Abstract
Minichromosomes of wild-type polyomavirus were previously shown to be highly acetylated on histones H3 and H4 compared either to bulk cell chromatin or to viral chromatin of nontransforming hr-t mutants, which are defective in both the small T and middle T antigens. A series of site-directed virus mutants have been used along with antibodies to sites of histone modifications to further investigate the state of viral chromatin and its dependence on the T antigens. Small T but not middle T was important in hyperacetylation at major sites in H3 and H4. Mutants blocked in middle T signaling pathways but encoding normal small T showed a hyperacetylated pattern similar to that of wild-type virus. The hyperacetylation defect of hr-t mutant NG59 was partially complemented by growth of the mutant in cells expressing wild-type small T. In contrast to the hypoacetylated state of NG59, NG59 minichromosomes were hypermethylated at specific lysines in H3 and also showed a higher level of phosphorylation at H3ser10, a modification associated with the late G2 and M phases of the cell cycle. Comparisons of virus growth kinetics and cell cycle progression in wild-type- and NG59-infected cells showed a correlation between the phase of the cell cycle at which virus assembly occurred and histone modifications in the progeny virus. Replication and assembly of wild-type virus were completed largely during S phase. Growth of NG59 was delayed by about 12 h with assembly occurring predominately in G2. These results suggest that small T affects modifications of viral chromatin by altering the temporal coordination of virus growth and the cell cycle.
Histone modifications play important roles in regulating cellular DNA transcription, replication, and repair. Histones undergo a variety of reversible covalent modifications including acetylation, methylation, and phosphorylation which contribute to the control of these processes (37, 52, 57). While much has been learned about the chemistry and enzymology of histone modifications, knowledge of their functional importance and how they are regulated is lagging. The mouse polyomavirus and other members of the polyomavirus group package their ∼5-kb circular DNA as a minichromosome utilizing cellular histones ordered into roughly 24 nucleosomes (42). We have used the minichromosomes of wild-type (WT) and mutant polyomaviruses as a model system to study the regulation of histone modifications by the viral T antigens.
Minichromosomes of polyomavirus and simian virus 40 (SV40) show an unusually high degree of acetylation of histones H3 and H4 compared to normal cell chromatin (43). Interestingly, minichromosomes of polyomavirus hr-t mutants (43), a class of host range mutants that are defective in cell transformation, were not hyperacetylated (3). These mutants are altered in sequences shared by the small T (sT) and middle T (mT) antigens but encode a normal large T (6, 22). Large T mutants of the ts-A class of both viruses were highly acetylated (43). Recent studies with SV40 have revealed a dynamic state of the viral minichromosome during replication. Extensive remodeling of viral chromatin occurs during replication in terms of the distribution and state of acetylation of nucleosomes, coinciding with the early versus late phases of viral gene expression and distribution of RNA polymerase II (2).
Because polyomavirus hr-t mutants are defective in both sT and mT, it was not possible in the earlier investigation to determine which of the two T antigens (or both) is responsible for H3 and H4 hyperacetylation. Nor did the earlier study address whether WT and hr-t mutant viral chromatin differ with respect to other modifications. mT is the major transforming protein of the virus. It resides principally in the plasma membrane, where it serves as both activator and substrate for tyrosine protein kinases of the src family. In this capacity it serves to activate several signaling pathways that promote mitogenesis and block apoptosis (reviewed in reference 25). sT serves as a regulatory subunit of the heterotrimeric protein phosphatase 2A (PP2A) (39), modulating its activity either positively or negatively toward numerous substrates including components of the mitogen-activated protein kinase pathway (41, 50). Both mT and sT play a positive role in viral DNA replication and transcription in NIH 3T3 cells (9-11). Using a series of site-directed mutants that selectively alter mT functions without altering sT, we show that expression of the sT antigen is sufficient for hyperacetylation of viral chromatin. In contrast to its hypoacetylated state, hr-t mutant viral chromatin is hypermethylated and hyperphosphorylated on H3 compared to WT virus. Evidence is presented that sT differentially modulates levels of acetylation and phosphorylation by altering the temporal coordination between virus growth and the virus-driven cell cycle.
MATERIALS AND METHODS
Viruses and cells.
hr-t and site-directed mutants have been described previously (3-5, 7, 31, 53) and are presented schematically in Fig. 1. WT and mutants NG59, NG18, 808A, 250YS, 315YF, 322YF, and 1387T were amplified in primary baby mouse kidney (BMK) cells prepared as described previously (14). To complement the loss of sT from NG59, the virus was cultured on NIH 3T3 cells expressing sT under a tetracycline-repressible promoter. NIH 3T3-sT (3T3-sT) cells were grown in Dulbecco modified Eagle medium containing 50 μg per ml Geneticin and 4 μg/ml tetracycline along with sodium bicarbonate, penicillin-streptomycin, and 5% calf serum. One day before viral inoculation, the 3T3-sT cells were split to Geneticin and tetracycline-free medium. Except for the single mT mutants 250YS, 315YF, and 322YF, results for all mutant and WT viruses were derived from independently amplified stocks, and assays were performed independently by at least two investigators with similar results. Assays with mT-defective mutants 808A and 1387T were done multiple times.
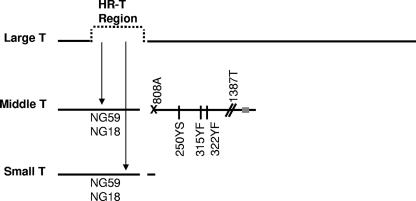
FIG. 1.
Schematic of polyomavirus T-antigen mutants NG59, NG18, 808A, 250YS, 315YF, 322YF, and 1387T used in this study. The three major early mRNAs, differing in splicing patterns, are denoted by horizontal lines. hr-t mutants NG59 and NG18 have mutations (insertion and substitution in NG59, deletion in NG18) in the large-T exon, resulting in nonfunctional mT and sT. A mutation at the 3′ slice site for mT in mutant 808A is depicted by an “X.” Single-site mutations at tyrosine phosphorylation sites in mT (250YS, 315YF, and 322YF) are depicted with vertical lines. The introduction of a stop codon upstream of the membrane binding domain near the C terminus of mT (gray rectangle) in mutant 1387T is depicted by slanted lines (//).
Virus purification.
Crude virus was prepared from infected-cell lysates by two rounds of freeze-thaw, centrifugation at 8,000 × g for 30 min at 4°C, and treatment of the cell debris with 0.5 unit/ml of neuraminidase for 30 min at 37°C. The treated cell debris was centrifuged at 8,000 × g for 30 min at 4°C. Virus was purified from the combined supernatants by pelleting the supernatants at 104,000 × g for 1.5 h at 4°C; resuspending them in 10 mM HEPES, pH 7.5, 1 mM CaCl2, and 150 mM NaCl (buffer A); and treating them with 0.5% deoxycholate at 37°C for 30 min. The deoxycholate-treated virus was centrifuged at 8,000 × g for 20 min at 20°C. Virus in the supernatant was pelleted at 104,000 × g for 2 h at 4°C and resuspended in buffer A containing 0.45 g/ml of cesium chloride. Gradients were centrifuged to equilibrium at 207,000 × g for 24 h at 10°C. Cesium chloride was removed by dialysis against buffer A. Purified virus was quantitated by UV spectroscopy using the Warburg-Christiansen formula. A 260/280 ratio of 1.28 to 1.33 was deemed an acceptable level of purification. Virus was stored at −80°C until use.
Total cell protein preparation.
BMK cells were infected with WT or NG59 virus at a multiplicity of infection of 5 to 10 PFU/cell. Total cellular protein was prepared with boiling 2× sodium dodecyl sulfate buffer (10 mM Tris, pH 6.8, 4% sodium dodecyl sulfate, 8% glycerol, 0.04% bromophenol blue, 1% β-mercaptoethanol). All samples were stored at −80° until use.
TAU gels and Western blotting.
Triton acid-urea (TAU) gels were prepared and run using the procedure found in Current Protocols in Molecular Biology (14a). Briefly, purified virus samples contained 90 μg of protein per well. Gels were silver stained using a protein silver staining kit from Amersham Pharmacia. Western blot assays were carried out on 12% NUPAGE Bis-Tris gels from Invitrogen (Carlsbad, CA) and transferred to Immobilon-PSQ (Millipore Corp., Bedford, MA) by semidry transfer. T antigens were analyzed as described previously (14).
Abs.
The antibodies (Abs) for Western blotting were anti-AcH3 Ab (06-599), anti-AcH3K9 Ab (06-942), anti-AcH3K14 Ab (06-911), anti-H3 Ab (05-499), anti-AcH4 Ab (06-598), anti-AcH4K5 Ab (07-327), and anti-AcH4K8 Ab (06-760). Anti-AcH4K12 Ab (06-761), anti-AcH4K16 Ab (06-762), anti-H4 Ab (05-858), anti-diMeH3K4 Ab (07-030), anti-triMeH3K9 Ab (07-442), anti-diMeH3K79 Ab (07-366), and anti-H3Pser10 Ab (06-570) were all from Upstate Cell Signaling Solutions, Lake Placid, NY. Secondary Abs were IRDye800 donkey anti-rabbit immunoglobulin G heavy-plus-light chain (611-732-127; Rockland) and Alexa Fluor 680 goat anti-mouse immunoglobulin G heavy-plus-light chain from Molecular Probes. Blots were analyzed using a Licor Odyssey infrared imager. Quantitation was performed using ImageJ or Licor Odyssey software. The concentration of modified histone was divided by the concentration of total histone, and the resulting ratios were compared to determine the relative levels of modification.
Cell cycle analysis.
BMK cells were cultured in medium containing 0.1% newborn calf serum for 24 h and infected at a multiplicity of infection of 5 to 10, giving >80% T-antigen-positive cells by 36 h postinfection. Cells were prepared and fluorescence-activated cell sorting (FACS) analysis was performed as described previously (14).
Assay for virus growth.
BMK cells were plated at 90% confluence in 24-well plates. Infection was done at a multiplicity of infection of 5 to 10 PFU per cell. At timed intervals plates were frozen in duplicate at −20°C and total virus yield was determined by plaque assay as described previously (14).
RESULTS
Hyperacetylation of histones H3 and H4 is dependent on sT.
A schematic of the WT and mutant polyomaviruses used in this study is shown in Fig. 1. hr-t mutants are altered in sequences within the large T intron. They are therefore defective in both sT and mT antigens but encode a normal large T. Histones H3 and H4 were previously shown to be hyperacetylated in WT virus compared to hr-t mutants (43). This is illustrated again here in Fig. 2A, showing the region of silver-stained TAU gels where the multiply acetylated species of H4 are well resolved. There is a clear shift toward the higher-acetylated forms in the WT compared to hr-t mutants NG59 and NG18. This is seen by noting the increases in the monoacetylated and diacetylated forms compared to unmodified H4 in the WT virus compared to the decreasing amounts of these forms relative to unacetylated H4 in the hr-t mutants. H3 shows similar results, although the acetylated species are less well resolved (not shown). Figure 2B shows results with three mT mutants with tyrosine substitutions that block different signal transduction pathways: 250YS is blocked in the Shc/ras/mitogen-activated protein kinase pathway (4), 315YF in the phosphatidylinositol 3-kinase pathways (5), and 322YF in the phospholipase Cγ pathway (53). Each of these mutants retains a normal sT (Fig. 1) and shows a distribution of acetylated H4 similar to that seen in the WT virus. Similarly, a triple mutant altered at all three sites was found to be hyperacetylated (not shown). Mutant 1387T encodes a mT with a premature termination codon upstream of the membrane binding domain (Fig. 1) (7). The 1387T mT accumulates in the cytoplasm and fails to associate with src kinases. This mutant encodes a normal sT along with its cytoplasmic mT and shows a slightly higher degree of H4 acetylation than does WT virus (Fig. 2C). Like hr-t mutants, 1387T is unable to transform cells but unlike these mutants shows a WT pattern of acetylation.
FIG. 2.
Hyperacetylation of histone H4 in purified virus is dependent on expression of normal sT. Purified virus was separated on TAU gels and visualized by silver staining. The numbers on the right of each gel indicate the acetylation state of H4. The bar graphs represent the percent distribution of the bands obtained from scanned images using ImageJ software. (A) TAU gels of WT virus and hr-t mutants NG18 and NG59. (B) TAU gels of WT virus and mT mutants 250YS, 315YF, and 322YF. (C) TAU gels of WT virus and mT truncation mutant 1387T. (D) TAU gels of WT virus and mT mutant 808A, hr-t mutant NG59, and NG59 grown on 3T3-sT cells.
Two additional approaches were taken to test whether sT alone is sufficient to induce hyperacetylation. Mutant 808A is altered at the mT splice acceptor site and expresses no mT (Fig. 1). This mutant expresses sT but at a reduced level compared to WT virus (31). The level of H4 acetylation in 808A was clearly higher than in NG59 though slightly less than in WT virus, consistent with a quantitative dependence of H4 acetylation on the level of sT (Fig. 2D). To test whether restoring sT function alone could complement the hyperacetylation defect of NG59, the mutant was grown on either normal cells or cells expressing sT. The level of H4 acetylation was higher in NG59 grown on NIH 3T3 cells rendered permissive by expression of an inducible sT (3T3-sT) than in NG59 grown on normal primary BMK cells (Fig. 2D).
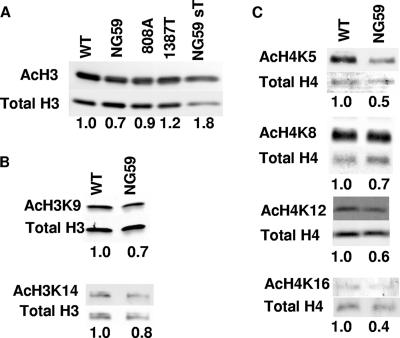
Western blotting with Abs to acetylated H3 and H4 peptides was used to further compare the state of acetylation in WT and mutant viral minichromosomes. The ratios of acetylated to total H3 and H4 for each of the mutants were normalized to the ratio found for WT virus. Using antibody to diacetylated (K9 and K14) H3, mutant NG59 was clearly underacetylated compared to the WT virus (Fig. 3A). Mutant 808A showed a value approaching that of the WT virus while 1387T and NG59 grown on 3T3-sT cells showed values equivalent to or slightly higher than those of the WT. These results support a role for sT in hyperacetylation of H3.
FIG. 3.
Hyperacetylation of viral histone H3 and acetylation of H3 and H4 at specific acetylation sites is dependent on expression of normal sT. (A) Western blot of dissociated purified virus with Abs to hyperacetylated H3 (AcH3) and total H3. (B) Western blots of dissociated purified virus with Abs specific for H3 acetylated at lysine 9 or 14 (AcH3K9 or AcH3K14, respectively) and total H3. (C) Western blots of dissociated purified virus with Abs specific for H4 acetylated at lysine 5, 8, 12, or 16 (AcH4K5, AcH4K8, AcH4K12, or AcH4K16, respectively) and total H4. The number below each lane represents the ratio of the acetylated histone to total histone normalized to one for WT virus.
Specific histone modifications have been assigned to specific cellular processes such as replication and transcription although assignments are not absolute. For instance, acetylations of histone H4 at lysines 5 and 12 and histone H3 at lysine 9 have been linked to replication whereas acetylations of histone H3 at lysine 14 and histone H4 at lysines 8 and 16 are associated with actively transcribing genes (52). Acetylation of lysines 9 and 14 in H3 was reduced in NG59 compared to WT virus (Fig. 3B). More dramatic reductions in NG59 were found at all major sites in H4 (Fig. 3C). The results in Fig. 2 and 3 point to the importance of sT and not mT in hyperacetylation of histones H3 and H4.
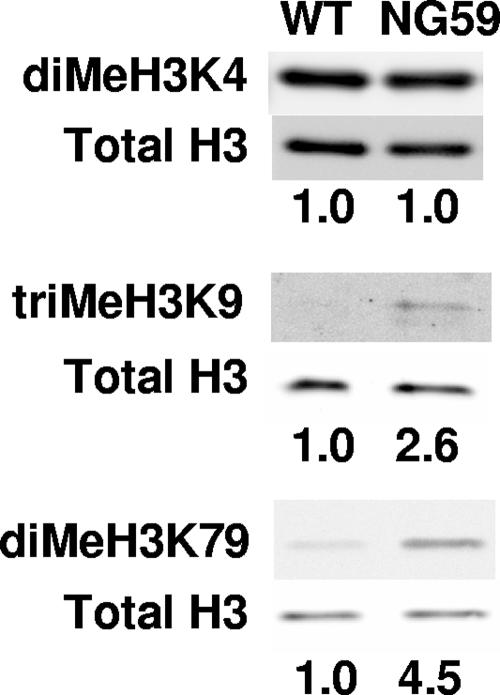
NG59 shows increased methylation at specific sites in H3.
WT and NG59 showed abundant and equal levels of dimethylated K4 of H3, a histone species associated with activation of transcription (36) (Fig. 4, top panel). In contrast, levels of triMeK9 in H3, a species associated with heterochromatin (29, 35), and diMeK79 were very low in WT viral chromatin. Unexpectedly, these two modifications were found at higher levels in NG59. Loss of hr-t gene function in the virus thus leads to opposite effects on acetylation and methylation of viral chromatin. Hypoacetylation of H3 and H4 in NG59 has been linked specifically to the loss of sT function. Hypermethylation of specific sites in H3 in the mutant may also reflect loss of sT, although a role of mT in these modifications has not been ruled out. It is clear, nevertheless, that the effects of sT and/or mT on viral chromatin are specific with respect to the site and kinds of modification.
FIG. 4.
Selective methylation of histone H3 is higher in NG59 mutant virus. Shown are Western blots of dissociated purified virus with Abs specific for H3 dimethylated on lysine 4 (diMeH3K4), H3 trimethylated on lysine 9 (triMeH3K9), or H3 dimethylated on lysine 79 (diMeH3K79) or total H3. Ratios of methylated H3 to total H3 normalized to one for WT virus are given below the lanes.
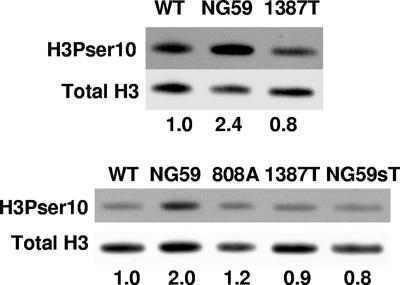
NG59 shows higher levels of phosphorylation of histone H3 at serine 10 than does WT virus.
PP2A is the only known cellular target of sT, suggesting that histone phosphorylation may be affected by the presence or absence of this viral protein. Phosphorylation of histone H3 at serine 10 has been linked to actively transcribing genes (12) but is also associated with chromatin condensation in mitosis (23, 37). PP2A has been shown elsewhere to dephosphorylate H3Pser10 in vitro and in vivo (38). NG59 showed a higher level of H3ser10 phosphorylation than did WT virus. Elevated levels of phosphorylation at this site were not seen in mutant 1387T (Fig. 5, top panel), nor in mutant 808A or NG59 grown on cells expressing WT sT (Fig. 5, bottom panel). These results point to a role of sT in controlling phosphorylation at this site. The fact that mutant 1387T exceeds the WT in its effects on H3 and H4 hyperacetylation (Fig. 2 and 3) as well as on downregulating H3ser10 phosphorylation (Fig. 5) is consistent with its truncated mT acting synergistically with sT, augmenting the binding of PP2A in the cytosol (54).
FIG. 5.
Hyperphosphorylation of histone H3 at serine 10 is independent of sT in NG59 virus. Shown are Western blots of dissociated purified virus with Abs specific for H3 phosphorylated at serine 10 (H3Pser10) or total H3. Ratios of phosphorylated H3 to total H3 normalized to one for WT virus are given below the lanes.
Differences in viral chromatin modification between WT and NG59 correlate with differences in rates of virus growth and cell cycle progression.
WT sT may conceivably act directly to inhibit PP2A's ability to dephosphorylate H3Pser10 (38). sT may also act indirectly to affect viral chromatin modifications at the level of posttranslational regulation of the modifying enzymes themselves, including histone acetyltransferases or histone deacetylases (HDACs). Attempts were made to determine whether the activities of these enzymes were altered in BMK cells infected with WT virus versus NG59. Nuclear extracts were prepared at roughly 45 h postinfection. No significant differences were seen in the bulk histone acetyltransferase or HDAC activities between mutant- and WT-infected cells (data not shown). Similarly, nuclear extracts were used to assay H3 dephosphorylation using as substrate an extract of HeLa cells arrested in mitosis and accumulating H3Pser10. No significant differences were seen in the rate of H3 dephosphorylation using extracts from WT- versus NG59-infected cells (data not shown).
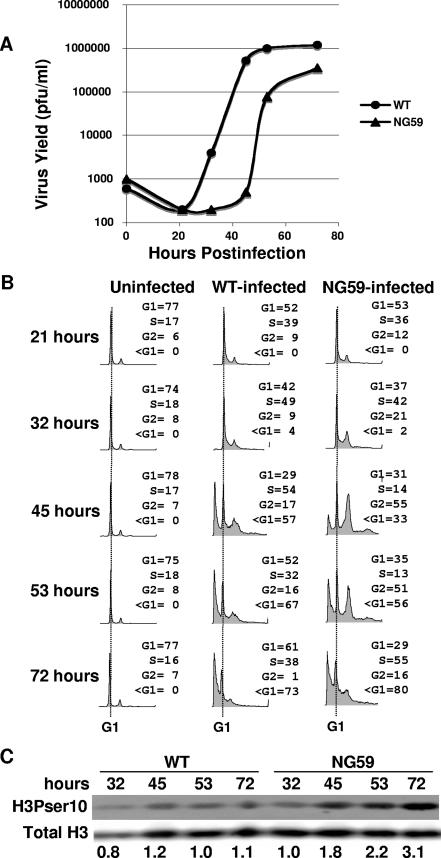
Levels of acetylation and phosphorylation of histones and various modifying enzymes are known to fluctuate during the cell cycle (1, 13, 16, 21, 24, 28, 30, 33). Notably, H4 acetylation is high during S phase (27) and phosphorylation of H3ser10 is high in late G2 and M (23). Differences in levels of these modifications in viral chromatin may therefore reflect possible differences between NG59 and WT virus in the rates of virus growth and maturation with respect to the cell cycle. To test this possibility, BMK cells were infected with NG59 or WT virus and followed for kinetics of virus growth and progression through the cell cycle (Fig. 6). hr-t mutants have previously been shown to replicate their DNA more slowly and to produce lower virus yields (11, 19). In primary BMK cells, growth of NG59 was delayed by 12 to 15 h compared to WT virus (Fig. 6A).
FIG. 6.
NG59-infected cells fail to pause in S phase but progress to G2/M and show a delay in virus production. (A) Virus growth curves for WT and NG59 virus. Virus yield in PFU per ml represents the averages of duplicate data points that varied no more than twofold. (B) FACS analysis of virus-infected cells. The G1 peak is marked with a dotted line with the <G1 peak to the left and the G2 peak to the right. (C) Western blot of whole-cell protein from virus-infected cells with Abs specific for H3Pser10 or total H3. Ratios of phosphorylated H3 to total H3 are given below the lanes.
The significance of the growth delay of NG59 with respect to its different pattern of chromatin modifications can be seen in relation to the progression of the cell cycle in WT- versus mutant-infected cells. FACS analysis showed roughly equal percentages of infected cells entering S phase by 32 h postinfection. Thereafter, WT- and mutant-infected cells diverged in their behavior (Fig. 6B). WT-infected cells continued to accumulate in S phase. Cytopathic effects (CPEs), estimated visually and by the sub-G1 peak in the FACS, occur at the late phase of the virus growth cycle concurrent with virus assembly. In WT-infected cells CPE set in around 40 h and increased until roughly 60 h, i.e., a period when cells are largely still in S phase and have not reached G2. In contrast, NG59-infected cells emerged from S phase after 32 h and progressed to G2. CPE was delayed compared to that for WT-infected cells. Over 50% of NG59-infected cells reached G2 at the time of widespread CPE compared to roughly 16% for the WT. Mutant 1387T grew with WT kinetics and with effects on the cell cycle similar to those of WT virus. Mutant 808A showed a FACS profile intermediate between those of NG59 and WT with more cells in S phase and fewer accumulating in G2 than found for NG59 (not shown). Cells in the WT- and NG59-infected cultures were followed over the same time course for H3 phosphorylation on serine 10. H3Pser10 in total chromatin from infected cells was higher in NG59-infected cells than in WT-infected cells (Fig. 6C). Differences in modeling of viral chromatin thus reflect the differences in timing of virus growth and assembly with respect to the cell cycle.
DISCUSSION
The present investigation was undertaken to better understand the basis of polyomavirus chromatin modifications and their dependence on T antigens. Previous results uncovered an unusually high degree of acetylation in viral chromatin from WT virus and a much lower degree in a class of host range transformation-defective (hr-t) mutants altered in both sT and mT antigens (43). Site-directed mutants that selectively alter mT without affecting sT and Abs to variously modified histones have provided new tools to investigate viral chromatin modeling. Results have shown that sT antigen alone suffices to induce hyperacetylation at major sites of H3 and H4. Thus, mT-defective mutants that express normal sT were normally hyperacetylated. hr-t mutant NG59, which is defective in both sT and mT, became hyperacetylated when grown on cells expressing WT sT. These results show that sT (along with large T) is sufficient to induce hyperacetylation of viral chromatin and that mT and retention of transforming functions of the virus are not required. The possibility that mT in the absence of sT might also lead to hyperacetylation has not been ruled out.
In contrast to acetylation of H3 and H4, methylation of histone H3 is found at higher levels in NG59 than in WT virus. H3 hypermethylation is selective for certain sites and modifications. triMeH3K9 and diMeH3K79 are more abundant in NG59 than in the WT while diMeH3K4 is equally abundant in the two viruses. NG59 also shows higher levels of phosphorylation of H3 on serine 10 than does WT virus. sT function underlies the difference in H3 phosphorylation, suggesting that it exerts a negative effect on phosphorylation of H3ser10. Modifications of viral chromatin appear to depend quantitatively on the level of expression of sT. This conclusion is supported by two observations with virus mutants. Mutant 808A underexpresses sT compared to WT virus (31). It shows a phenotype intermediate between those of NG59 and WT virus with respect to levels of H3 and H4 acetylation (Fig. 2 and 3) and H3 phosphorylation (Fig. 5). Mutant 1387T expresses a cytoplasmic mT which is expected to bolster the function of its normal sT (54). Its effects are enhanced compared to those of WT virus with respect to both acetylation (Fig. 2 and 3) and phosphorylation (Fig. 5).
We suggest that differences in viral chromatin modifications between WT virus and hr-t mutant NG59 can be understood as arising from differences in the rates of virus growth and progression through the cell cycle. Histone acetylation of H3 and H4, phosphorylation of H3 on serine 10, and methylation of triMeH3K9 and diMeH3K79 in bulk cell chromatin are all known to vary through the normal cell cycle (15, 21, 28, 32, 33). Global histone acetylation decreases sharply during mitosis whereas phosphorylation of histone H3 on serine 10 increases in late G2 and M (23). Levels of triMeH3K9 and diMeH3K79 also rapidly increase as cells enter mitosis (15, 32). Both WT virus and hr-t mutant NG59 drive resting BMK cells into S phase through large T-pRb interaction (17), but they differ with respect to rates of virus growth and subsequent progression through the cell cycle. WT virus induces and utilizes an ATM pathway of DNA repair to prolong S phase and promote virus replication (14). WT-infected BMK cells remain in S phase until virus replication is complete. Encapsidation of WT minichromosomes thus occurs when H3 and H4 acetylation are high and H3 phosphorylation is low. NG59 replicates its DNA more slowly and produces lower virus yields than does WT (11, 18). NG59-infected cells leave S phase and enter G2 before completing virus replication. Another hr-t mutant has been shown to induce a G2/M-phase arrest in NIH 3T3 cells (51). Assembly of NG59 thus occurs mainly in G2 when acetylation is low and H3 phosphorylation is high.
There are many points where sT acting through PP2A might affect cell cycle regulation and virus growth kinetics. For example, PP2A negatively regulates entry into mitosis (26). It also antagonizes ATM and ATR in the DNA damage checkpoint (20, 40), which is induced and utilized by the virus (14). Both SV40 large T and polyomavirus large T are phosphorylated at SQ motifs which could be potential sites of phosphorylation by ATM/ATR (8, 45). SV40 DNA replication is regulated by phosphorylation of large T in an sT/PP2A-dependent manner (44-46, 55). Polyomavirus DNA replication is also regulated by multiple phosphorylation sites on large T (8). SV40 sT regulates cell cycle progression at least in part through induction of cyclins D1 and A (49, 56). Polyomavirus sT has been shown to promote cell cycle progression (34) and to synergize with large T for S-phase induction in Swiss 3T3 mouse fibroblasts (47, 48). sT/PP2A may also affect rates and efficiency of encapsidation through phosphorylation of VP1 (18). Identities of the critical PP2A substrates at possible points of intervention in the virus growth cycle and cell cycle progression and how they may be affected by sT remain unknown.
Acknowledgments
This work has been supported by grant RO1 CA 90992. T.L.B. is a Virginia and D. K. Ludwig Professor of Cancer Research and Teaching. H.I.C. and M.G. were supported by the Proctor Fund summer program.
The technical assistance of John You is gratefully acknowledged.
Footnotes
Published ahead of print on 11 July 2007.
REFERENCES
- 1.Ait-Si-Ali, S., S. Ramirez, F. X. Barre, F. Dkhissi, L. Magnaghi-Jaulin, J. A. Girault, P. Robin, M. Knibiehler, L. L. Pritchard, B. Ducommun, D. Trouche, and A. Harel-Bellan. 1998. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature 396:184-186. [DOI] [PubMed] [Google Scholar]
- 2.Balakrishnan, L., and B. Milavetz. 2007. Histone hyperacetylation in the coding region of chromatin undergoing transcription in SV40 minichromosomes is a dynamic process regulated directly by the presence of RNA polymerase II. J. Mol. Biol. 365:18-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin, T. L. 1970. Host range mutants of polyoma virus. Proc. Natl. Acad. Sci. USA 67:394-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bronson, R., C. Dawe, J. Carroll, and T. Benjamin. 1997. Tumor induction by a transformation-defective polyoma virus mutant blocked in signaling through Shc. Proc. Natl. Acad. Sci. USA 94:7954-7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmichael, G., B. S. Schaffhausen, G. Mandel, T. J. Liang, and T. L. Benjamin. 1984. Transformation by polyoma virus is drastically reduced by substitution of phenylalanine for tyrosine at residue 315 of middle-sized tumor antigen. Proc. Natl. Acad. Sci. USA 81:679-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carmichael, G. G., and T. L. Benjamin. 1980. Identification of DNA sequence changes leading to loss of transforming ability in polyoma virus. J. Biol. Chem. 255:230-235. [PubMed] [Google Scholar]
- 7.Carmichael, G. G., B. S. Schaffhausen, D. I. Dorsky, D. B. Oliver, and T. L. Benjamin. 1982. Carboxy terminus of polyoma middle-sized tumor antigen is required for attachment to membranes, associated protein kinase activities, and cell transformation. Proc. Natl. Acad. Sci. USA 79:3579-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, A., B. J. Bockus, O. V. Gjorup, and B. S. Schaffhausen. 1997. Phosphorylation sites in polyomavirus large T antigen that regulate its function in viral, but not cellular, DNA synthesis. J. Virol. 71:6472-6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., and M. M. Fluck. 2001. Role of middle T-small T in the lytic cycle of polyomavirus: control of the early-to-late transcriptional switch and viral DNA replication. J. Virol. 75:8380-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L., X. Wang, and M. M. Fluck. 2006. Independent contributions of polyomavirus middle T and small T to the regulation of early and late gene expression and DNA replication. J. Virol. 80:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, M. C., D. Redenius, F. Osati-Ashtiani, and M. M. Fluck. 1995. Enhancer-mediated role for polyomavirus middle T/small T in DNA replication. J. Virol. 69:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 13.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahl, J., J. You, and T. L. Benjamin. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 79:13007-13017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, Q., H. Wang, H. H. Ng, H. Erdjument-Bromage, P. Tempst, K. Struhl, and Y. Zhang. 2002. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12:1052-1058. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira, R., I. Naguibneva, M. Mathieu, S. Ait-Si-Ali, P. Robin, L. L. Pritchard, and A. Harel-Bellan. 2001. Cell cycle-dependent recruitment of HDAC-1 correlates with deacetylation of histone H4 on an Rb-E2F target promoter. EMBO Rep. 2:794-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Freund, R., P. H. Bauer, H. A. Crissman, E. M. Bradbury, and T. L. Benjamin. 1994. Host range and cell cycle activation properties of polyomavirus large T-antigen mutants defective in pRB binding. J. Virol. 68:7227-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcea, R. L., K. Ballmer-Hofer, and T. L. Benjamin. 1985. Virion assembly defect of polyomavirus hr-t mutants: underphosphorylation of major capsid protein VP1 before viral DNA encapsidation. J. Virol. 54:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcea, R. L., and T. L. Benjamin. 1983. Host range transforming gene of polyoma virus plays a role in virus assembly. Proc. Natl. Acad. Sci. USA 80:3613-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodarzi, A. A., J. C. Jonnalagadda, P. Douglas, D. Young, R. Ye, G. B. Moorhead, S. P. Lees-Miller, and K. K. Khanna. 2004. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 23:4451-4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hans, F., and S. Dimitrov. 2001. Histone H3 phosphorylation and cell division. Oncogene 20:3021-3027. [DOI] [PubMed] [Google Scholar]
- 22.Hattori, J., G. G. Carmichael, and T. L. Benjamin. 1979. DNA sequence alterations in Hr-t deletion mutants of polyoma virus. Cell 16:505-513. [DOI] [PubMed] [Google Scholar]
- 23.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 24.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 25.Ichaso, N., and S. M. Dilworth. 2001. Cell transformation by the middle T-antigen of polyoma virus. Oncogene 20:7908-7916. [DOI] [PubMed] [Google Scholar]
- 26.Janssens, V., and J. Goris. 2001. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353:417-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasencakova, Z., A. Meister, J. Walter, B. M. Turner, and I. Schubert. 2000. Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription. Plant Cell 12:2087-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kruhlak, M. J., M. J. Hendzel, W. Fischle, N. R. Bertos, S. Hameed, X. J. Yang, E. Verdin, and D. P. Bazett-Jones. 2001. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J. Biol. Chem. 276:38307-38319. [DOI] [PubMed] [Google Scholar]
- 29.Lachner, M., R. J. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117-2124. [DOI] [PubMed] [Google Scholar]
- 30.Li, Y., Y. Butenko, and G. Grafi. 2005. Histone deacetylation is required for progression through mitosis in tobacco cells. Plant J. 41:346-352. [DOI] [PubMed] [Google Scholar]
- 31.Liang, T. J., G. G. Carmichael, and T. L. Benjamin. 1984. A polyoma mutant that encodes small T antigen but not middle T antigen demonstrates uncoupling of cell surface and cytoskeletal changes associated with cell transformation. Mol. Cell. Biol. 4:2774-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McManus, K. J., V. L. Biron, R. Heit, D. A. Underhill, and M. J. Hendzel. 2006. Dynamic changes in histone H3 lysine 9 methylations: identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. J. Biol. Chem. 281:8888-8897. [DOI] [PubMed] [Google Scholar]
- 33.McManus, K. J., and M. J. Hendzel. 2006. The relationship between histone H3 phosphorylation and acetylation throughout the mammalian cell cycle. Biochem. Cell Biol. 84:640-657. [DOI] [PubMed] [Google Scholar]
- 34.Mullane, K. P., M. Ratnofsky, X. Cullere, and B. Schaffhausen. 1998. Signaling from polyomavirus middle T and small T defines different roles for protein phosphatase 2A. Mol. Cell. Biol. 18:7556-7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 36.Noma, K., and S. I. Grewal. 2002. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc. Natl. Acad. Sci. USA 99(Suppl. 4):16438-16445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20:214-220. [DOI] [PubMed] [Google Scholar]
- 38.Nowak, S. J., C. Y. Pai, and V. G. Corces. 2003. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 23:6129-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pallas, D. C., L. K. Shahrik, B. L. Martin, S. Jaspers, T. B. Miller, D. L. Brautigan, and T. M. Roberts. 1990. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell 60:167-176. [DOI] [PubMed] [Google Scholar]
- 40.Petersen, P., D. M. Chou, Z. You, T. Hunter, J. C. Walter, and G. Walter. 2006. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2- and Cdc7-independent DNA damage checkpoint. Mol. Cell. Biol. 26:1997-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodriguez-Viciana, P., C. Collins, and M. Fried. 2006. Polyoma and SV40 proteins differentially regulate PP2A to activate distinct cellular signaling pathways involved in growth control. Proc. Natl. Acad. Sci. USA 103:19290-19295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Ryan, C. A., and A. T. Annunziato. 1999. Separation of histone variants and post-translationally modified isoforms by triton/acetic acid/urea polyacrylamide gel electrophoresis, p. 21.2.1-21.2.10. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. John Wiley and Sons, Inc., New York, NY. [DOI] [PubMed]
- 42.Saragosti, S., G. Moyne, and M. Yaniv. 1980. Absence of nucleosomes in a fraction of SV40 chromatin between the origin of replication and the region coding for the late leader RNA. Cell 20:65-73. [DOI] [PubMed] [Google Scholar]
- 43.Schaffhausen, B. S., and T. L. Benjamin. 1976. Deficiency in histone acetylation in nontransforming host range mutants of polyoma virus. Proc. Natl. Acad. Sci. USA 73:1092-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheidtmann, K. H., M. Buck, J. Schneider, D. Kalderon, E. Fanning, and A. E. Smith. 1991. Biochemical characterization of phosphorylation site mutants of simian virus 40 large T antigen: evidence for interaction between amino- and carboxy-terminal domains. J. Virol. 65:1479-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheidtmann, K. H., B. Echle, and G. Walter. 1982. Simian virus 40 large T antigen is phosphorylated at multiple sites clustered in two separate regions. J. Virol. 44:116-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheidtmann, K. H., M. C. Mumby, K. Rundell, and G. Walter. 1991. Dephosphorylation of simian virus 40 large-T antigen and p53 protein by protein phosphatase 2A: inhibition by small-t antigen. Mol. Cell. Biol. 11:1996-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuchner, S., M. Nemethova, A. Belisova, B. Klucky, W. Holnthoner, and E. Wintersberger. 2001. Transactivation of murine cyclin A by polyomavirus large and small T antigens. J. Virol. 75:6498-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuchner, S., and E. Wintersberger. 1999. Binding of polyomavirus small T antigen to protein phosphatase 2A is required for elimination of p27 and support of S-phase induction in concert with large T antigen. J. Virol. 73:9266-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skoczylas, C., B. Henglein, and K. Rundell. 2005. PP2A-dependent transactivation of the cyclin A promoter by SV40 ST is mediated by a cell cycle-regulated E2F site. Virology 332:596-601. [DOI] [PubMed] [Google Scholar]
- 50.Sontag, E., S. Fedorov, C. Kamibayashi, D. Robbins, M. Cobb, and M. Mumby. 1993. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation. Cell 75:887-897. [DOI] [PubMed] [Google Scholar]
- 51.Spink, K. M., and M. M. Fluck. 2003. Polyomavirus hr-t mutant-specific induction of a G2/M cell-cycle arrest that is not overcome by the expression of middle T and/or small T. Virology 307:191-203. [DOI] [PubMed] [Google Scholar]
- 52.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 53.Su, W., W. Liu, B. S. Schaffhausen, and T. M. Roberts. 1995. Association of polyomavirus middle tumor antigen with phospholipase C-gamma 1. J. Biol. Chem. 270:12331-12334. [DOI] [PubMed] [Google Scholar]
- 54.Templeton, D., S. Simon, and W. Eckhart. 1986. Truncated forms of the polyomavirus middle T antigen can substitute for the small T antigen in lytic infection. J. Virol. 57:367-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Virshup, D. M., M. G. Kauffman, and T. J. Kelly. 1989. Activation of SV40 DNA replication in vitro by cellular protein phosphatase 2A. EMBO J. 8:3891-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe, G., A. Howe, R. J. Lee, C. Albanese, I. W. Shu, A. N. Karnezis, L. Zon, J. Kyriakis, K. Rundell, and R. G. Pestell. 1996. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc. Natl. Acad. Sci. USA 93:12861-12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]