Abstract
Active filamentous phage particles were isolated from the deep-sea bacterium Shewanella piezotolerans WP3. A putative single-stranded DNA binding protein of the phage was found to be overexpressed at 4°C compared to its expression at 25°C by two-dimensional polyacrylamide gel electrophoresis. Reverse transcription quantitative PCR further revealed that the key genes of the SW1 phage were significantly induced at low temperature.
The genus Shewanella comprises a group of gram-negative facultative anaerobic gammaproteobacteria which is known for its versatile respiratory capabilities (9). At present, more than 18 Shewanella strains have been selected for genome sequencing by different organizations (1, 3). We have finished the genome sequencing and gene annotation of a deep-sea Shewanella strain, Shewanella piezotolerans WP3 (F. P. Wang, J. B. Wang, S. K. Li, X. W. Zeng, F. Wang, D. H. Bartlett, J. Yu, S. N. Hu, and X. Xiao, submitted for publication). WP3 is psychrotrophic and piezotrophic, with the best growth temperature and pressure at about 20°C and 20 MPa, respectively (11). In the genome of WP3, a putative filamentous phage genome (7,718 bp; named SW1) was found. Here, we report that WP3 contains a functional filamentous phage, the first filamentous phage found in the genus Shewanella. In addition, we show that the filamentous phage is significantly induced at low temperature and that its regulation may be different from that of CTXφ, whose regulation is the best studied among the filamentous phages (5, 10).
Genomic organization.
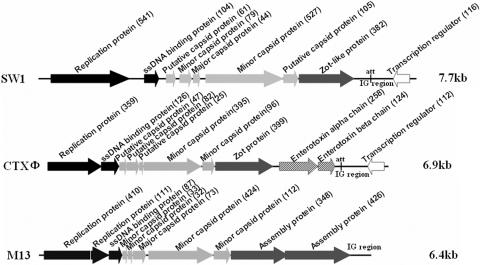
The SW1 phage genome is 7,718 nucleotides long and has nine putative open reading frames (ORFs) in a modular organization typical of filamentous phage (Fig. 1). SW1 contains four modular structures: the replication module, the structural module, the assembly module, and the regulation module (Fig. 1). The replication module contains ORF541 and ORF104. ORF61, ORF79, ORF44, ORF527, and ORF105 comprise the structural module of the phage SW1. They are of the same size and in the same relative positions as gVII, gIX, gVIII, gIII, and gVI of the capsid structural module of the Escherichia coli M13 phage (8), respectively, although with low sequence similarities, suggesting that the encoded proteins of these ORFs may have functions similar to those of the corresponding M13 phage. The putative assembly module of SW1 contains ORF382, whose corresponding N-terminal peptide is homologous to Zot; the rest of the sequence is not homologous to Zot, whose enterotoxic activity has been assigned to its C-terminal region. The SW1 phage has a putative regulator module to which ORF116 is assigned. ORF116 is transcribed in a direction opposite to all the other ORFs of SW1. The N-terminal peptide corresponding to ORF116 has a typical Cro/CI type helix-turn-helix DNA binding motif but lacks the putative peptidase domain, suggesting that the ORF116 product is a repressor protein but may not have the protein autoproteolysis function.
FIG. 1.
Genomic organization of phage SW1 and related filamentous phages CTXφ and M13. A linear map of SW1 is shown. ORFs are represented by arrows oriented in the direction of transcription. The putative gene product of each ORF is shown, and the numbers in brackets indicate the lengths of the proteins. The replication module is represented by black arrows, the structural module is represented by light gray arrows, the assembly and secretion modules are represented by dark gray arrows, and the regulatory module is represented by white arrows. The putative att site and the intergenic region are also indicated.
Phage purification and characterization.
The phage particles were successfully purified from WP3 cell cultures as described previously (4). WP3 was grown aerobically in 200 ml of 2216E medium (5 g/liter tryptone, 1 g/liter yeast extract, 0.1 g/liter FePO4, 34 g/liter NaCl) with shaking at 220 rpm at 4°C until an optical density at 600 nm of 1.2 was reached. The purified phage was negatively stained with 4% uranyl acetate and observed in a JEM2100HC transmission electron microscope (JEOL, Tokyo, Japan). Electron microscopic examination revealed that it is indeed a filamentous particle (Fig. 2). Replication form (RF) DNA was also isolated from the precipitated cells of WP3 by the standard alkaline pH extraction method (6). The purified genome and RF DNA of SW1 were treated with DNase I and RNase H. Both were digested by DNase but not by RNase (results not shown). RF DNA was digested by exonuclease III but not by mung bean nuclease, suggesting that the RF DNA is double stranded. DNA was electrophoresed on a 1% (wt/vol) agarose gel and transferred to a nylon membrane (Hybond-N+; Amersham). The SW1 forward and reverse single-stranded DNA (ssDNA) probes (each 100 bp) were synthesized by Sangon Inc. (Shanghai, China). Southern hybridization with the SW1 forward and reverse ssDNA probes showed that SW1 contains a single plus-chain genome, as the phage genome ssDNA could hybridize only with the reverse-direction ssDNA probe (photo not shown). The host range of SW1 was tested using Shewanella oneidensis (ATCC 700550T), Shewanella marinintestina (JCM11558T), Shewanella violacea (JCM10179T), and Shewanella psychrophila WP2 (CGMCC1.6159T). S. psychrophila, S. marinintestina, and S. violacea were incubated in marine 2216E medium. S. oneidensis was incubated in Luria-Bertani broth at 30°C. A 200-μl volume of host strain culture was mixed with 2 to 50 μl of purified phage and then injected into 3 ml of top agar, which was kept warm at 48°C. The mixture was overlaid on 2216E agar plates and incubated at 4 to 30°C for 1 to 3 days depending on the growth speed of the different host strains. Among them, WP2 is the only host capable of supporting plaque formation. Therefore, WP2 can be used as the test host for phage SW1. Phage SW1 was incubated at 60°C and at 70°C to check its stability under high temperatures. The plaque-forming activity of phage SW1 was stable at 60°C for 10 min but was inhibited completely by heating to 70°C. This temperature is 10°C lower than that inhibiting many other filamentous phages, such as fs-2 (4).
FIG. 2.
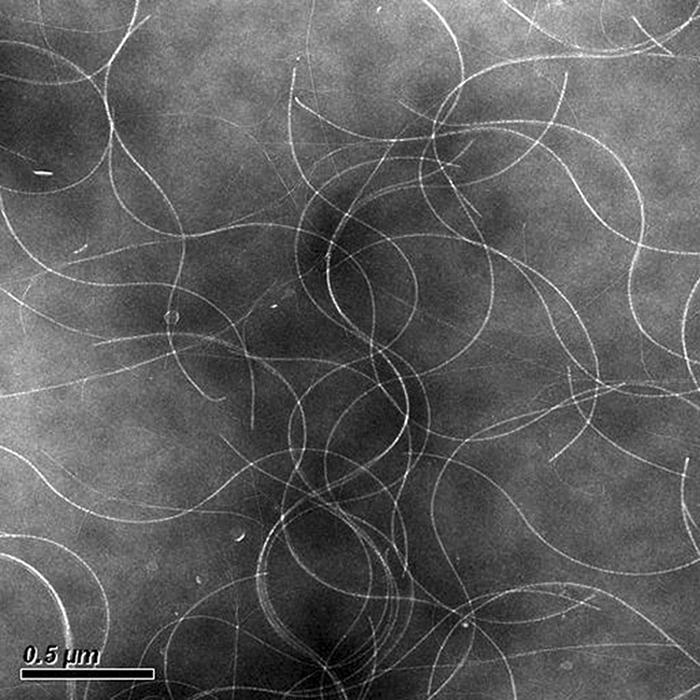
Transmission electron microscopy map of phage SW1.
Phage induction at low temperature.
The expressed protein patterns of Shewanella piezotolerans WP3 at 4°C and 25°C were compared by two-dimensional polyacrylamide gel electrophoresis (2, 7). Only a few proteins were found to be differentially expressed at these two different temperatures (results not shown). The differentially expressed proteins were later identified by mass spectrometry at GeneCore BioTechnologies Co. Ltd. (Shanghai, China). One protein which had an extremely high expression level at 4°C was identified as the ORF104 protein of filamentous phage SW1. This protein is the putative ssDNA binding protein that is involved in the integration of the phage. ssDNA binding protein is a key protein in phage assembly. The large increase in the ssDNA binding protein at low temperature suggested that phage SW1 assembled and extruded from the host cell in a large quantity at low temperature. Phage particles were isolated from WP3 cultures incubated at different temperatures (4°C, 10°C, 15°C, 20°C, and 25°C). The phage particles were enumerated by counting the phage plaques formed on WP2 cell lawns on the agar plates. No or few active phage particles were isolated from cultures incubated at 20 or 25°C as no phage plaques were observed. Active phage particles were isolated from cultures of strain WP3 at 4°C, 10°C, and 15°C. The WP3 cell culture yielded a higher number of SW1 phage particles at 4°C [(9.6 ± 1.8) × 104 PFU (mean ± standard deviation)] than at 10°C [(3.4 ± 0.8) × 104 PFU] or 15°C [(1.1 ± 0.3) × 104 PFU]. These data, representing averages from three repeat experiments, clearly demonstrate that the phage production is temperature regulated and that low temperature can induce phage production in strain WP3.
Confirmation of phage induction by quantitative PCR.
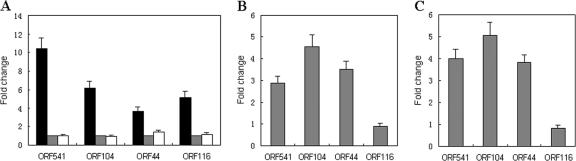
The transcription of some major genes of SW1 at different temperatures was assayed by real-time reverse transcription (RT)-PCR. Total RNA was isolated from WP3 cultures growing under different conditions with a TRI reagent-RNA/DNA/protein isolation kit (Molecular Research Center, Inc.) according to the manufacturer's instructions. The RNA samples were treated with DNase I at 37°C for 1 h and then purified with an RNeasy mini kit (QIAGEN, Germany). The purified RNA samples were used to synthesize cDNA with the RevertAid first-strand cDNA synthesis kit (MBI, Fermentas), in accordance with the manufacturer's instructions. The primer pairs of selected genes for real-time PCR were designed using Primer Express software (Applied Biosystems, San Francisco, CA). PCR cycling was conducted using ABI 7500 system software with reaction mixtures in total volumes of 50 μl containing 1× SYBR green I universal PCR master mix (ABI), a 0.5 μM concentration of each primer, and 1 μl cDNA template. By this method, the amount of target was normalized to that of the reference gene relative to the calibrator. The transcription levels of ORF541 (replication protein), ORF104 (ssDNA binding protein), ORF44 (major coat protein), and ORF116 (repressor) were all induced in a range of 3- to 12-fold at low temperature (4°C) compared with those at higher temperatures (15°C or 25°C) (Fig. 3A). The transcription levels of these genes with mitomycin C (MMC) and UV exposure treatment were also checked. For both MMC and UV treatments, overnight cultures were diluted 1:200 and grown to an optical density at 600 nm of 0.3. MMC was then added to a final concentration of 500 ng/ml, and the cells were grown in the presence of this reagent for 3 h. For all the experiments in which UV treatment was used, cells were harvested by centrifugation at room temperature, resuspended in 2216E medium, and placed in a plastic petri dish. The dish was placed without its cover into a Stratagene UV cross-linker, and the cells were irradiated with 25 J/m2 of UV. The cells were then harvested by centrifugation, resuspended in 2216E medium, and grown for 1 h. RNA was isolated from the MMC- and UV-treated and untreated WP3 cell cultures and subjected to real-time RT-PCR analysis as described above. It was shown that the transcription of ORF541, ORF104, and ORF44 was significantly induced by MMC (Fig. 3B) and UV irradiation (Fig. 3C), suggesting that the phage production may be under the cellular SOS control. Surprisingly, along with the higher production of the SW1 phage at lower temperature, the transcription of the putative repressor gene ORF116 is significantly induced, but the transcription of ORF116 is not induced by MMC or UV irradiation (Fig. 3). This suggests that government of phage SW1 production may be different from that of CTXφ, for which phage production depends on the inactivation of the repressor RstR (5). The regulation system of phage SW1 will be further investigated.
FIG. 3.
Gene transcription levels of phage SW1 at different temperatures (A) and under MMC (B) and UV irradiation (C) treatments. (A) The transcription levels of ORF541, ORF104, ORF44, and ORF116 at 4, 15, and 25°C were measured by quantitative RT-PCR. The mean values and standard deviations for three repeat experiments are given. Gray bars represent gene transcription levels at 15°C (set as the control), black bars represent gene transcription levels at 4°C, and white bars represent gene transcription levels at 25°C. (B and C) The transcription levels of ORF541, ORF104, ORF44, and ORF116 before and after MMC and UV treatment were measured by quantitative RT-PCR. Mean values and standard deviations of three repeat experiments are shown. The transcription levels of these genes before treatment were set as the control.
Acknowledgments
This work was financially supported by the China COMRA Foundation (DYLY02-2-03) and an NSF grant (40625016).
Footnotes
Published ahead of print on 27 July 2007.
REFERENCES
- 1.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2006. GenBank Nucleic Acids Res. 34:D16-D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorg, A., W. Weiss, and M. J. Dunn. 2004. Current two-dimensional electrophoresis technology for proteomics. Proteomics 4:3665-3685. [DOI] [PubMed] [Google Scholar]
- 3.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 4.Ikema, M., and Y. Honma. 1998. A novel filamentous phage, fs-2, of Vibrio cholerae O139. Microbiology 144:1901-1906. [DOI] [PubMed] [Google Scholar]
- 5.Quinones, M., H. H. Kimsey, and M. K. Waldor. 2005. LexA cleavage is required for CTX prophage induction. Mol. Cell 17:291-300. [DOI] [PubMed] [Google Scholar]
- 6.Sambrook, J. F., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor Laboratory, NY.
- 7.Shevchenko, A., O. N. Jensen, A. V. Podtelejnikov, F. Sagliocco, M. Wilm, O. Vorm, P. Mortensen, A. Shevchenko, H. Boucherie, and M. Mann. 1996. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. USA 93:14440-14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Wezenbeek, P. M., T. J. Hulsebos, and J. G. Schoenmakers. 1980. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene 11:129-148. [DOI] [PubMed] [Google Scholar]
- 9.Venkateswaran, K., D. P. Moser, M. E. Dollhopf, D. P. Lies, D. A. Saffarini, B. J. MacGregor, D. B. Ringelberg, D. C. White, M. Nishijima, H. Sano, J. Burghardt, E. Stackebrandt, and K. H. Nealson. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Int. J. Syst. Bacteriol. 49:705-724. [DOI] [PubMed] [Google Scholar]
- 10.Waldor, M. K., E. J. Rubin, G. D. Pearson, H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTXphi are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 11.Xiao, X., P. Wang, X. Zeng, M. X. Chen, D. H. Bartlett, and F. P. Wang. 2007. Shewanella psychrophila sp. nov. and Shewanella piezotolerans sp. nov., isolated from west Pacific deep-sea sediment. Int. J. Syst. Evol Microbiol. 57:60-65. [DOI] [PubMed] [Google Scholar]