG312 protein are essential for terminal organelle assembly and attachment functions. However, to determine whether some of these domains of the MG312 protein are required for normal cell gliding, we also examined MG_312ΔEAGR, MG_312ΔRep, and MG_312ΔN-t transformants by time-lapse cinematography (Table 3) and colony morphology (Fig. 4). Transformants harboring the full MG_312 recombinant gene moved at 98.4% relative to the WT gliding speed, with a mean gliding frequency of 84.9% of that of the WT strain. Likewise, MG_312ΔRep transformants glided at approximately WT velocities (85.7% of the WT level), with a mean gliding frequency of 73.1% of the WT levels. In contrast, when MG_312ΔN-t and MG_312ΔEAGR transformants were analyzed by time-lapse cinematography, we observed a significant reduction in their gliding velocities: 40.1% and 59% of the WT levels, respectively. Furthermore, while MG_312ΔEAGR transformants exhibited a mean gliding frequency of 72.7% of the WT levels, the MG_312ΔN-t transformants exhibited a significant reduction in their gliding frequencies (55.4% of the WT levels).
These results were in agreement with the colony morphology exhibited by the different ΔMG_312 derivative mutants when grown covered with SP-4 medium containing 0.5% low-melting-point agarose (Fig. 4). The full MG_312 recombinant gene or the MG_312ΔRep derivative restored the formation of satellite microcolonies. In contrast, despite the fact that some satellite microcolonies were observed in the MG_312ΔEAGR transformants, a compact morphology was still apparent in the center of the colony. Finally, MG_312ΔN-t transformants showed a more severe gliding deficiency phenotype, displaying compact and granulate colonies without satellite formation.
Plastic binding properties of the ΔMG_312 derivatives.
Plastic adherence is a prior and necessary step in order for cells to glide. To rule out whether the gliding deficiency phenotypes observed were due to a poor attachment to the plastic surface not visible microscopically, the plastic binding activity of the ΔMG_312 derivative mutants was further examined by quantitative analyses. All ΔMG_312 derivative mutants analyzed displayed plastic binding activities between 82.5% and 90.3% of the WT level, except for the MG_312ΔRep transformant, which exhibited an enhanced binding activity of 109.9% of the WT level (Table 3). Although several clones were screened, the plastic binding activity exhibited by transformants expressing the MG_312 recombinant gene was not completely restored. This could be explained by differences between the promoter sequence used in this study and the natural promoter and the possible existence of regulatory sequences in the MG_312 operon. However, the plastic binding activity exhibited by MG_312ΔN-t and MG_312ΔEAGR transformants was comparable to that obtained with transformants harboring the MG_312 recombinant gene. In consequence, the deficiencies in gliding motility observed in MG_312ΔN-t and MG_312ΔEAGR transformants are not derived from a reduced plastic binding activity in these mutants. These results indicate that the N terminus of the MG312 protein, and in particular the EAGR box, is involved in the gliding motility of M. genitalium.
Cell location of the MG312 protein.
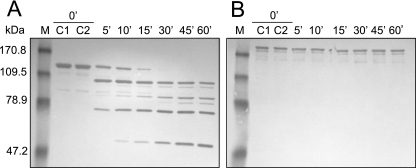
To further investigate the role of the MG312 protein in gliding motility, we have examined the possible surface exposure of this protein by limited proteolysis of intact cells using proteinase K (Fig. 7). Immunoblot analysis with anti-HMW1 serum of treated cells showed that the MG312 protein, even after 1 h of proteinase K incubation, is resistant to cleavage, suggesting an intracellular location of this protein. However, we cannot exclude the possibility that small amounts of MG312 are accessible to proteinase K since the corresponding proteolysis fragments are not detectable with the heterologous antisera used. We also cannot exclude the possibility that the MG312 protein is found on the cell surface tightly associated with other proteins and inaccessible to protease cleavage. In contrast, the P140 adhesin, a surface-exposed protein used as a positive control, is completely cleaved after 30 min of digestion.
FIG. 7.
Partial time-dependent surface proteolysis of M. genitalium WT strain. Intact cells were treated with proteinase K, and reactions were stopped by adding 1 mM PMSF after 5, 10, 15, 30, 45, and 60 min. Protein profiles from cells that underwent proteolysis were analyzed by Western blotting using anti-P140 (A) and the HMW1 antiserum (B). C1 corresponds to the protein profile of untreated cells, and C2 corresponds to the protein profile resulting when PMSF was added to the cells prior to proteinase K. M, apparent molecular mass prestained marker (Invitrogen).
DISCUSSION
Mycoplasmas have attracted the interest of many researchers since they are considered to be among the simplest self-replicating organisms (8, 9). However, if we examine in depth the genome from M. genitalium and related mycoplasmas (8, 17), we notice that, despite their minimal genomes, an important set of genes are coding for proteins exhibiting novel features not found in any other protein known. Interestingly, most of these novel proteins are components of the terminal organelle, reflecting the exceptionality of this fascinating structure in the living world. The role of the different components of the terminal organelle of M. genitalium remains mostly uncharacterized. To date, only the major adhesins P140 and P110 and the cytadherence-related protein MG218 (homologue to M. pneumoniae HMW2) have been characterized at the functional level (2, 5, 24). Since previous attempts to isolate spontaneous or transposon-induced M. genitalium mutants in genes coding for additional proteins of this structure have been unsuccessful, we have taken advantage of the possibility of deleting target genes by homologous recombination. This genetic tool is becoming an alternative and powerful approach to gain insight into the mechanisms of M. genitalium pathogenesis.
Here we demonstrate the involvement of MG312 in M. genitalium cytadherence, as previously suggested for the M. pneumoniae HMW1 orthologue protein (10). However, the intermediate-cytadherence phenotype exhibited by the ΔMG_312 mutant differs from the complete HA− phenotype exhibited by the M. pneumoniae M6 strain (10). This fact suggests differences regarding the role of MG312/HMW1 in the cytadherence properties of M. genitalium and M. pneumoniae or the presence of additional mutations in the M. pneumoniae M6 strain (10). On the other hand, the cytadherence activity remaining in the ΔMG_312 mutants justifies the absence of M. genitalium MG312 mutants after screening for nonadherent mutants (24, 28).
The loss of MG312 is also correlated with the loss of the terminal organelle and reduced levels of MG218, MG317, and MG217 proteins, as has been previously reported for M. pneumoniae (10, 45). In addition, our results also show reduced levels of P140, P110, and MG386 proteins. All these proteins are components of the attachment organelle, clearly indicating the importance of MG312 as a structural element maintaining the integrity of the M. genitalium terminal organelle and its components. In addition, we have previously shown that P140 and P110 adhesins are essential for cytadherence and MG386 stability but not for MG312 stability (2). Taken together, these data suggest that the partial-cytadherence phenotype and the reduced levels of MG386 observed in the ΔMG_312 mutants might be the result of the instability of P140 and P110 rather than a direct consequence of the loss of the MG312 protein. Although the loss of HMW1 in M. pneumoniae is not correlated with reduced levels of P140 and P110 homologues (P1 and B/C proteins, respectively), P1 is not properly localized and it is found scattered along the cell surface (10). If the MG312 protein is likewise involved in P140 localization, this could explain the low levels of P140 and P110 found in the ΔMG_312 mutant. As has been reported for other terminal organelle components (21), P140 and P110 adhesins could be subjected to an accelerated proteolytic turnover when not incorporated into the M. genitalium cytoskeletal framework.
We also found that ΔMG_312 mutants are unable to glide. Similar results were found for several M. pneumoniae (12, 14) and M. genitalium (O. Q. Pich, unpublished results) mutants with defects in cytadherence-related proteins. However, these findings may not be surprising given the unusual cell morphology exhibited by these mutants, which were characterized by lacking defined terminal organelles. The terminal organelle, in addition to mediating cell adhesion, is the leading end of gliding cells, and recent studies have provided clear evidence that this structure constitutes the gliding motor (13). Therefore, we must assume that those cytoskeletal components required for the assembly of the terminal organelle are in consequence necessary to obtain proper cell adhesion and gliding motility. In this way, MG312 (HMW1), which has been traditionally considered a cytadherence-associated protein (20), should also be considered a gliding-motility-related protein. However, some evidence, like the presence of an EAGR box, supports the idea that, independently from downstream events affecting the terminal organelle architecture as a whole, the MG312 protein may play a specific function in gliding motility. Analyses of the different deletion derivative mutants have allowed us to identify two important and separate functional domains in the MG312 protein. In accordance with previously described results for the M. pneumoniae HMW1 protein (45), we found that the C-terminal domain of the MG312 protein is required for adhesion and terminal organelle assembly functions. In contrast, the N-terminal domain is not required for these functions but seems to contribute itself to M. genitalium cell locomotion. It could be argued that the motility defects observed in the absence of the N-terminal region are a consequence of the decreased protein levels detected in the corresponding derivative. This possibility seems unlikely, since the observed levels of the MG312 N terminus deletion derivative are sufficient to restore adherence and terminal organelle assembly functions. Furthermore, when we examined the specific implication of the EAGR box in cell gliding, MG_312ΔEAGR derivative mutants also showed motility defects and expressed normal levels of the MG312 derivative. Since the EAGR box is located at the N terminus, these results suggest a direct role for the MG312 N terminus in motility and indicate a specific contribution of the EAGR box to mycoplasma cell locomotion. The EAGR boxes may contribute to the interactions among gliding-motility-related proteins. In fact, aromatic residues are commonly found in the interfaces of protein-protein interactions (19). However, further investigation is needed to elucidate the specific role of this motif in gliding motility.
Examination of the MG312 N terminus reveals the presence of the conserved ATP binding motif GDLYDGKS (residues 24 to 31), known as a P-loop or Walker A box (44). This motif is defined as a GXXXXGKT/S consensus and is present in most of the proteins that bind ATP, including regulatory kinases and motor proteins such as myosin. Although it remains to be determined whether the MG312 N terminus can actually bind and hydrolyze ATP, this finding is consistent with the idea that the MG312 protein, in addition to its role as a scaffold protein, also plays an active role in the gliding mechanism. Published evidence suggests that the attachment organelle is where the motive force is generated (13) and that therefore some component of the core must consume energy, as has been previously suggested (16). In addition, it has been proposed for M. pneumoniae that conformational changes in the electron-dense core could push forward the cell tip by an inchworm-like mechanism, driving in this way cell gliding (16). Since the absence of the MG312 N-terminal region reduces but does not abolish gliding motility, this region could be one of the elements contributing to the conformational changes driving cell gliding. Alternatively, this region could play a regulatory role, for instance, coordinating some of the events in the sequence of the steps needed for gliding. Such active roles argue for an intracellular location of the MG312 protein, consistent with the resistance of this protein to the surface proteolysis found in this work. However, studies using different immunological approaches for M. pneumoniae revealed the presence of two HMW1 pools, a cell surface-exposed pool and a second protein pool, which lies in the cell interior (1, 6). Although the origin of this divergence is unclear, the intracellular residence of the MG312 protein is consistent both with its role as a structural element of the terminal organelle and with the specific implication of the MG312 N-terminal region in gliding motility.
Dirksen and colleagues (6) suggested that HMW1 might exist with globular domains at each end separated by an extended central domain. The same structure is found in the MG312 protein, the N-terminal and C-terminal domains being well conserved between the two proteins. However, their central domains are completely different. The M. genitalium central domain is composed of two large repeats of 196 amino acids, which are not found in M. pneumoniae HMW1. An interesting finding of this study is the observation that ΔMG_312 mutants expressing the MG_312ΔRep derivative exhibited increased levels of plastic binding. The specific length of this central domain could be important to maintain the appropriate location of MG312 in the terminal organelle, making possible the proper function of its globular domains. Alternatively, the length of this central domain might be a modulator for the fine-tuning of M. genitalium cytadherence properties, as suggested by our results. Interestingly, the adjustment of the length of the MG312 central domain is potentially possible in a WT population by postreplicative recombination between the repetitive sequences in the recently replicated chromosomes (Fig. 6). In addition, the smallest repeats found downstream of the EAGR box could also undergo similar recombination events, contributing to the flexible and dynamic properties of the MG312 protein. However, it remains to be determined whether these rearrangements take place in the course of a natural infection.
In conclusion, the functional analysis performed on the MG312 protein has shown that this structural protein is not only involved in cell adhesion and terminal organelle architecture but also implicated in gliding motility. In particular, we found that the MG312 N terminus, which includes a Walker A box and an EAGR box, is specifically involved in gliding motility. The implication of the EAGR box in cell locomotion has been also demonstrated in this study. Future studies including the structural and biochemical characterization of the MG312 N terminus could contribute to the better understanding of M. genitalium cell locomotion, providing new insights into mycoplasma virulence and pathogenicity.
Acknowledgments
This work was supported by grant BFU2004-06377-C02-01 to E.Q. R.B. acknowledges an FPU predoctoral fellowship from the Ministerio de Educación y Ciencia, and O.Q.P. acknowledges a predoctoral fellowship from the Centre de Referència en Biotecnologia.
We are grateful to J. B. Baseman for providing P140 monoclonal antibodies and to D. C. Krause for providing HMW1 and HMW3 antisera. We also thank the staff of the Servei de Microscòpia (UAB) for processing scanning electron microscopy samples.
Footnotes
Published ahead of print on 3 August 2007.
REFERENCES
- 1.Balish, M. F., T. W. Hahn, P. L. Popham, and D. C. Krause. 2001. Stability of Mycoplasma pneumoniae cytadherence-accessory protein HMW1 correlates with its association with the triton shell. J. Bacteriol. 183:3680-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgos, R., O. Q. Pich, M. Ferrer-Navarro, J. B. Baseman, E. Querol, and J. Pinol. 2006. Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal-organelle development. J. Bacteriol. 188:8627-8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, C. R., L. E. Manhart, E. A. Bukusi, S. Astete, R. C. Brunham, K. K. Holmes, S. K. Sinei, J. J. Bwayo, and P. A. Totten. 2002. Association between Mycoplasma genitalium and acute endometritis. Lancet 359:765-766. [DOI] [PubMed] [Google Scholar]
- 4.Dhandayuthapani, S., M. W. Blaylock, C. M. Bebear, W. G. Rasmussen, and J. B. Baseman. 2001. Peptide methionine sulfoxide reductase (MsrA) is a virulence determinant in Mycoplasma genitalium. J. Bacteriol. 183:5645-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhandayuthapani, S., W. G. Rasmussen, and J. B. Baseman. 1999. Disruption of gene mg218 of Mycoplasma genitalium through homologous recombination leads to an adherence-deficient phenotype. Proc. Natl. Acad. Sci. USA 96:5227-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dirksen, L. B., T. Proft, H. Hilbert, H. Plagens, R. Herrmann, and D. C. Krause. 1996. Sequence analysis and characterization of the hmw gene cluster of Mycoplasma pneumoniae. Gene 171:19-25. [DOI] [PubMed] [Google Scholar]
- 7.Falk, L., H. Fredlund, and J. S. Jensen. 2005. Signs and symptoms of urethritis and cervicitis among women with or without Mycoplasma genitalium or Chlamydia trachomatis infection. Sex. Transm. Infect. 81:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 9.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103:425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn, T. W., M. J. Willby, and D. C. Krause. 1998. HMW1 is required for cytadhesin P1 trafficking to the attachment organelle in Mycoplasma pneumoniae. J. Bacteriol. 180:1270-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harlow, E., and D. Lane. 1998. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 12.Hasselbring, B. M., J. L. Jordan, and D. C. Krause. 2005. Mutant analysis reveals a specific requirement for protein P30 in Mycoplasma pneumoniae gliding motility. J. Bacteriol. 187:6281-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasselbring, B. M., and D. C. Krause. 2007. Cytoskeletal protein P41 is required to anchor the terminal organelle of the wall-less prokaryote Mycoplasma pneumoniae. Mol. Microbiol. 63:44-53. [DOI] [PubMed] [Google Scholar]
- 14.Hasselbring, B. M., C. A. Page, E. S. Sheppard, and D. C. Krause. 2006. Transposon mutagenesis identifies genes associated with Mycoplasma pneumoniae gliding motility. J. Bacteriol. 188:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegermann, J., R. Herrmann, and F. Mayer. 2002. Cytoskeletal elements in the bacterium Mycoplasma pneumoniae. Naturwissenschaften 89:453-458. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, G. P., and G. J. Jensen. 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol. Microbiol. 60:376-385. [DOI] [PubMed] [Google Scholar]
- 17.Himmelreich, R., H. Hilbert, H. Plagens, E. Pirkl, B. C. Li, and R. Herrmann. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24:4420-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horner, P. J., C. B. Gilroy, B. J. Thomas, R. O. Naidoo, and D. Taylor-Robinson. 1993. Association of Mycoplasma genitalium with acute non-gonococcal urethritis. Lancet 342:582-585. [DOI] [PubMed] [Google Scholar]
- 19.Jones, S., and J. M. Thornton. 1997. Analysis of protein-protein interaction sites using surface patches. J. Mol. Biol. 272:121-132. [DOI] [PubMed] [Google Scholar]
- 20.Krause, D. C., and M. F. Balish. 2004. Cellular engineering in a minimal microbe: structure and assembly of the terminal organelle of Mycoplasma pneumoniae. Mol. Microbiol. 51:917-924. [DOI] [PubMed] [Google Scholar]
- 21.Krause, D. C., and M. F. Balish. 2001. Structure, function, and assembly of the terminal organelle of Mycoplasma pneumoniae. FEMS Microbiol. Lett. 198:1-7. [DOI] [PubMed] [Google Scholar]
- 22.Layh-Schmitt, G., H. Hilbert, and E. Pirkl. 1995. A spontaneous hemadsorption-negative mutant of Mycoplasma pneumoniae exhibits a truncated adhesin-related 30-kilodalton protein and lacks the cytadherence-accessory protein HMW1. J. Bacteriol. 177:843-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manhart, L. E., C. W. Critchlow, K. K. Holmes, S. M. Dutro, D. A. Eschenbach, C. E. Stevens, and P. A. Totten. 2003. Mucopurulent cervicitis and Mycoplasma genitalium. J. Infect. Dis. 187:650-657. [DOI] [PubMed] [Google Scholar]
- 24.Mernaugh, G. R., S. F. Dallo, S. C. Holt, and J. B. Baseman. 1993. Properties of adhering and nonadhering populations of Mycoplasma genitalium. Clin. Infect. Dis. 17(Suppl. 1):S69-S78. [DOI] [PubMed] [Google Scholar]
- 25.Pich, O. Q., R. Burgos, M. Ferrer-Navarro, E. Querol, and J. Pinol. 2006. Mycoplasma genitalium mg200 and mg386 genes are involved in gliding motility but not in cytadherence. Mol. Microbiol. 60:1509-1519. [DOI] [PubMed] [Google Scholar]
- 26.Pich, O. Q., R. Burgos, R. Planell, E. Querol, and J. Pinol. 2006. Comparative analysis of antibiotic resistance gene markers in Mycoplasma genitalium: application to studies of the minimal gene complement. Microbiology 152:519-527. [DOI] [PubMed] [Google Scholar]
- 27.Popham, P. L., T. W. Hahn, K. A. Krebes, and D. C. Krause. 1997. Loss of HMW1 and HMW3 in noncytadhering mutants of Mycoplasma pneumoniae occurs post-translationally. Proc. Natl. Acad. Sci. USA 94:13979-13984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reddy, S. P., W. G. Rasmussen, and J. B. Baseman. 1996. Isolation and characterization of transposon Tn4001-generated, cytadherence-deficient transformants of Mycoplasma pneumoniae and Mycoplasma genitalium. FEMS Immunol. Med. Microbiol. 15:199-211. [DOI] [PubMed] [Google Scholar]
- 29.Regula, J. T., G. Boguth, A. Gorg, J. Hegermann, F. Mayer, R. Frank, and R. Herrmann. 2001. Defining the mycoplasma ‘cytoskeleton’: the protein composition of the Triton X-100 insoluble fraction of the bacterium Mycoplasma pneumoniae determined by 2-D gel electrophoresis and mass spectrometry. Microbiology 147:1045-1057. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, R. J. 2004. Identifying protein function—a call for community action. PLoS Biol. 2:E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robertson, J. A., and G. W. Stemke. 1995. Measurement of mollicute growth by ATP-dependent luminometry, p. 65-71. In S. Razin and J. G. Tully (ed.), Molecular and diagnostic procedures in mycoplasmology, vol. I. Academic Press, Inc., San Diego, CA. [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 33.Seto, S., T. Kenri, T. Tomiyama, and M. Miyata. 2005. Involvement of P1 adhesin in gliding motility of Mycoplasma pneumoniae as revealed by the inhibitory effects of antibody under optimized gliding conditions. J. Bacteriol. 187:1875-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto, S., G. Layh-Schmitt, T. Kenri, and M. Miyata. 2001. Visualization of the attachment organelle and cytadherence proteins of Mycoplasma pneumoniae by immunofluorescence microscopy. J. Bacteriol. 183:1621-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seto, S., A. Uenoyama, and M. Miyata. 2005. Identification of a 521-kilodalton protein (Gli521) involved in force generation or force transmission for Mycoplasma mobile gliding. J. Bacteriol. 187:3502-3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seybert, A., R. Herrmann, and A. S. Frangakis. 2006. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J. Struct. Biol. 156:342-354. [DOI] [PubMed] [Google Scholar]
- 37.Stevens, M. K., and D. C. Krause. 1991. Localization of the Mycoplasma pneumoniae cytadherence-accessory proteins HMW1 and HMW4 in the cytoskeletonlike Triton shell. J. Bacteriol. 173:1041-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens, M. K., and D. C. Krause. 1992. Mycoplasma pneumoniae cytadherence phase-variable protein HMW3 is a component of the attachment organelle. J. Bacteriol. 174:4265-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Svenstrup, H. F., J. S. Jensen, K. Gevaert, S. Birkelund, and G. Christiansen. 2006. Identification and characterization of immunogenic proteins of Mycoplasma genitalium. Clin. Vaccine Immunol. 13:913-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taylor-Robinson, D. 2002. Mycoplasma genitalium—an up-date. Int. J. STD AIDS 13:145-151. [DOI] [PubMed] [Google Scholar]
- 41.Tully, J. G., D. L. Rose, R. F. Whitcomb, and R. P. Wenzel. 1979. Enhanced isolation of Mycoplasma pneumoniae from throat washings with a newly-modified culture medium. J. Infect. Dis. 139:478-482. [DOI] [PubMed] [Google Scholar]
- 42.Uenoyama, A., A. Kusumoto, and M. Miyata. 2004. Identification of a 349-kilodalton protein (Gli349) responsible for cytadherence and glass binding during gliding of Mycoplasma mobile. J. Bacteriol. 186:1537-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uenoyama, A., and M. Miyata. 2005. Identification of a 123-kilodalton protein (Gli123) involved in machinery for gliding motility of Mycoplasma mobile. J. Bacteriol. 187:5578-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willby, M. J., M. F. Balish, S. M. Ross, K. K. Lee, J. L. Jordan, and D. C. Krause. 2004. HMW1 is required for stability and localization of HMW2 to the attachment organelle of Mycoplasma pneumoniae. J. Bacteriol. 186:8221-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]