Abstract
Resistance of Mycobacterium tuberculosis to pyrazinamide (PZA) derives mainly from mutations in the pncA gene. We developed a reverse hybridization-based line probe assay with oligonucleotide probes designed to detect mutations in pncA. The detection of PZA resistance was evaluated in 258 clinical isolates of M. tuberculosis. The sensitivity and specificity of PZA resistance obtained by this new assay were both 100%, consistent with the results of conventional PZA susceptibility testing. This assay can be used with sputa from tuberculosis patients. It appears to be reliable and widely applicable and, given its simplicity and rapid performance, will be a valuable tool for diagnostic use.
Pyrazinamide (PZA) is an important first-line antituberculosis drug used clinically for short-course chemotherapy because of its effectiveness against semidormant bacilli sequestered within macrophages (6, 10). The intracellular sterilizing activity of PZA allows the treatment period to be reduced to 6 months, whereas 9 months of treatment is required when PZA is not used (19). PZA is a prodrug. It requires conversion to pyrazinoic acid by bacterial pyrazinamidase (PZase) to affect tuberculosis bacilli (7, 17). Recent reports have established that mutations in the PZase gene (pncA) lead to the loss of PZase activity and constitute the primary mechanism of PZA resistance in Mycobacterium tuberculosis (11, 21, 22).
The time required for in vitro drug susceptibility testing of M. tuberculosis is constrained by the organism's relatively slow growth. Conventional drug susceptibility testing takes 7 to 28 days, depending on the culture system used (15). For most antituberculosis drugs, conventional methods produce reliable results, although PZA susceptibility testing with such methods is impaired by the poor bacterial growth under acidic conditions (7, 9). However, new culture methods were developed recently to resolve this problem (2, 13).
Previously, we described a DNA sequencing-based method to detect mutations in the genome of drug-resistant strains, including PZA-resistant M. tuberculosis (18). However, the use of this method in ordinary-scale clinical laboratories can be difficult. Therefore, we developed and describe here a new hybridization-based line probe assay (LiPA) for the rapid detection of pncA mutations in M. tuberculosis that is easily applied to clinical use. This assay can be used to evaluate PZA resistance, particularly in multidrug-resistant organisms, analyze PZA-resistant genes, and identify epidemic strains.
MATERIALS AND METHODS
Bacterial strains.
Two hundred twenty-five clinical isolates of M. tuberculosis were obtained from patients with pulmonary tuberculosis in Japan, and 33 were obtained from patients in Poland. The other bacterial strains used in this study are listed in Table 1.
TABLE 1.
Species specificity of the LiPA for detecting M. tuberculosis pncA
| Species | Straina | Nested PCR resultb | Hybridization signal with probes:
|
|
|---|---|---|---|---|
| 1-24 | 25-47 | |||
| M. tuberculosis | H37Rv (ATCC 27294) | + | All positive | All positive |
| M. tuberculosis | H37Ra (ATCC 25177) | + | All positive | All positive |
| M. bovis | BCG Japanese strain 172c | + | Δ16d | All positive |
| M. avium | ATCC 25291 | −* | All negative | All negative |
| M. fortuitum | RIMD 1317004 (ATCC 6841) | −* | All negative | All negative |
| M. gastri | GTC 610 (ATCC 15754) | −* | All negative | All negative |
| M. intracellulare | JCM 6384 (ATCC 13950) | −* | All negative | All negative |
| M. kansasii | JCM 6379 (ATCC 124878) | − | All negative | All negative |
| M. marinum | GTC 616 (ATCC 927) | −* | All negative | All negative |
| M. nonchromogenicum | JCM 6364 (ATCC 19530) | −* | All negative | All negative |
| M. phlei | RIMD 1326001 (ATCC 19249) | − | All negative | All negative |
| M. scrofulaceum | JCM 6381 (ATCC 19981) | − | All negative | All negative |
| M. simiae | GTC 620 (ATCC 25275) | − | All negative | All negative |
| M. smegmatis | ATCC 19420 | − | All negative | All negative |
| M. szulgai | JCM 6383 (ATCC 35799) | − | All negative | All negative |
| M. terrae | GTC 623 (ATCC 15755) | − | All negative | All negative |
| Escherichia coli | ATCC 8739 | − | All negative | All negative |
| Haemophilus influenzae | IID 984 (ATCC 9334) | − | All negative | All negative |
| Klebsiella pneumoniae | IID5209 (ATCC 15380) | − | All negative | All negative |
| Legionella pneumophila | GTC 745 | − | All negative | All negative |
| Mycoplasma pneumoniae | IID 817 | − | All negative | All negative |
| Pseudomonas aeruginosa | ATCC 27853 | − | All negative | All negative |
| Rhodococcus equi | ATCC 33710 | − | All negative | All negative |
| Staphylococcus aureus | N315 | − | All negative | All negative |
| Streptococcus pneumoniae | GTC 261 | − | All negative | All negative |
ATCC, American Type Culture Collection, Manassas, VA; RIMD, Research Institute for Microbial Diseases, Osaka University, Osaka, Japan; GTC, Gifu Type Culture Collection, Department of Microbiology-Bioinformatics, Regeneration and Advanced Medical Science, Gifu University, Graduate School of Medicine, Bacterial Genetic Resources, Gifu, Japan; JCM, Japan Collection of Microorganisms, Institute of Physical and Chemical Research (RIKEN), Saitama, Japan; IID, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
Approximately 100 ng of genomic DNA was used in the first PCR. Amplification results were determined by agarose gel electrophoresis. +, presence of amplification product of the expected; −, absence of amplification products; −*, presence of amplification products, but the sizes of the products were different from that of M. tuberculosis.
From Japan BCG Laboratory, Tokyo, Japan.
Absence of hybridization signal with one of the probes (probe 16).
Clinical samples.
Fifty-three sputum samples were collected from patients with tuberculosis or suspected tuberculosis. These samples were treated with an N-acetyl-l-cysteine-NaOH solution according to the procedure provided with the BBL MycoPrep Mycobacterial System Digestion/Decontamination kit (BD Diagnostic Systems, Franklin Lakes, NJ). Each sample was suspended in 1.5 ml of phosphate buffer. One milliliter of the suspension was transferred into a 1.5-ml tube. The remaining suspension was inoculated onto Ogawa medium and into MGIT 960 broth (BD BACTEC MGIT 960; BD Biosciences) and cultured for mycobacterial examination. One milliliter of the suspension was centrifuged for 15 min at 13,000 × g, and the supernatant was removed with a pipette. Tris-EDTA (TE) buffer (100 μl) was added to the sediment, and the solution was again centrifuged for 15 min at 13,000 × g. The sediment was suspended in 50 μl of TE buffer (50 μl), resuspended by vortexing, and incubated at 95°C for 30 min followed by incubation at 100°C for 10 min. The sample was vortexed again, allowed to cool, and centrifuged at 12,000 × g for 5 min to clarify the supernatant, which was transferred into another 1.5-ml tube. Each aliquot of the supernatant (5 μl) was used for each of the LiPA or Cobas Amplicor assays (Roche Diagnostic Systems, Basel, Switzerland).
PZA susceptibility testing and assay for PZase activity.
All clinical isolates of M. tuberculosis and M. tuberculosis strains H37Rv and H37Ra were tested for PZA susceptibility. Susceptibility to PZA was assessed by the broth method (MGIT 960). Nontuberculous Mycobacterium spp. were also tested for PZA susceptibility with the MGIT 960 method. However, standard methods for susceptibility testing are not available for nontuberculous Mycobacterium spp. (5). PZase activity was determined as described previously (20). In brief, a heavy loopful of mycobacterial culture freshly grown on Ogawa medium was inoculated onto 5 ml of Middlebrook 7H11 agar supplemented with pyrazinecarboxamide (0.812 mM; Wako Pure Chemical Industries, Osaka, Japan), sodium pyruvate (18.18 mM; Nacalai Tesque, Kyoto, Japan), and glycerol (0.5%, vol/vol; Nacalai) in a glass tube with a screw cap. After incubation at 37°C for 4 days, 1 ml of freshly prepared ferrous ammonium sulfate solution (25.5 mM; Sigma Chemical, St. Louis, MO) was added to each tube, and the presence of a pink band was assessed. M. tuberculosis strain H37Rv, which is susceptible to PZA and positive for PZase, was used as a positive control for the assay. M. bovis strain BCG, which is resistant to PZA and negative for PZase, was used as a negative control.
DNA extraction.
Two different methods were applied to extract genomic DNA. One method was described previously (12). The other method was performed as follows. Mycobacterial cells and other bacterial cells were collected from Ogawa medium and broth medium, respectively. A loopful of cells was suspended in 0.5 ml 1× TE buffer and inactivated at 100°C for 10 min. Cellular debris was pelleted at 13,000 × g for 5 min, and the supernatant with genomic DNA was used for PCR. Mycobacterial DNA in sputa was extracted with a cell lysis solution contained in a diagnosis kit (Amplicor respiratory specimen preparation kit; Roche Molecular Systems, Inc., Branchburg, NJ) or extracted by heating at 95°C for 30 min followed by freezing and thawing.
Preparation of oligonucleotide probes and strips.
Forty-seven oligonucleotide probes were designed to cover the entire pncA gene of wild-type H37Rv (Fig. 1). Two oligonucleotide probes were designed to compensate for silent mutations of C to T at nucleotide positions 180 and 195. A total of 49 probes were synthesized. These probes were immobilized on two strips. One strip contained 24 probes (probes 1 to 24) plus two probes to compensate for silent mutations. The other contained 23 probes (probes 25 to 47).
FIG. 1.
Locations of 49 oligonucleotide probes designed to cover the pncA gene of M. tuberculosis H37Rv.
LiPA.
The LiPA described here was developed on the basis of the same principle as that of the commercially available INNO-LiPA Rif. TB kit (Innogenetics, Ghent, Belgium) for the detection of rifampin resistance (14). The LiPA was conducted as described previously (14). In brief, biotinylated PCR products from test samples were hybridized to the immobilized probes and washed under strict conditions (1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] buffer containing 0.1% sodium dodecyl sulfate at 62°C). The presence or absence of bands on all strips was judged independently by three different observers. The classifications by the three observers were identical. Genomic DNA from the PZA-susceptible H37Rv strain was used as a positive control. Results for all samples were compared to those for the positive control. DNA from M. tuberculosis H37Rv was diluted into TE buffer (final concentrations, 4.84 pg/μl, 484 fg/μl, 242 fg/μl, 48.4 fg/μl, 24.2 fg/μl, and 2.42 fg/μl), and 1 μl of each solution was used to determine the sensitivity of the LiPA.
PCR and DNA sequencing.
Unless otherwise indicated, approximately 2 to 5 ng of genomic DNA was used for the amplification of a 670-bp fragment that includes the complete open reading frame of the pncA gene. To increase the sensitivity, nested PCR was performed with unlabeled external primers PR9-1 (5′-GGC GTC ATG GAC CCT ATA TCT G-3′) and PR10-1 (5′-CTT GCG GCG AGC GCT C-3′) for the first PCR and biotin-labeled internal primers IP-F (5′-GCT GCG GTA GGC AAA CTG C-3′) and IP-R (5′-CCA ACA GTT CAT CCC GGT TCG-3′) for the second PCR. The amplification conditions for the first and second PCRs were the same and consisted of 5 min of denaturation at 95°C followed by 40 cycles of 1 min at 95°C, 1 min at 55°C, and 1 min at 72°C. In some experiments, only the second PCR was done. Sequencing of pncA and its promoter region (nucleotides −80 to 572 relative to the initiation codon) was performed as described previously (18) for M. tuberculosis H37Rv and H37Ra, Mycobacterium bovis BCG, and all 258 clinical M. tuberculosis isolates tested regardless of the LiPA results.
COBAS Amplicor assays.
COBAS Amplicor assays, including the Amplicor MTB test, Amplicor M. avium test, and Amplicor M. intracellulare test (Roche Diagnostic Systems), were performed according to the instructions provided by the manufacturer.
RESULTS AND DISCUSSION
Among the 25 bacterial strains listed in Table 1, three strains, M. tuberculosis H37Rv, M. tuberculosis H37Ra, and M. bovis BCG, yielded PCR products of the expected length (approximately 600 bp). The other 13 strains of nontuberculous Mycobacterium spp. and nine strains of nonmycobacterial spp. yielded products of different lengths or were not amplified to a level detectable in agarose gels (Table 1). In the LiPA, PCR products from M. tuberculosis strains hybridized with all of the pncA probes (Table 1). LiPA results for strain H37Rv are shown in Fig. 2 (lane 1). The M. bovis product hybridized with all probes except for probe 16 (Table 1 and Fig. 2, lane 22). All other bacteria tested showed no hybridization with any of the probes (data not shown). These data indicate that the LiPA is specific for M. tuberculosis and M. bovis.
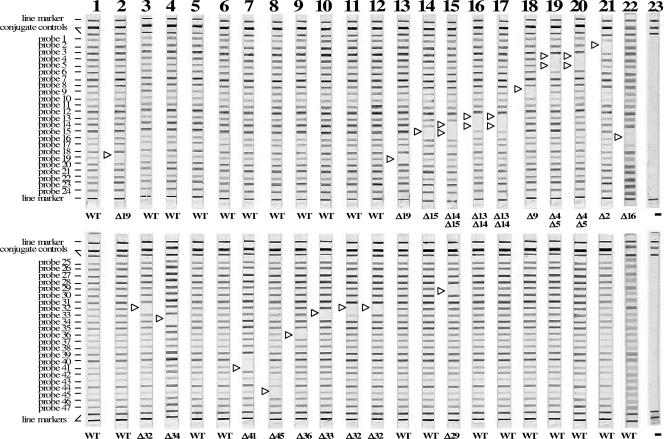
FIG. 2.
Representative patterns of isolates that had a mutation(s) according to the LiPA for detection of pncA mutations. Positions of the oligonucleotides, conjugate control lines, and marker lines are shown. A negative signal is shown by an open triangle. LiPA patterns of M. tuberculosis strains are shown in lanes 1 to 21. Lanes: 1, H37Rv; 2, IMCJ(130); 3, IMCJ904III; 4, IMCJ(66); 5, IMCJ(85); 6, IMCJ(67); 7, 13229; 8, IMCJ.M22; 9, P26; 10, 13243; 11, IMCJ850; 12, IMCJ835; 13, IMCJ501; 14, IMCJ844; 15, IMCJ479; 16, IMCJ843; 17, P12; 18, IMCJ901III; 19, IMCJ(80); 20, P10; 21, IMCJ.K1; 22, M. bovis BCG Japanese strain 172; 23, no DNA. WT, wild-type pncA.
Sensitivity of the LiPA with the nested PCR was 24.2 fg of M. tuberculosis DNA, which is equivalent to five copies of the pncA gene (data not shown), whereas when only the second PCR was done, the sensitivity of the LiPA was 484 fg, which is equivalent to 1,000 copies of pncA (data not shown). These data suggest that nested PCR is needed to yield the higher sensitivity.
Of 258 clinical isolates of M. tuberculosis tested with the LiPA, 228 were wild type, and the other 30 showed at least one mutation pattern (Table 2). Representative LiPA patterns are shown in Fig. 2. Hybridization signals visualized as violet bands on the strips were strong and readily discernible, with low background, although there was variability in bands intensities, and some strains yielded less intense bands than others. The 228 isolates hybridized to all probes, as shown by the data for strain H37Rv in Fig. 2 (lane 1). The products of the other 30 M. tuberculosis strains containing a mutation(s) did not hybridize to the probes corresponding to the position of the mutation(s) but did hybridize to the others. These results were fully consistent with the DNA sequencing results (Table 2). DNA sequencing confirmed that pncA of BCG had a C-to-G point mutation at codon 59, causing histidine to become aspartic acid (Table 2). These data are consistent with our previous findings and those of others (17, 18) showing that M. bovis BCG is naturally resistant to PZA. In addition, of the 13 other nontuberculous Mycobacterium spp. listed in Table 1, M. gastri was susceptible to PZA. All others were resistant to PZA (data not shown).
TABLE 2.
Identification of pncA mutations by LiPA among 258 clinical isolates of M. tuberculosis and M. bovis BCG
| Strain | LiPA profilea | Mutation
|
PZA susceptibility | PZase activity | |
|---|---|---|---|---|---|
| Amino acid change | Variant nucleotides | ||||
| IMCJ.K1 | Δ2 | 3 Ala→Glu | GCG→GAG | R | − |
| P10 | Δ4, Δ5 | 10 Gln→Pro | CAG→CCG | R | − |
| IMCJ(80) | Δ4, Δ5 | 12 Asp→Ala | GAC→GCC | R | − |
| IMCJ693 | Δ4, Δ5 | 12 Asp→Ala | GAC→GCC | R | − |
| IMCJ(120) | Δ4, Δ5 | 12 Asp→Ala | GAC→GCC | R | − |
| IMCJ901III | Δ8, Δ9 | 27 Leu→Pro | CTG→CGC | R | − |
| IMCJ918III | Δ8, Δ9 | 27 Leu→Pro | CTG→CGC | R | − |
| P12 | Δ13, Δ14 | 51 His→Gln | CAC→CAG | R | − |
| IMCJ843 | Δ13, Δ14 | 51 His→Gln | CAC→CAA | R | − |
| IMCJ846 | Δ13, Δ14 | 51 His→Gln | CAC→CAA | R | − |
| IMCJ479 | Δ14, Δ15, Δ29 | 53 Asp→Asn frameshift | GAC→AAC, 349 insertion CACTG | R | − |
| IMCJ844 | Δ15 | 54 Pro→Leu | CCG→CTG | R | − |
| IMCJ695 | Δ15 | 54 Pro→Leu | CCG→CTG | R | − |
| IMCJ838 | Δ15 | 54 Pro→Leu | CCG→CTG | R | − |
| IMCJ29 | Δ15 | 54 Pro→Leu | CCG→CTG | R | − |
| IMCJ907III | Δ15 | 54 Pro→Leu | CCG→CTG | R | − |
| IMCJ908III | Δ15 | 54 Pro→Leu | CCG→CTG | R | − |
| IMCJ501 | Δ19 | 72 Cys→Trp | TGC→TGG | R | − |
| IMCJ(133) | Δ19 | 72 Cys→Trp | TGC→TGG | R | − |
| IMCJ(130) | Δ19 | Frameshift | 218 insertion CGCATTGCCG | R | − |
| IMCJ835 | Δ32 | 132 Gly→Ser | GGT→AGT | R | − |
| IMCJ849 | Δ32 | 132 Gly→Ser | GGT→AGT | R | − |
| IMCJ850 | Δ32 | 133 Ile→Thr | ATT→ACT | R | − |
| IMCJ837 | Δ32 | 133 Ile→Thr | ATT→ACT | R | − |
| IMCJ904III | Δ32 | Frameshift | 386-388 deletion ATG | R | − |
| 13243 | Δ33 | 136 Asp→Tyr | GAT→TAT | R | ND |
| IMCJ(66) | Δ34 | Frameshift | 420 insertion G | R | − |
| P26 | Δ36 | 148 Arg→Ser | CGC→AGC | R | − |
| 13229 | Δ41 | Frameshift | 493 insertion C | R | ND |
| IMCJ.M22 | Δ45 | 175 Met→Val | ATG→GTG | R | − |
| IMCJ(67) | Wild type (Δ16)b | (60 Gly→Gly) | (GGC→GGT silent) | S | + |
| IMCJ(134) | Wild type (Δ16) | (60 Gly→Gly) | (GGC→GGT silent) | S | + |
| IMCJ(85) | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| IMCJ(75) | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| IMCJ851 | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| IMCJ839 | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| IMCJ(90) | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| IMCJ(96) | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| IMCJ(125) | Wild type (Δ17) | (65 Ser→Ser) | (TCC→TCT silent) | S | + |
| H37Rv | Wild type | No change | No change | S | + |
| BCGc | Δ16 | 59 His→Asp | CAC→GAC | R | − |
Δ indicates the negative signal at any of the probes.
Δ in parentheses indicates the negative signal at any of the probes unless the probe to compensate for the silent mutation was used.
M. bovis BCG Japanese strain 172.
Regarding the PZA resistance profile, the LiPA yielded results that were 100% in agreement with those obtained by the culture-based susceptibility method (Table 3). In addition, all PZase-positive bacilli were sensitive to PZA, and all PZase-negative bacilli were resistant to PZA (Table 2). These data were consistent with those of previously published reports (18, 22). The LiPA correctly identified PZA susceptibility and resistance in all strains in which a mutation(s) occurred. All of the 30 PZA-resistant M. tuberculosis isolates were correctly identified as being PZA resistant by the LiPA, and all of the 228 PZA-susceptible isolates were identified as being PZA susceptible. The specificity, sensitivity, and positive and negative predictive values of the LiPA for the detection of PZA resistance were all 100% (Table 3). In addition, we determined PZase activities of all Mycobacterium spp. listed in Table 1 and all clinical isolates of M. tuberculosis tested. Strains with PZase activity were sensitive to PZA, and those without PZase activity were resistant to PZA (data not shown).
TABLE 3.
Diagnostic performance of the LiPA compared to PZA susceptibility testing
| Result by PZA susceptibility test | No. of M. tuberculosis clinical isolates (n = 258) | LiPA result
|
Sensitivity (%) | Specificity (%) | Predictive value (%)
|
||
|---|---|---|---|---|---|---|---|
| No. resistant | No. susceptible | Positive | Negative | ||||
| Resistant | 30 | 30 | 0 | 100 | 100 | 100 | 100 |
| Susceptible | 228 | 0 | 228a | ||||
Includes nine isolates with a silent mutation in pncA.
To examine whether the LiPA can be applied to clinical samples from patients with tuberculosis, sputum samples obtained from 53 patients with suspected tuberculosis were tested (data not shown). All samples were positive for acid-fast staining. Among these 53 samples, 46 were positive for tuberculosis by the Amplicor MTB test and/or mycobacterial culture, and six were positive for M. avium by the Amplicor M. avium test. The remaining sample was positive for acid-fast staining but PCR and culture negative. In 45 of the 46 samples, M. tuberculosis pncA was detected with the LiPA. However, no mutation of pncA was found in these 45 samples, suggesting that all of the samples contained PZA-sensitive organisms. These results were later confirmed by drug susceptibility testing when M. tuberculosis isolates were obtained from the samples. Hybridization was not detected on the strips in the six M. avium-positive samples as well as the remaining sample that was positive for acid-fast staining but negative for PCR and culture. All samples were culture positive for mycobacteria. These results indicate that the LiPA is applicable to clinical samples. However, further studies of clinical samples containing PZA-resistant M. tuberculosis are necessary.
It appears that nested PCR rarely introduces additional mutations that may lead to false-positive results for the LiPA. Taq DNA polymerase is reported to make one error every 120 bases, and it was reported that these errors occur randomly (3). To affect LiPA results, the error must occur very early in the amplification process and at a specific site causing false-positive results in almost all PCR products. The frequency may be 3 × 10−11 [(1/120)5] when there are five copies of the template. In fact, nested PCR is used for other LiPA assays (1, 8) and for single-strand conformation polymorphism analysis (4).
We showed the usefulness of the LiPA for PZA susceptibility testing of M. tuberculosis. This assay can detect M. tuberculosis in smear-positive sputa from patients. This LiPA can rapidly and efficiently assess the resistance of M. tuberculosis to PZA. It is simple, convenient, and highly reliable when run in parallel with a convenient M. tuberculosis diagnostic algorithm in laboratories. However, the LiPA has some limitations. First, this assay does not have an internal control. In addition, this assay cannot correctly identify mixed PZA-resistant and -susceptible isolates. This assay cannot detect novel silent mutations; however, it can detect known mutations. Finally, genes other than pncA may be associated with PZA resistance. Scorpio et al. (16) previously reported PZase-positive PZA- resistant M. tuberculosis strains that were very rare and usually showed a low level of resistance. Nevertheless, our LiPA is a valuable tool for the detection of resistant M. tuberculosis strains within one working day and can easily be included in the routine workflow.
Acknowledgments
We thank M. Nakano (Jichi Medical School, Tochigi, Japan) for comments on the manuscript.
This study was supported by Health Sciences Research grants from the Ministry of Health, Labor, and Welfare of Japan (H15-SHINKO-3 and H18-SHINKO-IPPAN-012) and by the Program of Founding Research Centers for Emerging and Reemerging Infectious Diseases of the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Footnotes
Published ahead of print on 27 June 2007.
REFERENCES
- 1.Beenhouwer, H. D., Z. Lhiang, G. Jannes, W. Mijs, L. Machtelinckx, R. Rossau, H. Traore, and F. Portaels. 1995. Rapid detection of rifampicin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber. Lung Dis. 76:425-430. [DOI] [PubMed] [Google Scholar]
- 2.Bemer, P., T. Bodmer, J. Munzinger, M. Perrin, V. Vicent, and H. Drugeon. 2004. Multicenter evaluation of the MB/BACT system for susceptibility testing of Mycobacterium tuberculosis. J. Clin. Microbiol. 42:1030-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, B. D., and B. A. Connolly. 2004. Low-fidelity Pyrococcus furiosus DNA polymerase mutants useful in error-prone PCR. Nucleic Acids Res. 32:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bum-joon, K., K. H. Lee, B. N. Park, S. J. Kim, E. M. Park, Y. G. Park, G. H. Bai, S. J. Kim, and Y. H. Kook. 2001. Detection of rifampin-resistant Mycobacterium tuberculosis in sputa by nested PCR-linked single-strand conformation polymorphism and DNA sequencing. J. Clin. Microbiol. 39:2610-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2000. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes. Approved standard M24-A. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed]
- 6.Heifets, L., and P. Lindholm-Levy. 1992. Pyrazinamide sterilizing activity in vitro against semidormant Mycobacterium tuberculosis bacterial populations. Am. Rev. Respir. Dis. 145:1223-1225. [DOI] [PubMed] [Google Scholar]
- 7.Konno, K., F. M. Feldmann, and W. McDermott. 1967. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am. Rev. Respir. Dis. 95:461-469. [DOI] [PubMed] [Google Scholar]
- 8.Marttila, H. J., H. Soini, E. Vyshnevskaya, B. I. Vishnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljamen. 1999. Line probe assay in the rapid detection of rifampin-resistant Mycobacterium tuberculosis directly from clinical specimens. Scand. J. Infect. Dis. 31:269-273. [DOI] [PubMed] [Google Scholar]
- 9.McDermott, W., and R. Tompsett. 1954. Activation of pyrazinamide and nicotinamide in acidic environments in vitro. Am. Rev. Tuberc. 70:748-754. [DOI] [PubMed] [Google Scholar]
- 10.Mitchison, D. A. 1985. The action of antituberculosis drugs in short-course chemotherapy. Tubercle 66:219-225. [DOI] [PubMed] [Google Scholar]
- 11.Morlock, G. P., J. T. Crawford, W. R. Butler, S. E. Brim, D. Sikes, G. H. Mazurek, C. L. Woodley, and R. C. Cooksey. 2000. Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 44:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otsuka, Y., P. Parniewski, Z. Zwolska, M. Kai, T. Fujino, F. Kirikae, E. Toyota, K. Kudo, T. Kuratsuji, and T. Kirikae. 2004. Characterization of a trinucleotide repeat sequence (CGG)5 and potential use in restriction fragment length polymorphism typing of Mycobacterium tuberculosis. J. Clin. Microbiol. 42:3538-3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfyffer, G. E., F. Palicova, and S. Rusch-Gerdes. 2002. Testing of susceptibility of Mycobacterium tuberculosis to pyrazinamide with the nonradiometric BACTEC MIGIT 960 system. J. Clin. Microbiol. 40:1670-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossau, R., H. Traore, H. De Beenhouwer, W. Mijs, G. Jannes, P. De Rijk, and F. Portaels. 1997. Evaluation of the INNO-LiPA Rif. TB assay, a reverse hybridization assay for the simultaneous detection of Mycobacterium tuberculosis complex and its resistance to rifampin. Antimicrob. Agents Chemother. 41:2093-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salfinger, M., and G. E. Pfyffer. 1994. The new diagnostic mycobacteriology laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 13:961-979. [DOI] [PubMed] [Google Scholar]
- 16.Scorpio, A., P. Lindholm-Levy, L. Heifets, R. Gilman, S. Siddiqi, M. Cynamon, and Y. Zhang. 1997. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 41:540-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scorpio, A., and Y. Zhang. 1996. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat. Med. 2:662-667. [DOI] [PubMed] [Google Scholar]
- 18.Sekiguchi, J., T. Miyoshi-Akiyama, E. Augustynowicz-Kopec, Z. Zwolska, F. Kirikae, E. Toyota, I. Kobayashi, K. Morita, K. Kudo, S. Kato, T. Kuratsuji, T. Mori, and T. Kirikae. 2007. Detection of multidrug resistance in Mycobacterium tuberculosis. J. Clin. Microbiol. 45:179-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snider, D. E., Jr., J. Rogowski, M. Zierski, E. Bek, and M. W. Long. 1982. Successful intermittent treatment of smear-positive pulmonary tuberculosis in six months: a cooperative study in Poland. Am. Rev. Respir. Dis. 125:265-267. [DOI] [PubMed] [Google Scholar]
- 20.Wayne, L. G. 1974. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am. Rev. Respir. Dis. 109:147-151. [DOI] [PubMed] [Google Scholar]
- 21.Zhang, Y., and D. Mitchison. 2003. The curious characteristics of pyrazinamide: a review. Int. J. Tuberc. Lung Dis. 7:6-21. [PubMed] [Google Scholar]
- 22.Zhang, Y., and A. Telenti. 2000. Genetics of drug resistance in Mycobacterium tuberculosis, p. 235-256. In G. F. Hatfull and W. R. Jabobs (ed.), Molecular genetics of mycobacteria. American Society for Microbiology, Washington, DC.