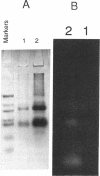
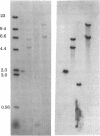
Abstract
Nitrosomonas europaea, a chemolithotrophic bacterium, was found to contain two copies of the gene coding for the presumed active site polypeptide of ammonia monooxygenase, the 32-kDa acetylene-binding polypeptide. One copy of this gene was cloned, and its complete nucleotide sequence is presented. Immediately downstream of this gene, in the same operon, is the gene for a 40-kDa polypeptide that copurifies with the ammonia monooxygenase acetylene-binding polypeptide. The sequence of the first 692 nucleotides of this structural gene, coding for about two-thirds of the protein, is presented. These sequences are the first sequences of protein-encoding genes from an ammonia-oxidizing autotrophic nitrifying bacterium. The two protein sequences are not homologous with the sequences of any other monooxygenase. From radioactive labelling of ammonia monooxygenase with [14C]acetylene it was determined that there are 23 nmol of ammonia monooxygenase per g of cells. The kcat of ammonia monooxygenase for NH3 in vivo was calculated to be 20 s-1.
Full text
PDF


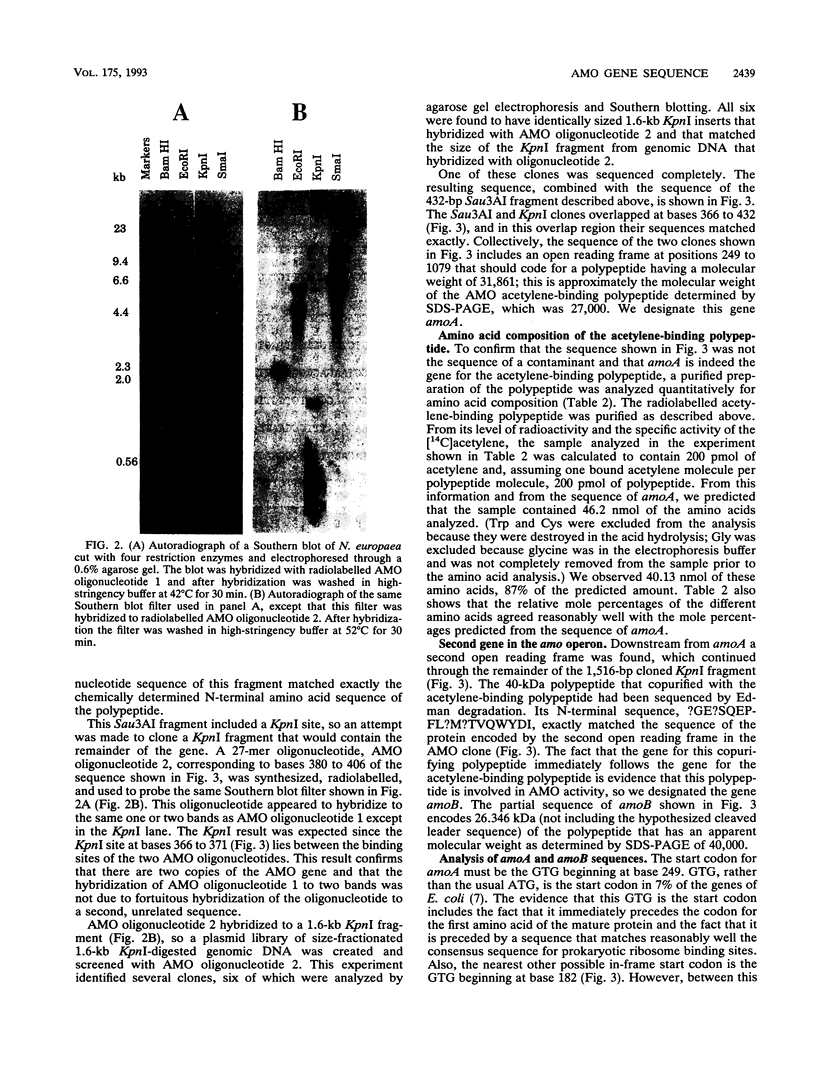


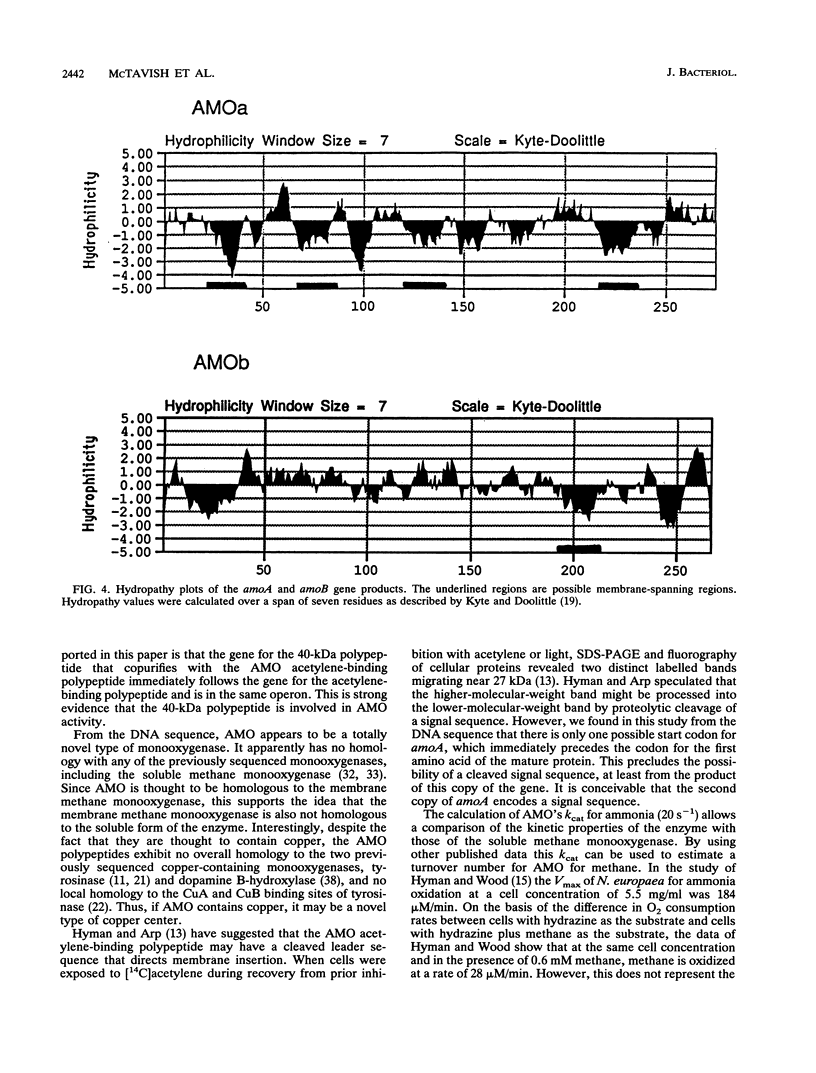


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arciero D. M., Balny C., Hooper A. B. Spectroscopic and rapid kinetic studies of reduction of cytochrome c554 by hydroxylamine oxidoreductase from Nitrosomonas europaea. Biochemistry. 1991 Dec 3;30(48):11466–11472. doi: 10.1021/bi00112a014. [DOI] [PubMed] [Google Scholar]
- Bédard C., Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989 Mar;53(1):68–84. doi: 10.1128/mr.53.1.68-84.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby J., Stirling D. I., Dalton H. The soluble methane mono-oxygenase of Methylococcus capsulatus (Bath). Its ability to oxygenate n-alkanes, n-alkenes, ethers, and alicyclic, aromatic and heterocyclic compounds. Biochem J. 1977 Aug 1;165(2):395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dispirito A. A., Lipscomb J. D., Hooper A. B. Cytochrome aa3 from Nitrosomonas europaea. J Biol Chem. 1986 Dec 25;261(36):17048–17056. [PubMed] [Google Scholar]
- Fox B. G., Lipscomb J. D. Purification of a high specific activity methane monooxygenase hydroxylase component from a type II methanotroph. Biochem Biophys Res Commun. 1988 Jul 15;154(1):165–170. doi: 10.1016/0006-291x(88)90665-1. [DOI] [PubMed] [Google Scholar]
- Gren E. J. Recognition of messenger RNA during translational initiation in Escherichia coli. Biochimie. 1984 Jan;66(1):1–29. doi: 10.1016/0300-9084(84)90188-3. [DOI] [PubMed] [Google Scholar]
- Hollocher T. C., Tate M. E., Nicholas D. J. Oxidation of ammonia by Nitrosomonas europaea. Definite 18O-tracer evidence that hydroxylamine formation involves a monooxygenase. J Biol Chem. 1981 Nov 10;256(21):10834–10836. [PubMed] [Google Scholar]
- Hooper A. B., Maxwell P. C., Terry K. R. Hydroxylamine oxidoreductase from Nitrosomonas: absorption spectra and content of heme and metal. Biochemistry. 1978 Jul 25;17(15):2984–2989. doi: 10.1021/bi00608a007. [DOI] [PubMed] [Google Scholar]
- Hooper A. B., Terry K. R. Specific inhibitors of ammonia oxidation in Nitrosomonas. J Bacteriol. 1973 Aug;115(2):480–485. doi: 10.1128/jb.115.2.480-485.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber M., Hintermann G., Lerch K. Primary structure of tyrosinase from Streptomyces glaucescens. Biochemistry. 1985 Oct 22;24(22):6038–6044. doi: 10.1021/bi00343a003. [DOI] [PubMed] [Google Scholar]
- Hyman M. R., Arp D. J. 14C2H2- and 14CO2-labeling studies of the de novo synthesis of polypeptides by Nitrosomonas europaea during recovery from acetylene and light inactivation of ammonia monooxygenase. J Biol Chem. 1992 Jan 25;267(3):1534–1545. [PubMed] [Google Scholar]
- Hyman M. R., Arp D. J. The small-scale production of [U-14C]acetylene from Ba14CO3: application to labeling of ammonia monooxygenase in autotrophic nitrifying bacteria. Anal Biochem. 1990 Nov 1;190(2):348–353. doi: 10.1016/0003-2697(90)90206-o. [DOI] [PubMed] [Google Scholar]
- Hyman M. R., Murton I. B., Arp D. J. Interaction of Ammonia Monooxygenase from Nitrosomonas europaea with Alkanes, Alkenes, and Alkynes. Appl Environ Microbiol. 1988 Dec;54(12):3187–3190. doi: 10.1128/aem.54.12.3187-3190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman M. R., Wood P. M. Methane oxidation by Nitrosomonas europaea. Biochem J. 1983 Apr 15;212(1):31–37. doi: 10.1042/bj2120031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman M. R., Wood P. M. Suicidal inactivation and labelling of ammonia mono-oxygenase by acetylene. Biochem J. 1985 May 1;227(3):719–725. doi: 10.1042/bj2270719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Meulenberg C. H., Kingma J., Witholt B. The Pseudomonas oleovorans alkBAC operon encodes two structurally related rubredoxins and an aldehyde dehydrogenase. J Biol Chem. 1989 Apr 5;264(10):5442–5451. [PubMed] [Google Scholar]
- Kok M., Oldenhuis R., van der Linden M. P., Raatjes P., Kingma J., van Lelyveld P. H., Witholt B. The Pseudomonas oleovorans alkane hydroxylase gene. Sequence and expression. J Biol Chem. 1989 Apr 5;264(10):5435–5441. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LEES H. The biochemistry of the nitrifying organisms. I. The ammonia oxidizing systems of Nitrosomonas. Biochem J. 1952 Sep;52(1):134–139. doi: 10.1042/bj0520134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch K. Amino acid sequence of tyrosinase from Neurospora crassa. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3635–3639. doi: 10.1073/pnas.75.8.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Moos M., Jr, Nguyen N. Y., Liu T. Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988 May 5;263(13):6005–6008. [PubMed] [Google Scholar]
- Porzio M. A., Pearson A. M. Improved resolution of myofibrillar proteins with sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochim Biophys Acta. 1977 Jan 25;490(1):27–34. doi: 10.1016/0005-2795(77)90102-7. [DOI] [PubMed] [Google Scholar]
- Shears J. H., Wood P. M. Spectroscopic evidence for a photosensitive oxygenated state of ammonia mono-oxygenase. Biochem J. 1985 Mar 1;226(2):499–507. doi: 10.1042/bj2260499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. D., Dalton H. Solubilisation of methane monooxygenase from Methylococcus capsulatus (Bath). Eur J Biochem. 1989 Jul 1;182(3):667–671. doi: 10.1111/j.1432-1033.1989.tb14877.x. [DOI] [PubMed] [Google Scholar]
- Stainthorpe A. C., Lees V., Salmond G. P., Dalton H., Murrell J. C. The methane monooxygenase gene cluster of Methylococcus capsulatus (Bath). Gene. 1990 Jul 2;91(1):27–34. doi: 10.1016/0378-1119(90)90158-n. [DOI] [PubMed] [Google Scholar]
- Stainthorpe A. C., Murrell J. C., Salmond G. P., Dalton H., Lees V. Molecular analysis of methane monooxygenase from Methylococcus capsulatus (Bath). Arch Microbiol. 1989;152(2):154–159. doi: 10.1007/BF00456094. [DOI] [PubMed] [Google Scholar]
- Suzuki I., Kwok S. C. A partial resolution and reconstitution of the ammonia-oxidizing system of Nitrosomonas europaea: role of cytochrome c554. Can J Biochem. 1981 Jul;59(7):484–488. doi: 10.1139/o81-067. [DOI] [PubMed] [Google Scholar]
- Suzuki I., Kwok S. C., Dular U., Tsang D. C. Cell-free ammonia-oxidizing system of Nitrosomonas europaea: general conditions and properties. Can J Biochem. 1981 Jul;59(7):477–483. doi: 10.1139/o81-066. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Hayakawa T., Shaw J. P., Rekik M., Harayama S. Primary structure of xylene monooxygenase: similarities to and differences from the alkane hydroxylation system. J Bacteriol. 1991 Mar;173(5):1690–1695. doi: 10.1128/jb.173.5.1690-1695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taljanidisz J., Stewart L., Smith A. J., Klinman J. P. Structure of bovine adrenal dopamine beta-monooxygenase, as deduced from cDNA and protein sequencing: evidence that the membrane-bound form of the enzyme is anchored by an uncleaved signal peptide. Biochemistry. 1989 Dec 26;28(26):10054–10061. doi: 10.1021/bi00452a026. [DOI] [PubMed] [Google Scholar]
- Vannelli T., Hooper A. B. Oxidation of Nitrapyrin to 6-Chloropicolinic Acid by the Ammonia-Oxidizing Bacterium Nitrosomonas europaea. Appl Environ Microbiol. 1992 Jul;58(7):2321–2325. doi: 10.1128/aem.58.7.2321-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannelli T., Logan M., Arciero D. M., Hooper A. B. Degradation of halogenated aliphatic compounds by the ammonia- oxidizing bacterium Nitrosomonas europaea. Appl Environ Microbiol. 1990 Apr;56(4):1169–1171. doi: 10.1128/aem.56.4.1169-1171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youvan D. C., Bylina E. J., Alberti M., Begusch H., Hearst J. E. Nucleotide and deduced polypeptide sequences of the photosynthetic reaction-center, B870 antenna, and flanking polypeptides from R. capsulata. Cell. 1984 Jul;37(3):949–957. doi: 10.1016/0092-8674(84)90429-x. [DOI] [PubMed] [Google Scholar]