Abstract
The growth and persistence of rhizobia and bradyrhizobia in soils are negatively impacted by drought conditions. In this study, we used genome-wide transcriptional analyses to obtain a comprehensive understanding of the response of Bradyrhizobium japonicum to drought. Desiccation of cells resulted in the differential expression of 15 to 20% of the 8,480 B. japonicum open reading frames, with considerable differentiation between early (after 4 h) and late (after 24 and 72 h) expressed genes. While 225 genes were universally up-regulated at all three incubation times in response to desiccation, an additional 43 and 403 up-regulated genes were common to the 4/24- and 24/72-h incubation times, respectively. Desiccating conditions resulted in the significant induction (>2.0-fold) of the trehalose-6-phosphate synthetase (otsA), trehalose-6-phosphate phosphatase (otsB), and trehalose synthase (treS) genes, which encode two of the three trehalose synthesis pathways found in B. japonicum. Gene induction was correlated with an elevated intracellular concentration of trehalose and increased activity of trehalose-6-phosphate synthetase, collectively supporting the hypothesis that this disaccharide plays a prominent and important role in promoting desiccation tolerance in B. japonicum. Microarray data also indicated that σ54- and σ24-associated transcriptional regulators and genes encoding isocitrate lyase, oxidative stress responses, the synthesis and transport of exopolysaccharides, heat shock response proteins, enzymes for the modification and repair of nucleic acids, and the synthesis of pili and flagella are also involved in the response of B. japonicum to desiccation. Polyethylene glycol-generated osmotic stress induced significantly fewer genes than those transcriptionally activated by desiccation. However, 67 genes were commonly induced under both conditions. Taken together, these results suggest that B. japonicum directly responds to desiccation by adapting to changes imparted by reduced water activity, such as the synthesis of trehalose and polysaccharides and, secondarily, by the induction of a wide variety of proteins involved in protection of the cell membrane, repair of DNA damage, stability and integrity of proteins, and oxidative stress responses.
Symbiotic biological nitrogen fixation (BNF) is often strongly inhibited in arid and semiarid soils due to the poor survival of rhizobia and bradyrhizobia under desiccation stress (27). Rhizobium strains were shown to be sensitive to desiccation both in soils and peat cultures (53) and on seed surfaces (47, 49). Although several drought-tolerant soybean cultivars have been selected and are currently in use (46), the absence of corresponding drought-tolerant Bradyrhizobium strains limits BNF. While several studies showed the influence of drought conditions on the survival of rhizobia in culture, many of these used changes in external solute concentrations, salts and sugars, to indirectly influence water potential (32). However, since many of the solutes examined are permeable to the cell membrane or are catabolized, these studies in essence examined the response of cells to changes in the osmotic potential of the growth medium. In order to avoid these confounding influences, some studies have used nonpermeating substrates, such as polyethylene glycol (PEG), to lower external water potential or water activity (7). Although both sets of conditions ultimately result in low external water activity, mechanisms for survival and growth under non-solute-mediated stress conditions of drought may significantly differ from mechanisms for survival under conditions of high osmotic stress or PEG-induced reduced water activity.
The capacity of prokaryotes to withstand significant desiccating conditions requires a wide array of physiological responses, one of which involves the synthesis or accumulation of the disaccharides sucrose and trehalose (38). It has been hypothesized that these sugars stabilize membranes and proteins in the dry state by glass formation (10). The intracellular concentration of trehalose is thought to have a major positive impact on the desiccation survival of rhizobia. The trehalose concentration in some rhizobia was shown to increase in response to increased extracellular salt concentrations and decreased levels of oxygen (17). The addition of exogenously supplied trehalose was also shown to enhance the survival of Bradyrhizobium japonicum strain USDA 110 in response to desiccation (47). Recently, Streeter and Gomez (48) reported that B. japonicum has three independent mechanisms for the synthesis of trehalose: trehalose synthase (TS), maltooligosyltrehalose synthase (MOTS), and trehalose-6-phosphate synthetase (TPS). Several additional mechanisms have been proposed to confer desiccation resistance in bacteria. Shifts in membrane fatty acid composition, resulting in an increase in cis-trans isomerization of monounsaturated fatty acids, are thought to maintain membranes in a liquid crystalline phase following significant reductions in osmotic potential (15). Boumahdi et al. (5) reported that reductions in the water activity (aw) of the growth medium and mild desiccation altered the degree of unsaturation of cellular fatty acids in S. meliloti and B. japonicum. Induction of oxidative-stress-responsive enzymes, such as peroxidases, catalases and superoxide dismutases, was also shown to be essential for the survival of cells exposed to long periods of desiccation (14, 39). It was suggested that enhanced DNA repair capacity that imparts radiation resistance in Deinococcus radiodurans may be incidental to this bacterium's adaptation to dehydration stress (28).
Tolerance of water stress varies widely between microorganisms (16), and several phyla of cyanobacteria were found to be highly tolerant of desiccation stress (21, 38, 50). Results from microarray analyses indicated that drought, salt, osmotic, and low-temperature stress induced similar genes in Anabaena sp. strain PCC 7120 (21). In contrast, Synechocystis sp. strain PCC 6803 differentially responds to stress induced by salt and reduced water activity (20).
Although desiccation is among the most significant environmental stresses encountered by terrestrial bacteria, which face both fluctuating-periodical and long-term exposure to drought conditions, the genetic mechanisms underlying sensing and providing resistance to this stress are still poorly understood. The objective of this study was to obtain a comprehensive understanding of the genetic mechanisms involved in desiccation tolerance in the bradyrhizobia. Genome-wide transcriptional analyses of desiccated versus fully hydrated cells indicate that several classes of B. japonicum genes are induced in response to desiccation stress. Moreover, we show that desiccation specifically induces genes and proteins involved in the synthesis of trehalose and that induction and repression of desiccation-responsive genes occurs in a time-dependent manner.
MATERIALS AND METHODS
Bacteria, media, and experimental growth conditions.
Bradyrhizobium japonicum strain USDA 110 was grown, with shaking, at 30°C in Bergersen's minimal salts medium (BMM) (3) supplemented with 0.5 g of yeast extract (Becton Dickenson, Sparks, MD) per liter. Desiccating and hydrating conditions were generated by using a modified version of the filter disk desiccation assay previously described by van de Mortel and Halverson (52). Briefly, 30 ml (optical density at 600 nm, 0.5 to 0.7) of cells were harvested by filtration onto 0.4-μm (47-mm-diameter) polycarbonate membrane filters (Sterlitech, Kent, WA). Filters were washed with BMM devoid of glycerol, immediately dried by application of a vacuum, aseptically placed into the bottom of perforated polystyrene petri dishes, and immediately transferred to perforation plates in 150-mm plastic desiccators (Nalgene, Rochester, NY). The bottoms of the desiccators under the perforation plates were filled with either double-distilled water, 100% relative humidity (RH), or a saturated potassium acetate solution, to give desiccated, 27% RH, conditions. The RH values of solutions in desiccators were measured with an Aqua Lab CX-2 water activity meter (Decon Devices, Pullman, WA). Desiccators were incubated in the dark at 30°C for 4, 24, or 72 h. Generally, nine filters were prepared for each desiccation level and incubation.
Mild, nonpermeating, solute-induced osmotic stress was achieved, as previously described by van de Mortel and Halverson (52), by adding 9.25 g of PEG 8000 (Sigma, St. Louis, MO) per liter to BMM supplemented with 0.5 g of yeast extract. This resulted in a calculated water potential of 0.228 MPa. B. japonicum strain USDA 110 was grown at 30°C in BMM with and without PEG, and microarray analyses were done as described above.
Preparation of cell samples.
RNA extractions for quantitative reverse transcription-PCR (qRT-PCR) and microarray analyses were done with cells from three filters, which constituted one biological replicate. Cells were rapidly washed from filters and immediately combined in a single tube containing 50 ml of ice-cold BMM without glycerol and supplemented with 10% stop solution (5% H2O-phenol, pH 4.3, in 100% ethanol). Cells were centrifuged for 15 min at 8,000 × g at 4°C, flash-frozen in liquid N2, and stored at −80°C until use. Cells for bacterial quantification, nuclear magnetic resonance (NMR) analyses, and enzyme assays were washed from filters as described above, resuspended in 5 ml of BMM devoid of glycerol, and kept continuously on ice.
Quantification of hydrated and desiccated cells.
The viability of desiccated and hydrated B. japonicum cells at each time point was quantified, in triplicate, by plate counting and direct quantification using a LIVE/DEAD BacLight bacterial viability kit (Molecular Probes, Eugene, OR). Viable counts for the 4- and 72-h incubations were obtained by plating 10-fold serial dilutions onto BMM agar plates supplemented with 0.5 g yeast extract per ml. Direct cell counts were done on cell suspensions from the 4-, 24-, and 72-h incubation samples, according to the protocol provided by the manufacturer. For each sample, 1,000 cells were counted in order to determine the live/dead cell ratio. The viability of B. japonicum wild-type and σ54 mutant strains (rpoN1 and rpoN2 mutant strains and an rpoN1 rpoN2 double-mutant strain), incubated under desiccating (27% RH) and hydrating (100% RH) conditions, was assessed, in duplicate, by direct plating onto BMM agar plates supplemented with 0.5 g yeast extract per ml, as described above. The statistical significance of results was determined by using the analysis of variance (ANOVA) and Tukey-Kramer honestly significant difference (HSD) subroutines of R program, version 2.0.1 (http://www.r-project.org/) at α of 0.05.
NMR analyses.
1H-NMR analyses were done to assess intracellular trehalose concentrations in hydrated versus desiccated cells from 24- and 72-h incubation samples. The NMR analyses were done with the procedures described by Xu et al. (57), with slight modification. Cell suspensions from filters were centrifuged for 10 min at 6,000 × g at 4°C and resuspended in 70% ethanol. Cells were subjected to five 30-s sonification cycles with a Heat Systems-Ultrasonics (Plainview, NY) cell disruptor at full power and a 40% duty cycle, followed by vigorous shaking for 1 h. Extracts were dried in vacuo by using a speedvac concentrator (Thermo Company, Waltham, MA), resuspended in D2O, redried, and resuspended in a final volume of 0.8 ml D2O (Sigma, St. Louis, MO). Prior to analysis, TSP [3-(trimethylsilyl)-propionic acid-D4 sodium salt] was added to samples to a final concentration of 10 μM as an internal reference standard. Analysis was performed using a Varian Inova (Palo Alto, CA) 500-MHz NMR spectrometer. Sixty-four scans were obtained at ambient temperature, using an acquisition time of 5 s, a relaxation delay of 10 s, and a 45° pulse. Trehalose produces five specific peaks by NMR. The concentration of trehalose in samples was calculated by integration of the area under the 5.2-ppm peak and by comparison to authentic trehalose standards and TSP.
Assays for enzymes involved in trehalose synthesis.
Enzyme assays were done on duplicate biological samples obtained from the 4- and 72-h desiccated and hydrated treatments. Replicate biological samples were derived from 12 pooled polycarbonate filters, each containing 30 ml of cells per filter. Cells from filters were resuspended in ice-cold BMM, devoid of organic amendment, by using 10- to 30-s vortex cycles to detach cells. Cells were immediately centrifuged for 10 min at 6,000 × g at 4°C and stored at −80°C prior to analysis. Enzyme assays for trehalose synthase and TPS were done as previously described (44), with some modifications. In the current studies, the sonication volume and time were limited to 3.0 ml and 10 min, respectively. Moreover, since at low protein concentrations TS was slightly inhibited by blue dextran (used for gel filtration), assays for this enzyme were done using crude protein preparations. Assays for TPS were done after partial purification by gel filtration chromatography. Since B. japonicum cells contain some trehalose, enzyme activity calculations were corrected by subtracting the amount of trehalose found in boiled control reaction mixtures. To prevent precipitation of protein during secondary reactions in TPS assays (i.e., the analysis of UDP), mixtures were incubated for ∼15 min at −80°C following the boiling step and prior to centrifugation of the primary mixtures. Pyruvate kinase (P-1506; Sigma) and lactate dehydrogenase (L-1500; Sigma) were used in the analysis of UDP and gave the most consistent results. Enzyme activity values were normalized to the number of live cells in each treatment, determined with the BacLight bacterial viability assay.
Microarray fabrication.
The GeneMark (4) and Glimmer2 (41) gene models were used to generate a list of unique, nonoverlapping, predicted open reading frames (ORFs) from the B. japonicum strain USDA 110 genome (19). The genome was reannotated prior to array development. Seventy-mer oligonucleotides were designed and synthesized (QIAGEN Operon, Valencia, CA) for 8,480 predicted B. japonicum ORFs. The oligonucleotides were printed onto Corning epoxysilane-coated slides (Corning Inc., Lowell, MA) at the Genome Sequencing Center, Washington University School of Medicine. Each slide contains the predicted ORFs from the genome annotation and 36 control genes, printed in duplicate. More-detailed information about the arrays is available through the Bradygenome website (http://bradygenome.org/).
RNA extraction, RT, labeling, and hybridization conditions.
RNA was extracted from frozen pellets by a modified hot phenol extraction protocol (6), and cDNA was synthesized from 20 μg of total RNA template. Random hexamers (Invitrogen Corporation, Carlsbad, CA) were added to 17 μl of RNA solution to a final concentration of 0.35 μg per μl, and the mixture was heated to 90°C for 10 min and flash-frozen for 30 s in liquid N2. Six microliters of 5× first-strand synthesis buffer, 3 μl of 0.1 M dithiothreitol, 1.2 μl of 25× aa-dNTP mix (containing 12.5 mM concentrations each of dATP, dCTP, and dGTP; 7.5 mM dTTP; and 10 mM aa-dUTP [Ambion Inc., Austin, TX]) and 2 μl (200 units per μl) of Superscript III reverse transcriptase (Invitrogen Corporation, Carlsbad, CA) were added to the solution, and the reaction was incubated overnight at 42°C. The unincorporated nucleotides were removed with a Micron YM-30 column (Millipore, Bedford, MA), and the cDNA was dried with a speedvac concentrator. Dehydrated cDNA samples were resuspended in 4.5 μl of 0.1 M NaCO3, pH 9.0, labeled with Amersham Cy3 and Cy5 monoreactive dyes (GE Healthcare, Buckinghamshire, United Kingdom), and incubated for 90 min at 25°C. Then, 35 μl of 100 mM Na acetate, pH 5.2, was added, and unincorporated dyes were removed with the QuickPCR purification kit (QIAGEN Inc., Valencia, CA). Alternatively labeled (Cy3 and Cy5) cDNAs from control (hydrated) and experimental (desiccated) samples were combined and dried to completion. Microarray slides were prehybridized in a solution containing 25% 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), and 1% bovine serum albumin (BSA) at 42°C for 45 min, washed thoroughly in distilled water, dipped in isopropanol, and immediately dried by centrifugation. Labeled cDNA samples were resuspended in 70 μl of hybridization solution (25% formamide, 25% 20× SSC, 0.2% SDS, 0.7 μl of 10-mg/ml salmon sperm DNA [Invitrogen Corporation, Carlsbad, CA]), incubated at 95°C for 3 min, and cooled on ice. The labeled cDNA was applied to slides, and the microarrays were incubated at 42°C for 18 h. Slides were washed as previously described (6) and scanned with an Axon GenePix 4000B scanner (Molecular Devices Corp., Sunnyvale, CA).
Statistical analysis of microarray data.
Signal intensities for each spot on scanned image files were determined with GenePix Pro 6.0 software (Molecular Devices Corp., Sunnyvale, CA). Data were normalized for spot and slide abnormalities with the spatial Lowess algorithm and analyzed by mixed-effect ANOVA (MAANOVA). Lowess and MAANOVA are part of the R/MAANOVA microarray statistical package, available at http://www.jax.org/staff/churchill/labsite/software/Rmaanova/index.html. Values representing the variety by gene interaction between log2-transformed values of the control and experimental spot intensities were combined with the residual noise from each spot to obtain filtered and adjusted expression values (22). Duplicate spots from each array were averaged, and values from MAANOVA were subsequently subjected to significance analysis of microarray (SAM) data with the SAM package (51). Significantly up- and down-regulated genes were identified based on a 1.5- and 2.0-fold cutoff threshold, a global false discovery rate lower than 5%, and α of 0.05. Six arrays representing a total of 12 replicates of each ORF were analyzed for each of the three desiccation times and for treatments with PEG.
qRT-PCR.
qRT-PCR analysis was done to specifically assess the induction of trehalose synthesis and σ54 (rpoN1 and rpoN2) genes under desiccation and full hydration conditions. Each 50-μl reaction mixture contained 3 μg of total RNA, 17.5 μl of distilled water, 25 μl of Power SYBR green PCR Master Mix (Applied Biosystems, Warrington, United Kingdom), 0.5 μg/μl BSA, and 0.2 μM concentrations each of forward and reverse gene-specific primers (Table 1). Primers producing amplicon sizes of 80 to 200 bp were designed online with Primer 3 software (40), available at http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi. The qRT-PCRs were run on an Applied Biosystems (Foster City, CA) 7500 Real Time PCR system using Sequence Detection System, version 1.3, software. The PCR program consisted of an initial denaturation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. Expression values for three biological replicates for each treatment were normalized to the expression level of a housekeeping control gene in the B. japonicum genome, the chromosome partitioning protein A gene (parA; bll0631). This gene did not show differential expression under a wide array of conditions in comparative microarray analyses, including desiccation, salt, and nutrient symbiotic and autotrophic growth conditions (E. J. Cytryn, unpublished data; Chang and Frank, personal communication).
TABLE 1.
qRT-PCR primers used in these studies
| Primer | Sequence (5′→3′) | Amplicon size (bp) | Gene description |
|---|---|---|---|
| bll0322F | GGGGACGACCTGTGAACTTA | 251 | Trehalose 6-phosphate synthetase (otsA) |
| bll0322R | CTTCGTAATAGCCGCCGTAA | ||
| blr6767F | TTCTACTGGCACCGCTTCTT | 173 | Trehalose synthase (treS) |
| blr6767R | GAGGTTCTCGTTCGAAGTGC | ||
| blr6771F | CAAGCTGATCGACGATACCA | 218 | Maltooligosyltrehalose synthase (treY) |
| blr6771R | AATTGGTAGAACGCGGTGTC | ||
| blr0723F | CTTCGTGGAAGAGGAACTCG | 182 | rpoN2 |
| blr0723R | CCTTGCTCATCCATTCTTCC | ||
| blr1883F | ACGGCTTCTTTACCCTTGGT | 167 | rpoN1 |
| blr1883R | GATCGATGCCGTGAAGAAAT | ||
| bll0631F | GGACGCCGATTACACCTATG | 282 | Chromosome partitioning protein A, cppA (for normalization) |
| bll0631R | GTAGACCTTCTCGCCCATGA |
RESULTS
Desiccation-induced changes in cell viability.
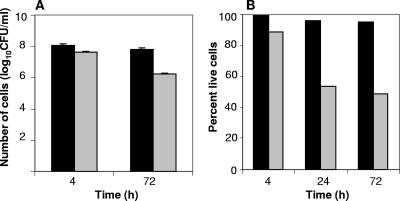
The viability of Bradyrhizobium japonicum cells under hydrated and desiccating conditions was assessed by plate count analysis, following incubation periods of 4 and 72 h (Fig. 1A), and by direct microscopic analyses using BacLight after 4, 24 and 72 h of incubation (Fig. 1B). Cell viability, as visualized by direct microscopic observation, decreased most significantly after 24 h and thereafter remained relatively stable. Live/dead counts of cells incubated for 72 h showed a 49% survival ratio of desiccated cells relative to hydrated cells. In contrast, plate count analysis showed only 17% survival of desiccated cells after 72 h of incubation at 27% RH, suggesting the presence of a large number of viable-but-nonculturable (VBNC) cells.
FIG. 1.
Viability of B. japonicum cells following incubation under desiccating (27% RH) and hydrated (100% RH) conditions. (A) Plate count data expressed as log10 CFU/ml for desiccated and hydrated cells at the 4- and 72-h incubation times. Error bars representing the standard deviation of three biological replicate samples are shown. (B) Percentages of live cells obtained by Baclight following 4-, 24-, and 72-h incubation periods.
Genome-wide transcriptional analyses of desiccated cells.
Microarray analyses were done to assess the global transcriptional responses of B. japonicum to desiccation stress. In order to specifically isolate transcriptional responses to desiccation, as opposed to genes involved in more general stress responses of starvation or transition to stationary phase, filtered B. japonicum cells incubated under severe desiccating conditions (27% RH) were compared to filtered cells concomitantly incubated under hydrated (control) conditions (100% RH). RNA from cells incubated under both conditions was extracted from cells following incubation for 4, 24, and 72 h.
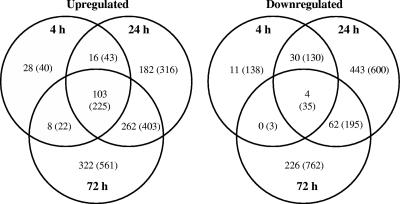
Microarray analyses indicated that of the 8,480 B. japonicum ORFs analyzed, 200 (636), 1,102 (1, 947), and 987 (2,206) genes were differentially expressed more than 2-fold (or 1.5-fold, as indicated in parentheses) in the 4-, 24-, and 72-h incubations, respectively (Fig. 2). This represented approximately 2% (7%), 13% (23%), and 12% (26%) of the coding sequences analyzed. One hundred three (225) genes were universally up-regulated at all three incubation times, and an additional 16 (43) and 262 (403) up-regulated genes were common to the 4/24-h and 24/72-h incubation times, respectively. Conversely, 28 (40), 182 (316), and 322 (516) genes were uniquely up-regulated in the three individual incubation periods. Commonly up-regulated expressed genes in the 24/72-h samples were referred to as late-induced genes, whereas those from the 4-h samples were termed early-induced genes. A substantially smaller number of common genes were down-regulated in samples from the three incubation times. Four (35) of these genes were universal to all three time points, and 30 (130) and 62 (195) genes were mutual to the 4/24- and 24/72-h incubation periods, respectively. In contrast, 11 (138), 443 (600), and 226 (762) genes were unique to the 4-, 24-, and 72-h incubation times, respectively.
FIG. 2.
Venn diagrams showing statistically significant (α = 0.05) up- and down-regulated genes in the desiccated versus hydrated treatments from B. japonicum microarray analyses. Values shown represent more than 2- and 1.5-fold (in parentheses) differential expression. Identified genes had a global false discovery rate of ≤5%. Six arrays representing a total of 12 replicates of each ORF were analyzed for each of the three desiccation times.
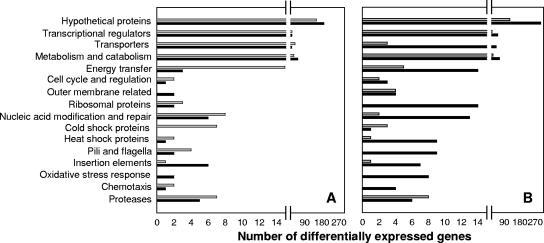
Categories of genes that were significantly and differentially up- and down-regulated in response to desiccation, in the early and late periods, could be divided into 16 functional groups (Fig. 3). The greatest number of induced and repressed genes encoded hypothetical proteins; thus, their specific physiological function could not be inferred. However, a large number of genes encoding transcriptional regulatory proteins and transposases and proteins involved in metabolism, transport, catabolism, and energy transfer were specifically induced and repressed in response to desiccation. In general, more late than early genes were induced following incubation under desiccating conditions (Fig. 3A and B). A complete list of genes significantly (α = 0.05) induced and repressed by incubation of cells under desiccated versus hydrated conditions is presented in Table S1 in the supplemental material.
FIG. 3.
Functional categories of statistically significant (α = 0.05), differentially expressed genes. (A) Genes expressed only in the early (4-h) desiccation period. (B) Genes commonly expressed in the 24- and 72-h desiccation periods. Black bars, up-regulated genes; gray bars, down-regulated genes.
Functional roles of genes responding to desiccation stress.
To more precisely define desiccation-responsive processes, the microarray data sets were evaluated for genes meeting at least one of the following criteria: (i) there was simultaneous and differential expression of genes having similar physiological function, (ii) there was induction of multiple genes in a specific biochemical pathway, or (iii) induction of the same gene occurred in more than one time period and with an especially high level of expression (more than threefold). Ten categories of genes potentially involved in the tolerance of B. japonicum to desiccation were defined with these criteria (Table 2). These included genes involved in sugar metabolism and transport, transcriptional regulation, oxidative stress and heat shock responses, modification and repair of nucleic acids, membrane modification, pyrroloquinoline synthesis, transport of cations, synthesis of pili and flagella, and formation of extracellular polysaccharides. Genes showing the greatest induction and meeting the above criteria encoded trehalose-6-phosphate synthetase (bll0322), a gluconolactone precursor protein (bll2817), an ABC transporter sugar binding protein (blr3200), nitrogen regulatory protein PII (blr0612), isocitrase lyase (blr2455), a cyclic AMP binding regulator (blr0536), a tryptophan-rich sensory protein (bll5190), σ54 (RpoN2) (blr0723), a σ54 modulation protein (blr0724), σ24 (RpoE), two-component response regulators (bll7795 and blr8144), an HspC2 (Hsp20) (blr4637), a probable DNA glycosylase protein (blr0857), an OpgC-like protein involved in the succinylation of osmoregulated periplasmic glucans (bll1464), putative alcohol dehydrogenases (bll0332 and bll0333), pyrroloquinoline quinone synthase protein A (bsr6735), pilus assembly proteins (bsl1442, bsl7141, bsl3118, bsl6587), flagellin (bll5845, and bll5846), a glycosyl transferase (bll2752), and a fasciclin I-like adhesin domain protein (bll5191).
TABLE 2.
Physiological processes and significant desiccation-induced genes in B. japonicum
| Physiological process | ORF | Gene name and/or function | Desiccation-induced stress (fold induction)a
|
||
|---|---|---|---|---|---|
| 4 h | 24 h | 72 h | |||
| Sugar metabolism and transport | |||||
| Trehalose synthesis | bll0322 | Trehalose-6-phosphate synthetase (otsA) | 4.6 | 9.2 | 4.5 |
| bll0323 | Trehalose-6-phosphate phosphatase (otsB) | 2.4 | 2.9 | 2.9 | |
| blr6767 | Trehalose synthase (treS) | — | 2.7 | 6.2 | |
| General sugar metabolism | bll6764 | Glycoside hydrolase | — | 1.8 | 2.9 |
| bll6765 | Putative 4-α-glucanotransferase | 2.2 | 4.1 | 3.4 | |
| blr6766 | Putative glucanohydrolase | 2.0 | 4.1 | 1.5 | |
| blr0901 | Putative oligo-1,6-glucosidase | 1.7 | 3.0 | 2.4 | |
| bll6177 | β-Glucosidase | 2.4 | 2.3 | 1.6 | |
| blr2754 | Putative glycogen debranching enzyme | 1.8 | 3.1 | 4.6 | |
| bll7185 | Probable glucose 1-dehydrogenase | — | 2.2 | 1.8 | |
| bll5979 | Putative CDP-glucose 4,6-dehydratase | 2.4 | 1.9 | ||
| blr6459 | Glycogen synthase (glgA) | — | 2.4 | 3.1 | |
| Pentose phosphate pathway | bll2817 | Gluconolactonase precursor | — | 5.6 | 6.3 |
| blr6758 | Probable transaldolase | — | — | 3.7 | |
| blr6759 | 6-Phosphogluconate dehydrogenase | — | 1.9 | 4.2 | |
| blr6760 | Glucose-6P-1-dehydrogenase (zwf) | — | 2.5 | 4.4 | |
| blr6761 | 6-Phosphogluconolactonase | 1.8 | 2.2 | 2.7 | |
| blr6762 | Gluconokinase | — | 2.2 | 3.3 | |
| Sugar transporters | bll0324 | MFS-like sugar transport protein | 5.2 | 5.6 | 3.0 |
| blr3200 | ABC transporter sugar-binding protein | 2.5 | 10.0 | 4.1 | |
| blr3201 | ABC transporter ATP-binding protein | — | 1.5 | 2.5 | |
| bll3208 | ABC transporter sugar-binding protein | — | 2.5 | 2.6 | |
| blr2271 | ABC transporter permease protein | — | 2.5 | 3.2 | |
| Phosphotransferase system | blr0725 | σ54-dependent nitrogen regulator IIA (ptsN) | — | 5.8 | 3.9 |
| blr4948 | Nitrogen regulatory protein PII (glnB) | — | 2.4 | 2.6 | |
| blr0612 | Nitrogen regulatory protein PII (glnK) | — | 12.7 | 1.6 | |
| blr8148 | Phosphocarrier protein hpr (ptsH) | — | 1.7 | 1.9 | |
| blr3575 | Phosphocarrier protein hpr (ptsH) | — | 1.6 | 2.0 | |
| blr8147 | PTS system IIA component | — | 2.5 | — | |
| EPS formation | bll2362 | Succinoglycan biosynthesis (exoP) | — | 5.2 | 4.8 |
| bll7574 | UDP-hexose transferase (exoM) | — | 1.6 | 3.1 | |
| blr1499 | UTP-G1P uridylyltransferase (exoN) | — | 3.2 | 2.4 | |
| bll2752 | Probable glycosyl transferase group I | 3.9 | 11.4 | 17.8 | |
| blr2358 | Putative LPS synthesis transferase | — | 3.8 | 3.7 | |
| Transcriptional regulation | |||||
| Sensory regulators | blr0536 | cAMP-binding transcriptional regulator | 5.6 | 15.5 | 19.3 |
| bll7795 | Two-component response regulator | 1.6 | 5.0 | 7.0 | |
| blr8144 | Two-component response regulator | — | 4.0 | 4.9 | |
| bll5190 | Tryptophan-rich sensory protein (tspO) | — | 10.8 | 5.1 | |
| blr2475 | Tryptophan-rich sensory protein (tspO) | 2.0 | 2.3 | 3.7 | |
| Sigma factors and σ54-associated regulators | blr7797 | rpoE (σ24) | — | 1.8 | 2.4 |
| blr0723 | RNA polymerase σ54 subunit (rpoN2) | 2.2 | 10.0 | 13.9 | |
| blr0724 | σ54 modulation protein | — | 6.6 | 18.0 | |
| blr0725 | σ54-dependent nitrogen regulator IIA (ptsn) | 1.8 | 5.8 | 3.9 | |
| blr2037 | NifA-like regulatory protein | — | 4.2 | 3.7 | |
| blr4489 | Two-component sensor (ntrY) | — | 2.8 | 2.9 | |
| blr4490 | Two-component sensor (ntrX) | — | 2.5 | 3.4 | |
| bsr6672 | Transcriptional regulator (flbD-like) | — | 3.4 | 3.4 | |
| Oxidative stress response mechanisms | bll0735 | Organic hydroperoxide resistance protein | — | 3.4 | 3.7 |
| bll4012 | Organic hydroperoxide resistance protein | — | 3.0 | 1.9 | |
| bll7559 | Fe/Mn superoxide dismutase (chrC) | — | 1.5 | 3.3 | |
| bll7774 | Superoxide dismutase (sodF) | — | 3.3 | 3.6 | |
| blr5308 | Antioxidant protein (prxS-like) | — | 2.1 | 3.6 | |
| bll1317 | Peroxiredoxin (prxS-like) | — | 1.7 | 2.4 | |
| Heat shock response systems | blr4637 | HspC2 (hsp20) heat shock protein | — | 9.1 | 12.4 |
| blr3152 | Hsp70 family molecular chaperone | 2.1 | 2.1 | 2.5 | |
| bll5226 | Heat shock protein (groES1) | — | 1.7 | 1.8 | |
| blr5625 | 10-kDa chaperonin (groES) | — | 2.3 | 2.0 | |
| bll5626 | 60-kDa chaperonin (groEL) | — | 1.7 | 1.7 | |
| bll7789 | Probable Hsp90 heat shock protein (htpG) | — | 2.3 | 1.7 | |
| blr7961 | Probable HspC2 heat shock protein | — | 2.6 | 1.6 | |
| blr6978 | Chaperonin (groES2) | — | 2.0 | 2.2 | |
| bsr7532 | 10-kDa chaperonin (groES) | — | 2.0 | 2.2 | |
| Nucleic acid modification and repair mechanisms | bll1447 | DEAD-box RNA helicase (rhlE) | 3.3 | 5.7 | 3.3 |
| bll0572 | Putative DNA topoisomerase I | 2.8 | 2.8 | 2.5 | |
| bll4696 | DNA gyrase subunit A (gyrA) | — | 2.8 | 5.5 | |
| bll7350 | DNA primase (dnaG) | 2.7 | 2.3 | 2.8 | |
| bll6175 | Histone-like protein HU (hupA) | — | 2.7 | 5.0 | |
| blr0857 | Probable DNA glycosylase protein | 3.5 | 5.9 | 4.0 | |
| blr7493 | DNA mismatch repair protein (mutL) | 1.9 | 1.8 | 2.3 | |
| bll0827 | DNA recomb. And repair protein (recF) | 1.5 | 1.7 | 2.1 | |
| bll8014 | Ku-like-ATP-dependent DNA helicase | — | 2.2 | 3.0 | |
| Membrane modification | blr0537 | Prolipoprotein diacylglyceryl transferase | 2.8 | 3.9 | 5.0 |
| blr0638 | Maf-like protein | 2.5 | 2.1 | 2.5 | |
| bll1464 | OpgC-like succinylation of glucans | 7.6 | 12.6 | 11.6 | |
| Pyrroloquinoline-associated factors | bsr6735 | Pyrroloquinoline synthesis protein (pqqA) | — | 1.5 | 6.0 |
| blr6736 | Pyrroloquinoline synthesis protein (pqqB) | — | 2.9 | 2.9 | |
| blr6738 | Pyrroloquinoline synthesis protein (pqqD) | — | 2.2 | 1.8 | |
| blr6739 | Pyrroloquinoline synthesis protein (pqqE) | — | 1.8 | 1.7 | |
| bll0332 | pqq methanol dehydrogenase-like | 5.3 | 12.9 | 2.6 | |
| bll0333 | Quinoprotein ethanol dehydrogenase | 7.7 | 13.1 | 3.4 | |
| bll6220 | Quinoprotein ethanol dehydrogenase | — | 4.6 | 4.3 | |
| Cation transporters | blr3803 | Potassium uptake protein | 2.6 | 2.1 | 2.5 |
| bll6262 | Osmotically inducible protein (osmC) | 1.9 | — | 3.5 | |
| blr2934 | Probable cation efflux protein | 2.0 | 2.3 | 2.6 | |
| blr4937 | Probable cation efflux system protein | — | 1.7 | 2.1 | |
| Pili and flagella | bsl1442 | Pilin subunit (ctpA) | — | 5.8 | 20.6 |
| bsl7141 | Pilin subunit (ctpA) | — | 4.4 | 18.7 | |
| bsl3118 | Pilin subunit (ctpA) | — | 5.2 | 13.9 | |
| bsl6587 | Pilin subunit (pilA) | — | 5.3 | 13.1 | |
| bsr1550 | Pilus assembly protein (pilA2) | 1.7 | 2.2 | 1.8 | |
| blr3695 | Flagellin (flaA) | 2.5 | 5.0 | 2.0 | |
| bll6866 | Flagellin | — | 4.9 | 2.6 | |
| bll6865 | Flagellin | — | 2.2 | — | |
| bll5845 | Putative flagellin, C terminus | — | 9.2 | 6.0 | |
| bll5846 | Putative flagellin, C terminus | — | 6.4 | 5.9 | |
| bll2207 | Flagellar biosynthesis protein (flhA) | — | 1.7 | 1.6 | |
| blr7002 | Flagellar motor switch protein (fliN) | — | 2.6 | 2.0 | |
| Other desiccation-induced ORFs | blr2455 | Isocitrate lyase (aceA) | 11.7 | 31.9 | 17.8 |
| blr0241 | ACC deaminase (acaS) | — | 5.2 | 15.1 | |
| bll5191 | Fasciclin I-like adhesion domain protein | — | 36.4 | 36.5 | |
Significance based on a ≥1.5-fold induction cutoff, a false discovery rate of ≤5%, and an α of 0.05. —, not significantly induced.
Induction of genes involved in trehalose synthesis.
Desiccation stress has previously been shown to result in accumulation of the disaccharide trehalose in B. japonicum (47). Consistent with this observation, we observed the induced transcription of genes encoding enzymes involved in two of the three B. japonicum trehalose synthesis pathways in desiccated cells (Table 2). Trehalose-6-phosphate synthase (bll0322) and trehalose 6-phosphate phosphatase (bll0323), participating in the OtsA/OtsB pathway in which glucose-6-phosphate and UDP glucose are transformed to trehalose, were significantly induced at all three incubation times assessed. Desiccation also resulted in the induction of trehalose synthase (blr6767) in the late period, which is responsible for transforming maltose to trehalose. A UTP-glucose-1-phosphate uridylyltransferase (blr1499) was induced 3.2- and 2.4-fold in the 24- and 72-h samples, respectively. This enzyme transforms UTP and α-d-glucose 1-phosphate to produce UDP-glucose, a precursor for the OtsA/OtsB trehalose synthesis pathway. Interestingly, three other sugar metabolism-related ORFs (bll6764, bll6765, and bll6766), located upstream of the trehalose synthase gene (bll6767), were also significantly induced in both late desiccation samples. These genes encode a glycoside hydrolase, an amylomaltase-like 4-α-glucanotransferase, and an α-amylase (distantly related to trehalose synthase), respectively.
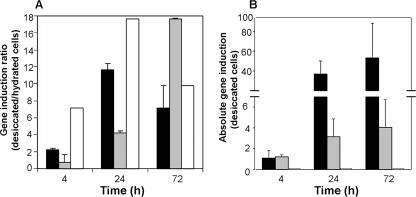
Real-time qRT-PCR was used to verify the induction of trehalose synthase genes observed in the microarray analyses. Results in Fig. 4A show the standardized ratio of the induction of the three B. japonicum trehalose synthesis genes (otsA, treS, and treY) in the desiccated versus hydrated cells for the three incubation times. Consistent with the microarray analyses, the trehalose 6-P-synthase (otsA) and trehalose synthase (treS) genes were significantly induced in 24- and 72-h samples, with maximal otsA and treS expression at 24 and 72 h postdesiccation, respectively (Fig. 4A). In contrast to the microarray data, qRT-PCR also showed a significant induction of the treY gene, encoding MOTS (Fig. 4A). However, the absolute value of treY expression, under both desiccating and hydrating conditions, normalized to the housekeeping gene parA, was extremely small (below 0.0001); thus, the expression of TreY may play a lesser role in desiccation tolerance than do the other two trehalose synthase enzymes (Fig. 4B). The absolute normalized ratios of otsA/parA increased considerably in the late incubation period for both the hydrated and desiccated samples, reaching values exceeding 20-fold higher than those measured in the 4-h samples (Fig. 4B). This implies that although the induction of otsA was significantly higher in the desiccated samples, it was also substantially induced in the hydrated samples.
FIG. 4.
qRT-PCR analysis of differentially expressed trehalose synthesis genes (otsA, treS, treY) in desiccated and hydrated B. japonicum cells normalized to the housekeeping gene parA. (A) Ratio of trehalose synthesis gene expression in desiccated versus hydrated samples. (B) Absolute expression of trehalose synthesis genes (otsA, treS, treY) in desiccated B. japonicum cells (values for treY relative to parA were <0.01; thus, the graph bars are difficult to see). Black bars, otsA; gray bars, treS; white bars, treY. Error bars representing the standard deviation of three biological replicates samples are shown.
Biochemical analysis of trehalose synthase activity.
The activity of trehalose 6-P synthase (OtsA) and trehalose synthase (TreS) was measured in desiccated and hydrated cells from the 4- and 72-h incubation periods. The results, normalized for cell viability with the LIVE/DEAD assay, indicated that the specific activity of trehalose 6-P synthase for desiccated (27% RH), relative to hydrated (100% RH), cells was 79.5 ± 21.7 versus 52.7 ± 15.6 nmol/h/mg protein for the 4-h postincubation samples and 101.8 ± 43.3 versus 54.3 ± 9.2 nmol/h/mg protein for the 72-h postincubation samples. These values are in agreement with the microarray expression and qRT-PCR data, which showed significant induction of otsA and otsB when B. japonicum was incubated under desiccating conditions. In contrast, significant induction of TreS was not detected in desiccated cells relative to hydrated cells, where values of 44.3 ± 7.2 versus 41.1 ± 3.5 and 74.8 ± 4.3 versus 67.8 ± 0.5 nmol/h/mg protein were measured for the 4- and 72-h incubation times, respectively. It should be noted, however, that these values were normalized based on live/dead counts and not on viability plate counting; had the latter been done, the apparent activity of both enzymes would have significantly increased under conditions of desiccation.
NMR profiles of intercellular trehalose pools.
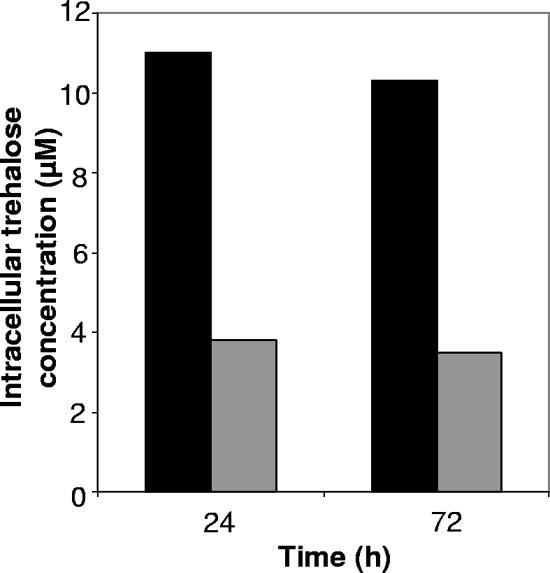
NMR analysis was done to determine if the induced transcription and activity of trehalose synthase genes, as seen in the transcriptional and biochemical assays, were consistent with an increase in the intracellular concentration of trehalose. Intracellular trehalose concentrations, based on the integrated value of the trehalose peak at 5.2 ppm, and the live/dead viability counts, were approximately 3.0 and 2.8 times greater in the desiccated samples, relative to the hydrated ones, following 24 and 72 h of incubation, respectively (Fig. 5).
FIG. 5.
Intracellular trehalose concentrations in desiccated and hydrated B. japonicum cells. Values are normalized based on cell viability determined by using LIVE/DEAD staining. Black bars, desiccated cells (27% RH); gray bars, hydrated cells (100% RH).
Effect of rpoN mutations on desiccation tolerance.
The B. japonicum genome contains two σ54-encoding genes, rpoN1 and rpoN2. Microarray analyses of the desiccated versus hydrated samples revealed significant induction of rpoN2 under desiccating conditions (2.2-, 10.0-, and 13.9-fold induction at the 4-, 24-, and 72-h times, respectively), but there was no significant induction of rpoN1. Real-time qRT-PCR of cDNA extracted from cells incubated for 72 h using primers targeting both rpoN1 and rpoN2 showed a significant induction of rpoN2 (2.1-fold) in the desiccated versus hydrated cells, with no significant induction of rpoN1.
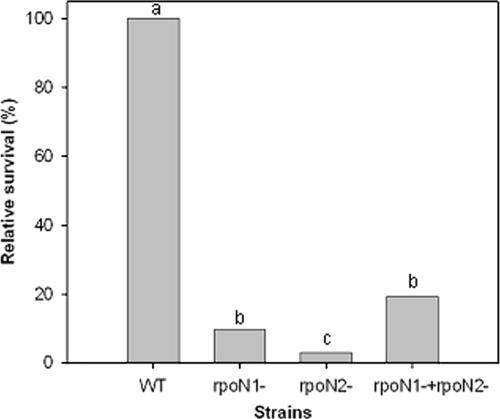
In order to assess the potential role of σ54 in the desiccation response, we compared the relative viability of rpoN1, rpoN2, and rpoN1 rpoN2 mutants (24) and wild-type B. japonicum cells grown under desiccating and hydrating conditions (Fig. 6). The viability of desiccated cells was significantly lower (P < 0.001) in all three B. japonicum rpoN mutants relative to the wild-type strain. In contrast, under hydrated conditions there was no significant difference (P < 0.001) in cell viability between the wild-type and the rpoN1 and rpoN2 mutant strains. However, under 100% RH conditions, the rpoN1 rpoN2 double mutant survived at a rate 8.5% lower than the mean survival rate of the rpoN1, rpoN2, and wild-type strains (data not shown).
FIG. 6.
Survival of B. japonicum rpoN mutant strains, relative to wild-type cells, following incubation under desiccating (27% RH) conditions. The significance of data was determined by comparison of means with ANOVA and Tukey-Kramer HSD. Columns followed by different letters are significantly different at P of 0.05. Survival rates of all strains at 100% RH were not significantly different (P < 0.001).
Influence of nonpermeating, solute-induced stress on the B. japonicum transcriptome.
Microarray analysis revealed that mild PEG-induced nonpermeating osmotic stress resulted in the differential expression of 184 and 43 up-regulated and 123 and 29 down-regulated genes, based on criteria of 1.5- and 2-fold induction, respectively. Of the 1.5-fold-induced genes, 30 were also induced in both early and later desiccated samples, and an additional 34 ORFs were upregulated only in the late (24- and 72-h) desiccated samples. Most notable were genes encoding trehalose 6-P-synthase (bll0322), an α-glucosidase (blr0901), potential EPS forming proteins (bll2362 and bll2752), transcriptional regulators (blr0536, bll7795, blr8144, and blr7797), a potential osmotic transport protein (blr7593), glutamine synthase II (blr4169), an organic hydroperoxide resistance protein (bll0735), and three pqq-associated alcohol dehydrogenases (bll0332, bll0333, and bll6220). Since many of the same proteins were produced under conditions of desiccation (Table 2), similar mechanisms are likely involved in tolerance to reduced water potential imparted by both conditions of incubation. Interestingly, 11 of the genes specifically induced by mild PEG-mediated nonpermeating osmotic stress were localized to the B. japonicum symbiosis island and included the symbiosis-related genes nolB, nolT, nolU, nolV, nolW, rhcR, and rhcS. While relatively mild osmotic stress was imparted by the PEG concentration used in our studies, none of these genes were induced by desiccation stress. Reduced water activity also resulted in down-regulation of 16 flagellar genes and 8 heat shock proteins.
DISCUSSION
Viability assays.
A large discrepancy was observed between the survival of cells of desiccation stress assessed by microscopic live/dead counts (∼50%) and dilution series plate counts (∼17%). This is likely due to the presence of cells in the VBNC state, which microorganisms often enter following environmental stress. Recently, it was reported that storage of inoculants leads to the VBNC state in B. japonicum (29) and that oxygen deficiency and exposure to soil were found to induce the VBNC state in Sinorhizobium meliloti cells (1).
Genes involved in the synthesis of trehalose.
Microarray data, qRT-PCR analyses, enhanced activity of trehalose-6-phosphate synthetase, and elevated intracellular concentrations of trehalose in cells incubated under desiccating conditions collectively support the hypothesis that trehalose plays a prominent and important role in promoting desiccation tolerance in B. japonicum. This is also supported by data showing that exogenously supplied trehalose enhances the survival of B. japonicum strain USDA 110 in response to desiccation (47). The otsA, otsB, and treS genes were significantly up-regulated in the desiccated versus hydrated cells, and these results were supported by data obtained from qRT-PCR analyses. McIntyre et al. (30) recently demonstrated that Rhizobium leguminosarum bv. trifolii otsA treY double mutants were more sensitive to the effects of drying (at both 5 and 32% RH) than were wild-type strains. However, and in contrast to our results which show a significant induction in expression of otsA following desiccation, studies of R. leguminosarum showed that the otsA and treY genes were expressed constitutively, regardless of the growth phase. This implies that trehalose regulation may behave differently in phylogenetically and physiologically distinct root nodule bacteria. While trehalose accumulation in B. japonicum is regulated at the transcriptional level, in R. leguminosarum trehalose production may be controlled at the posttranscriptional level or by control of trehalose degradation rates. The quantitative expression levels of otsA, otsB, and treS, relative to the housekeeping gene parA (Fig. 4B), and enzyme activity analyses indicate that the trehalose-6-phosphate synthetase and trehalose-6-phosphate phosphatase enzymes most likely constitute the prominent trehalose synthesis pathway involved in the desiccation response in B. japonicum. An ABC-type sugar transport protein (blr0324), located directly upstream from the otsA/otsB genes, was also significantly induced in the desiccation treatments and had a pattern of expression similar to those of otsA (blr0322) and otsB (blr0323), suggesting that it also may be involved in trehalose accumulation in Bradyrhizobium. A highly homologous sugar transporter is similarly positioned directly downstream from the trehalose 6-P-synthase gene on the chromosomes of Rhodopseudomonas palustris CGA009 (RPA4662), Bradyrhizobium sp. strain BTAi1 (BBTA_0576), and Bradyrhizobium sp. strain ORS278 (BRADO 6954). These ORFs have 80, 82, and 83% amino acid identity, respectively, with blr0323 from B. japonicum USDA 110.
Transcriptional regulators and desiccation stress response.
A total of 46 and 70 transcriptional regulators were differentially expressed during the early and late periods of desiccation, respectively, indicating that B. japonicum uses diverse regulatory pathways to respond to water stress conditions. The significant induction of two tryptophan-rich, TspO-like sensory proteins (bll5190 and blr2475) at the late incubation times suggests the importance of these proteins in the desiccation response. Similar TspO-like sensors have been correlated to nutrient stress in Sinorhizobium meliloti (11) and light response in Rhodobacter sphaeroides (58).
The gene encoding σ54 (blr0723) was found to be one of the most significantly (α = 0.05) induced transcriptional regulators at all three incubation times. While originally found to be responsible for the regulation of N metabolism (37), σ54 was shown to be involved in the regulation of disparate stress-related functions, including growth on alternative C sources (9), polar flagellar synthesis (37, 56), biodegradation of recalcitrant hydrocarbons (36), and growth under high pressure (34). Although the B. japonicum genome contains two σ54 genes, rpoN1 (blr1883) and rpoN2 (blr0723) (24), desiccation stress significantly induced expression only of rpoN2. Moreover, the concomitant induction of additional genes with defined σ54 interaction sites—those encoding a σ54 modulation protein (blr0724), a σ54-dependent nitrogen regulatory IIA protein (blr0725), the NifA-like regulatory protein (blr2037), a ntrXY two-component sensor histidine kinase (blr4489, blr4490), and a transcriptional regulatory protein (bsr6672) with high sequence identity to the σ54-regulated flagellar transcriptional regulator (flbD) (Table 2)—further strengthens the argument that rpoN2 plays a significant role in regulating gene expression in response to desiccation.
Exposure of the B. japonicum rpoN2 mutant to prolonged (72 h) desiccation resulted in a substantial (>80%) reduction in viability relative to the wild-type cells subjected to identical conditions. This supports the notion that this alternative sigma factor plays a fundamental role in the desiccation stress response. Surprisingly, the rpoN1 mutant showed a similar reduction in viability following desiccation, despite the fact that rpoN1 (blr1883) did not show significant changes in differential expression (visualized both in microarray and qRT-PCR analyses) under desiccating versus hydrating conditions. This suggests that the involvement of RpoN1 in the desiccation response may be due to posttranscriptional regulation (modulation) or by differences in the degradation rates of this alternative sigma factor.
Oxidative Stress.
Genes encoding several oxidative stress response proteins, including reactive oxygen scavengers such as peroxidases and superoxide dismutases, were significantly induced following desiccation, most prominently during the late incubation periods. A number of studies showed that desiccation in aerobic bacteria results in the formation of reactive oxygen species and subsequent damage to proteins, membranes, and DNA (14, 44). Desiccation stress in B. japonicum resulted in the significant induction of two genes encoding superoxide dismutase, chrC (bll7559) and sodF (bll7774) (Table 2). Shirkey et al. (44) reported that active Fe-superoxide dismutase (SodF) was the third most abundant soluble protein in Nostoc commune CHEN/1986 cells after prolonged desiccation. Recently, Dombrecht et al. (13) indicated that rpoN2 may be involved in the oxidative stress response by regulating the prxS-like peroxidase, which confers enhanced survival and growth to R. etli in the presence of H2O2. Two B. japonicum ORFs encoding prxS-like proteins (bll1317 and blr5308) were induced under desiccating conditions, suggesting that RpoN2 is also involved in oxidative stress responses in B. japonicum.
Desiccation significantly induced expression of glucose-6-phosphate dehydrogenase (blr6760), 6-phosphoglucolactonase (blr6761), and 6-phosphogluconate dehydrogenase (blr6759) (Table 2), all of which encode enzymes in the oxidative pentose phosphate pathway (OPPP) of B. japonicum. Superoxide dismutase and the OPPP were reported to be complementary in the capacity to confer oxidative stress protection in yeast, and glucose-6-phosphate dehydrogenase mutants of yeast and Salmonella are sensitive to oxidative stress (26, 45). Furthermore, Zhang et al. (60) suggested that the NADPH+ generated from the OPPP is utilized for oxidative stress protection in Deinococcus radiodurans. Taken together, our data indicate that desiccation results in the induction of several ORFs likely to be involved in resistance to oxidative stress in Bradyrhizobium. Interestingly, Benaroudj et al. (2) showed that in addition to its structural function in desiccation tolerance, trehalose accumulation during cellular stress can also protect cells and proteins from damage by oxygen radicals.
The pyrroloquinoline synthesis protein genes pqqA, pqqB, pqqD, and pqqE, as well as the genes for three putative pyrroloquinoline quinone (PQQ)-containing alcohol dehydrogenases, were significantly induced under desiccating conditions. Misra et al. (31) showed that transgenic Escherichia coli expressing a Deinococcus radiodurans PQQ synthase gene had superior survival following oxidative stress and that PQQ in vitro reacted with reactive oxygen species producing nonreactive molecular products. This suggests that the desiccation-induced pqq-related ORFs may be directly involved in the B. japonicum desiccation response by reducing oxidative stress in cells.
Induction of genes involved in polysaccharide formation.
Extracellular polymers and biofilms may serve as protective mechanisms to reduce desiccation-induced stress in terrestrial bacteria (38, 50, 52). Microarray analyses revealed that several exopolysaccharide-associated genes in B. japonicum were significantly induced in the late stages of desiccation, implying that extracellular polymers may be involved in desiccation tolerance. These genes include bll7574, which encodes a UDP-hexose transferase (exoM); blr1499, encoding a UTP-glucose-1-phosphate uridylyltransferase (exoN); and bll2362, which codes for a succinoglycan biosynthesis protein (exoP), all involved in exopolysaccharide production in rhizobia (59). In addition to these better-characterized genes, we also observed induction of two glycosyl transferases (bll2752 and blr2358), which were induced up to 17.8-fold in the 24-h samples. The ORF blr2358 has 48% amino acid identity with an exopolysaccharide production protein, PSS (P10498), from R. leguminosarum bv. phaseoli, suggesting that it potentially plays a role in exopolymer formation. The function of ORF bll2752 could not be determined based on sequence similarity. Microarray analyses also indicated that B. japonicum responds to desiccation and mild PEG-mediated nonpermeating osmotic stress by inducing rpoE, encoding the alternative sigma factor σ24. The σ24 factor has been most studied in Pseudomonas, where it was shown to play a pivotal role in the regulation of biosynthesis of exopolysaccharide in response to desiccation and other environmental stresses (42).
The fasciclin domain-containing ORF bll5191 was one of the most desiccation-induced genes, increasing 26- and 38-fold in desiccated samples from 24- and 72-h samples, respectively. This ORF lies immediately downstream of the desiccation-induced TspO-like sensory protein (bll5190) and encodes a protein having 65% amino acid identity with Nex18, a symbiotically induced protein in Sinorhizobium meliloti 1021 (35). Fas1-type fasciclin domains are thought to be involved in mediating interactions between bacterial cells and extracellular matrices during pathogenesis (8). The nex18 gene product may mediate stress responses through tspO in rhizobia (12).
Microarray analyses indicated that four type IV-like pilin subunits (bsl1442, bsl7141, bsl3118, and bsl6587) were significantly induced, by 5- to 20-fold, in desiccated cells in the 24- and 72-h incubations. This result strongly suggests that pilin biosynthesis may play a role in desiccation tolerance. Previous studies have demonstrated that type IV pili are prevalent in biofilm matrices, where they play an integral role in both cell adhesion and bacterial agglutination during biofilm formation (18, 43). White et al. (55) demonstrated enhanced desiccation tolerance in Salmonella sp. morphotypes, a multicellular phenotype which is characterized by abundance of fimbriae and cellulose. They hypothesized that thin aggregative fimbriae play a critical role in organizing the extracellular matrix of the biofilm, leading to an optimal spatial arrangement of cells and increased survival. Our results suggest that pili along with EPS may indirectly enhance the desiccation tolerance of bradyrhizobia by supporting biofilm formation, leading to the formation of a protective niche for these bacteria.
Additional desiccation-induced genes.
Other B. japonicum genes induced under conditions of desiccation included isocitrate lyase (blr2455), which was induced up to 32-fold, and blr0241, encoding 1-aminocyclo-propane-1-carboxylate deaminase, which was induced up to 15-fold. It has been suggested that isocitrate lyase plays an important role in the survival of bacteria under different stresses (25). However, the specific mechanism(s) linking isocitrate lyase induction to stress has not been studied.
Several nucleic acid modification and repair proteins potentially involved in desiccation-induced DNA damage repair were also identified by transcription profiling. These included genes encoding a DEAD-box ATP-dependent RNA helicase (bll1447), a MutL DNA mismatch repair protein (blr7493), a HupA histone-like protein HU (bll6175), and a RecF DNA recombination and repair protein (bll0827). Salt stress was previously shown to significantly induce expression of deaD, an ATP-dependent RNA helicase in Desulfovibrio vulgaris (33), and Vashisht and Tuteja (54) suggested that there is a correlation between induction of DEAD-box RNA helicases and salinity and drought stress in plants. Taken together, these results suggest that both excess intracellular cations and drought influence RNA and DNA melting, resulting in subsequent damage to nucleic acids.
Microarray analyses indicated that 12 putative transposases in B. japonicum were significantly up-regulated in response to desiccation. These included ORFs blr7557 and blr2478, which showed 7.0- and 4.4-fold induction after 72 h of incubation, respectively. It has been suggested that stress-induced transposition may be a strategy by which microbial populations adapt to harsh environmental conditions (23).
Conclusions.
Desiccation is one of the most prevalent and extreme environmental stress factors encountered by bacteria in arid and semiarid terrestrial environments. Although trehalose appears to be essential for the survival of desiccated cells, it is clear that additional protective mechanisms are also required and that B. japonicum directly responds to desiccation by the induction of a wide variety of proteins involved in protection of the cell membrane, repair of DNA damage, stability and integrity of proteins, and oxidative stress responses.
Supplementary Material
Acknowledgments
We thank Hans-Martin Fischer and Hauke Hennecke for providing the rpoN mutant strains, Betsy Martinez-Vaz for technical assistance, and Arkady Khodursky for many helpful suggestions. We dedicate this research to the memory of John G. Streeter.
This work was supported, in part, by grant 2004-35604-14708 from the USDA/CSREES/NRI (to D.W.E., M.J.S., G.S., and D.X.).
Footnotes
Published ahead of print on 27 July 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Basaglia, M., S. Povolo, and S. Casella. 2007. Resuscitation of viable but not culturable Sinorhizobium meliloti 41 pRP4-luc: effects of oxygen and host plant. Curr. Microbiol. 54:167-174. [DOI] [PubMed] [Google Scholar]
- 2.Benaroudj, N., D. H. Lee, and A. L. Goldberg. 2001. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 276:24261-24267. [DOI] [PubMed] [Google Scholar]
- 3.Bergersen, F. J. 1961. The growth of rhizobia in synthetic media. Aust. J. Biol. Sci. 14:349-360. [Google Scholar]
- 4.Borodovsky, M., and J. McIninch. 1993. GeneMark: parallel gene recognition for both DNA strands. Comp. Chem. 17:123-133. [Google Scholar]
- 5.Boumahdi, M., P. Mary, and J. P. Hornez. 2001. Changes in fatty acid composition and degree of unsaturation of (brady)rhizobia as a response to phases of growth, reduced water activities and mild desiccation. Antonie Leeuwenhoek 79:73-79. [DOI] [PubMed] [Google Scholar]
- 6.Bowtell, D. D., and J. F. Sambrook. 2002. DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Busse, M. D., and P. J. Bottomley. 1989. Growth and nodulation responses of Rhizobium meliloti to water stress induced by permeating and nonpermeating solutes. Appl. Environ. Microbiol. 55:2431-2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr, M. D., M. J. Bloemink, E. Dentten, A. O. Whelan, S. V. Gordon, G. Kelly, T. A. Frenkiel, R. G. Hewinson, and R. A. Williamson. 2003. Solution structure of the Mycobacterium tuberculosis complex protein MPB70: from tuberculosis pathogenesis to inherited human corneal disease. J. Biol. Chem. 278:43736-43743. [DOI] [PubMed] [Google Scholar]
- 9.Cases, I., and V. de Lorenzo. 2000. Genetic evidence of distinct physiological regulation mechanisms in the σ54 Pu promoter of Pseudomonas putida. J. Bacteriol. 182:956-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowe, J. H., J. F. Carpenter, and L. M. Crowe. 1998. The role of vitrification in anhydrobiosis. Annu. Rev. Physiol. 60:73-103. [DOI] [PubMed] [Google Scholar]
- 11.Davey, M. E., and F. J. de Bruijn. 2000. A homologue of the tryptophan-rich sensory protein TspO and FixL regulate a novel nutrient deprivation-induced Sinorhizobium meliloti locus. Appl. Environ. Microbiol. 66:5353-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djordjevic, M. A., H. C. Chen, S. Natera, G. V. Noorden, C. Menzel, S. Taylor, C. Renard, O. Geiger, and G. F. Weiller. 2003. A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant-Microbe Interact. 16:508-524. [DOI] [PubMed] [Google Scholar]
- 13.Dombrecht, B., C. Heusdens, S. Beullens, C. Verreth, E. Mulkers, P. Proost, J. Vanderleyden, and J. Michiels. 2005. Defense of Rhizobium etli bacteroids against oxidative stress involves a complexly regulated atypical 2-Cys peroxiredoxin. Mol. Microbiol. 55:1207-1221. [DOI] [PubMed] [Google Scholar]
- 14.França, M. B., A. D. Panek, and E. C. Eleutherio. 2006. Oxidative stress and its effects during dehydration. Comp. Biochem. Physiol. A 146:621-631. [DOI] [PubMed] [Google Scholar]
- 15.Halverson, L. J., and M. K. Firestone. 2000. Differential effects of permeating and nonpermeating solutes on the fatty acid composition of Pseudomonas putida. Appl. Environ. Microbiol. 66:2414-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, R. F. 1981. Effect of water potential on microbial growth and activity, p. 23-95. In J. F. Parr, W. R. Gardner, and L. F. Elliott (ed.), Water potential relations in soil microbiology. Soil Science Society of America Special Publication no. 9. Soil Science Society of America, Madison, WI.
- 17.Hoelzle, I., and J. G. Streeter. 1990. Increased accumulation of trehalose in rhizobia cultured under 1% oxygen. Appl. Environ. Microbiol. 56:3213-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jurcisek, J. A., and L. O. Bakaletz. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 189:3868-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, I. Kumi, M. Iriguchi, K. Kawashima, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9:189-197. [DOI] [PubMed] [Google Scholar]
- 20.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290:339-348. [DOI] [PubMed] [Google Scholar]
- 21.Katoh, H., R. K. Asthana, and M. Ohmori. 2004. Gene expression in the cyanobacterium Anabaena sp. PCC7120 under desiccation. Microb. Ecol. 47:164-174. [DOI] [PubMed] [Google Scholar]
- 22.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 23.Kivisaar, M. 2003. Stationary phase mutagenesis: mechanisms that accelerate adaptation of microbial populations under environmental stress. Environ. Microbiol. 5:814-827. [DOI] [PubMed] [Google Scholar]
- 24.Kullik, I., S. Fritsche, H. Knobel, J. Sanjuan, H. Hennecke, and H. M. Fischer. 1991. Bradyrhizobium japonicum has two differentially regulated, functional homologs of the σ54 gene (rpoN). J. Bacteriol. 173:1125-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, S., X. Xiao, J. Li, J. Luo, and F. Wang. 2006. Identification of genes regulated by changing salinity in the deep-sea bacterium Shewanella sp. WP3 using RNA arbitrarily primed PCR. Extremophiles 10:97-104. [DOI] [PubMed] [Google Scholar]
- 26.Lundberg, B. E., R. E. Wolf, M. C. Dinauer, Y. Xu, and F. C. Fang. 1999. Glucose 6-phosphate dehydrogenase is required for Salmonella typhimurium virulence and resistance to reactive oxygen and nitrogen intermediates. Infect. Immun. 67:436-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mary, P., D. Ochin, and R. Tailliez. 1985. Rates of drying and survival of Rhizobium meliloti strains during storage and different relative humidities. Appl. Environ. Microbiol. 50:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maurice, S., P. Beauclair, J.-J. Giraud, G. Sommer, A. Hartmann, and G. Catroux. 2001. Survival and change in physiological state of Bradyrhizobium japonicum in soybean (Glycine max L. Merril) liquid inoculants after long-term storage. World J. Microbiol. Biotechnol. 17:635-643. [Google Scholar]
- 30.McIntyre, H. J., H. Davies, T. A. Hore, S. H. Miller, J. P. Dufour, and C. W. Ronson. 2007. Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance. Appl. Environ. Microbiol. 73:3984-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misra, H. S., N. P. Khairnar, A. Barik, K. Indira Priyadarsini, and H. Mohan. 2004. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 578:26-30. [DOI] [PubMed] [Google Scholar]
- 32.Mohamed, R. M., M. Khavankharazian, W. F. Campbell, and M. D. Rumbaugh. 1991. Identification of salt-tolerant and drought-tolerant Rhizobium meliloti strains. Plant Soil 134:271-276. [Google Scholar]
- 33.Mukhopadhyay, A., Z. He, E. J. Alm, A. P. Arkin, E. E. Baidoo, S. C. Borglin, W. Chen, T. C. Hazen, Q. He, H. Y. Holman, K. Huang, R. Huang, D. C. Joyner, N. Katz, M. Keller, P. Oeller, A. Redding, J. Sun, J. Wall, J. Wei, Z. Yang, H. C. Yen, J. Zhou, and J. D. Keasling. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068-4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakasone, K., A. Ikegami, H. Kawano, C. Kato, R. Usami, and K. Horikoshi. 2002. Transcriptional regulation under pressure conditions by RNA polymerase σ54 factor with a two-component regulatory system in Shewanella violacea. Extremophiles 6:89-95. [DOI] [PubMed] [Google Scholar]
- 35.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 36.Pérez-Martín, J., and V. de Lorenzo. 1995. The σ54-dependent promoter Ps of the TOL plasmid of Pseudomonas putida requires HU for transcriptional activation in vivo by XylR. J. Bacteriol. 177:3758-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poggio, S., A. Osorio, G. Dreyfus, and L. Camarena. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J. Biol. Chem. 281:27205-27215. [DOI] [PubMed] [Google Scholar]
- 38.Potts, M. 1994. Desiccation tolerance of prokaryotes. Microbiol. Rev. 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos, J. L., M. T. Gallegos, S. Marques, M. I. Ramos-Gonzalez, M. Espinosa-Urgel, and A. Segura. 2001. Responses of gram-negative bacteria to certain environmental stressors. Curr. Opin. Microbiol. 4:166-171. [DOI] [PubMed] [Google Scholar]
- 40.Rozen, S., and H. J. Skaletsky. 2000. Primer 3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 41.Salzberg, A., A. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnider-Keel, U., K. B. Lejbolle, E. Baehler, D. Haas, and C. Keel. 2001. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 67:5683-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shime-Hattori, A., T. Iida, M. Arita, K. S. Park, T. Kodama, and T. Honda. 2006. Two type IV pili of Vibrio parahaemolyticus play different roles in biofilm formation. FEMS Microbiol. Lett. 264:89-97. [DOI] [PubMed] [Google Scholar]
- 44.Shirkey, B., D. P. Kovarcik, D. J. Wright, G. Wilmoth, T. F. Prickett, R. F. Helm, E. M. Gregory, and M. Potts. 2000. Active Fe-containing superoxide dismutase and abundant sodF mRNA in Nostoc commune (cyanobacteria) after years of desiccation. J. Bacteriol. 182:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slekar, K. H., D. J. Kosman, and V. C. Culotta. 1996. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 271:28831-28836. [DOI] [PubMed] [Google Scholar]
- 46.Specht, J. E., K. Chase, M. Macrander, G. L. Graef, J. Chung, J. P. Markwell, M. Germann, J. H. Orf, and K. G. Lark. 2001. Soybean response to water: a QTL analysis of drought tolerance. Crop Sci. 41:493-509. [Google Scholar]
- 47.Streeter, J. G. 2003. Effect of trehalose on survival of Bradyrhizobium japonicum during desiccation. J. Appl. Microbiol. 95:484-491. [DOI] [PubMed] [Google Scholar]
- 48.Streeter, J. G., and M. L. Gomez. 2006. Three enzymes for trehalose synthesis in Bradyrhizobium cultured bacteria and in bacteroids from soybean nodules. Appl. Environ. Microbiol. 72:4250-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Streeter, J. G. Factors affecting the survival of Bradyrhizobium applied in liquid cultures to soya bean [Glycine max (L.) Merr.] seeds. J. Appl. Microbiol., in press. [DOI] [PubMed]
- 50.Tamaru, Y., Y. Takani, T. Yoshida, and T. Sakamoto. 2005. Crucial role of extracellular polysaccharides in desiccation and freezing tolerance in the terrestrial cyanobacterium Nostoc commune. Appl. Environ. Microbiol. 71:7327-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van de Mortel, M., and L. J. Halverson. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735-750. [DOI] [PubMed] [Google Scholar]
- 53.van Rensburg, J. H., and B. Strijdom. 1980. Survival of fast- and slow-growing Rhizobium spp. under conditions of relatively mild desiccation. Soil Biol. Biochem. 12:353-356. [Google Scholar]
- 54.Vashisht, A. A., and N. Tuteja. 2006. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance. J. Photochem. Photobiol. B 84:150-160. [DOI] [PubMed] [Google Scholar]
- 55.White, A. P., D. L. Gibson, W. Kim, W. W. Kay, and M. G. Surette. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, J., A. K. Benson, and A. Newton. 1995. Global regulation of a σ54-dependent flagellar gene family in Caulobacter crescentus by the transcriptional activator FlbD. J. Bacteriol. 177:3241-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, X., M. Abo, A. Okubo, and S. Yamazaki. 1998. Trehalose as osmoprotectant in Rhodobacter sphaeroides f. sp. denitrificans IL106. Biosci. Biotechnol. Biochem. 62:334-337. [DOI] [PubMed] [Google Scholar]
- 58.Yeliseev, A. A., and S. Kaplan. 1999. A novel mechanism for the regulation of photosynthesis gene expression by the TspO outer membrane protein of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 274:21234-21243. [DOI] [PubMed] [Google Scholar]
- 59.Zhan, H. J., and J. A. Leigh. 1990. Two genes that regulate exopolysaccharide production in Rhizobium meliloti. J. Bacteriol. 172:5254-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y. M., T. Y. Wong, L. Y. Chen, C. S. Lin, and J. K. Liu. 2000. Induction of a futile Embden-Meyerhof-Parnas pathway in Deinococcus radiodurans by Mn: possible role of the pentose phosphate pathway in cell survival. Appl. Environ. Microbiol. 66:105-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.