Abstract
Cytoplasmic processing bodies are sites where nontranslating mRNAs accumulate for different fates, including decapping and degradation, storage, or returning to translation. Previous work has also shown that the Lsm1-7p complex, Dhh1p, and Pat1p, which are all components of P bodies, are required for translation and subsequent recruitment to replication of the plant virus brome mosaic virus (BMV) genomic RNAs when replication is reproduced in yeast cells. To better understand the role of P bodies in BMV replication, we examined the subcellular locations of BMV RNAs in yeast cells. We observed that BMV genomic RNA2 and RNA3 accumulated in P bodies in a manner dependent on cis-acting RNA replication signals, which also directed nonviral RNAs to P bodies. Furthermore, the viral RNA-dependent RNA polymerase coimmunoprecipitates and shows partial colocalization with the P-body component Lsm1p. These observations suggest that the accumulation of BMV RNAs in P bodies may be an important step in RNA replication complex assembly for BMV, and possibly for other positive-strand RNA viruses.
The life cycle of viruses in eukaryotic cells requires that the virus complete its life cycle in the context of the host physiology. One interesting aspect of this process is how important steps in viral replication and packaging interface with translation. Since positive-strand RNA viruses, double-stranded RNA viruses, and reverse-transcribing viruses use the same viral genomic RNA as substrates for translation, encapsidation, and replication, mechanisms are required to segregate packaging and replicative events away from translation of the RNAs, thereby avoiding competition between elongating ribosomes and the packaging or replicative machineries. Although they are of significant importance to the viral replicative process, the mechanisms by which viruses segregate translation from replication and assembly are not well understood.
One class of viruses in which the interplay between translation and replication is important is the positive-strand RNA viruses, which encompass over one-third of all virus genera and include numerous well-known pathogens, such as hepatitis C virus and West Nile virus. Despite their differences, all positive-strand RNA viruses share similar life cycles. Following infection, the positive-strand RNA first serves as mRNA to produce viral replication factors, and then the transcript exits translation and is selectively recruited to a membrane-associated replication complex (27). An unresolved issue is the mechanism(s) bringing the viral RNAs and proteins together and thus facilitating the assembly of these replication complexes.
Insight into the host factors required for the assembly of replication complexes has come from the study of brome mosaic virus (BMV), which has a tripartite segmented genome consisting of RNA1, RNA2, and RNA3 and normally infects plants. Viral RNA replication requires the RNA-dependent RNA polymerase (RdRp), encoded by RNA2, and the 1a protein, which is encoded by RNA1 and functions in the assembly of endoplasmic reticulum (ER)-bound replication complexes (1, 33). RNA3 is dispensable for RNA replication and encodes cell-to-cell movement and capsid proteins, with the latter translated from a subgenomic RNA that is produced during replication (RNA4). The BMV life cycle has been shown to occur in the yeast Saccharomyces cereviase, which has allowed the use of genetic strategies to identify host factors required for viral functions (15, 20, 21, 24, 26, 28, 29). This work has shown that the host factors Lsm1p, Lsm6p, and Lsm7p, which are subunits of a larger Lsm1-7p heteroheptameric complex, as well as Pat1p, and Dhh1p, are required both for translation of the BMV genomic RNAs and independently for their entry into the replication complex (15, 24, 26, 28, 29). However, the mechanisms by which these proteins affect the 1a-mediated recruitment of viral RNAs to the replication complex are unknown.
Possible ways in which the Lsm1-7p complex, Pat1p, and Dhh1p might affect BMV replication can be suggested based on the roles of these proteins in controlling the translation and degradation of host mRNAs. In eukaryotes, two general mRNA decay pathways have been identified, both of which begin with shortening of the 3′ poly(A) tail in a process referred to as deadenylation (reviewed in reference 31). Following deadenylation, mRNAs can be degraded by a 3′-to-5′ exonuclease complex termed the exosome. Alternatively, after deadenylation, the mRNA can be decapped by the Dcp1p/Dcp2p decapping enzyme complex, followed by 5′-to-3′ degradation by the exonuclease Xrn1p.
An important point is that targeting an mRNA for decapping involves the formation of a translationally repressed mRNA-protein complex, which can also aggregate into specific cytoplasmic foci referred to as P bodies (reviewed in references 2, 16, and 30). mRNAs targeted to P bodies can be decapped and degraded or stored for subsequent return to translation (7, 8, 13, 35). P bodies appear to be a ubiquitous feature of eukaryotic cells and have been described in fungi, mammals, insects, nematodes, and trypanosomes, as well as in plants, which are the natural hosts for BMV infection (13, 16, 17, 18, 19, 35, 43). Interestingly, Dhh1p, Pat1p, and the Lsm1-7p complex function in facilitating the movement of mRNAs into P bodies and in stimulating decapping and degradation of the mRNA once assembled in a P-body state (11, 12, 37, 39).
The roles of Pat1p, Dhh1p, and the Lsm1-7p complex in controlling the targeting of mRNAs into P bodies and/or their subsequent degradation suggests two possible models where these proteins might be required for recruitment of BMV RNAs into replication complexes. First, it could be that the perturbation in the control of translation and mRNA degradation in strains lacking Pat1p, Dhh1p, or the Lsm1-7p complex significantly alters the host physiology, thereby exerting an indirect effect on BMV replication. Alternatively, since P bodies represent a pool of nontranslating RNAs, an alternative model is that P bodies play a more direct role in the formation of BMV replication complexes. In this model, viral RNAs in P bodies might represent ideal substrates to use for steps in initiating the assembly of viral replication complexes.
In this work, we explored the relationship between P bodies and BMV replication using yeast cells. We observed that BMV genomic RNA2 and RNA3 accumulated in yeast P bodies in a manner dependent on cis-acting RNA replication signals. In addition, the viral RdRp coimmunoprecipitated with and partially colocalized in P bodies. These observations suggest that the accumulation of BMV RNAs in P bodies may be an important step in RNA replication complex assembly and that the Lsm1-7p complex, Dhh1p, and Pat1p have direct roles in the assembly of BMV replication complexes. Moreover, because all plus-strand RNA viruses share a required transition in which RNAs exit translation to form replication complexes, these results raise the possibility that P bodies may be functionally important in the replication of a range of viruses.
MATERIALS AND METHODS
Yeast strains and growth conditions.
The genotypes of all strains used in this study are listed in Table 1. The strains were grown in yeast extract/peptone medium or synthetic medium (SC) supplemented with appropriate amino acids and 2% glucose (Glu) or 2% galactose (Gal) as a carbon source.
TABLE 1.
Strains used in this study
Steady-state RNA analysis.
Wild-type yRP2065 cells were transformed with RNA2 or RNA3, either containing bacteriophage MS2 sequences (pRP1402 and pRP1403) or not (pRP1538 and pRP1539). The cells were grown in SC supplemented with appropriate amino acids and 2% Gal to mid-log phase. RNA was prepared by glass bead lysis/phenol-choloroform and ethyl alcohol precipitation. Twenty micrograms of RNA was loaded on a 1.25% formaldehyde agarose gel, transferred to nitrocellulose, and probed with a radiolabeled oligonucleotide: RNA2, 5′ CTG GAA CGT CCG CGT TTC GC, or RNA3, 5′ ATC GGA CAA TCA TAG CTA GGC GTC. Samples were normalized to the loading control SCR1. Quantitation was done using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager and ImageQuant 5.2 software.
Preparation of cells for fluorescence microscopy.
Cells were grown as described above to mid-log phase (optical density at 600 nm, 0.3 to 0.4), washed twice, and resuspended in SC plus amino acids supplemented with either 2% Gal or 2% Glu. Observations were made using a Nikon PCM 2000 confocal microscope using a 100× objective and 3× zoom with Compix software. All images shown are single Z sections.
Plasmids and constructs for analyzing subcellular locations of BMV RNA2 and RNA3.
The plasmids used for this study are listed in Table 2, and primers are listed in Table 3. pRP1399, which expresses RdRp-cherry RFP, was constructed in multiple stages. First, a fragment of the BMV RdRp was amplified from pB2YT5 (9) by primer extension using primers oAN130 and oAN194. Cherry RFP (34) was amplified using primers oAN193 and oAN195. The two PCR products were gel purified and extended by mutually primed synthesis using primers oAN130 and oAN193. The final PCR product was cut with KpnI and PacI and cloned into the KpnI and PacI sites of pB2YT5.
TABLE 2.
Plasmids used in this study
| Plasmid | Construct | Reference |
|---|---|---|
| pRP1094 | MS2-GFP | 6 |
| pRP1399 | RdRp-cherry RFP | This work |
| pRP1400 | Lsm1p-cherry RFP | This work |
| pRP1402 | RNA2-MS2 | This work |
| pRP1403 | RNA3-MS2 | This work |
| pRP1404 | RNA3-MFA2 | This work |
| pRP1405 | RNA3-MS2 (in 3′UTR) | This work |
| pRP1406 | RNA3 (GAL-RE-MFA2) | This work |
| pRP1407 | RNA3 (Box B) | This work |
| pRP1408 | RNA3 Δ32 5′ | This work |
| pRP1409 | RNA3 Δ45 5′ | This work |
| pRP1410 | RNA3 Δ79 5′ | This work |
| pRP1411 | RNA3 Δ52 3′ | This work |
| pRP1412 | RNA3 Δ73 3′ | This work |
| pRP1413 | RNA3 (GAL) | This work |
| pRP1414 | RNA3 (RE) | This work |
| pRP1415 | β-globin (MFA2) | This work |
| pRP1416 | β-globin (RE-MFA2) | This work |
| pRP1417 | β-globin (box B) | This work |
| pRP1418 | β-globin Δ32 5′ | This work |
| pRP1419 | β-globin Δ45 5′ | This work |
| pRP1420 | β-globin Δ79 5′ | This work |
| pRP1421 | β-globin Δ52 3′ | This work |
| pRP1422 | β-globin Δ73 3′ | This work |
TABLE 3.
PCR primers used to construct the plasmids in this study
| Primer | Sequence |
|---|---|
| oAN43 | 5′ CGC AAC GCA ATT AAT GTG AG 3′ |
| oAN60 | 5′ CCC GGG GGA TCC ACT AGT TCT AGA GCG GCC GCC ACC GCG GTT TTT AAC CTT AAC CAA AGG GC 3′ |
| oAN61 | 5′ CCG CGG TGG CGG CCG CTC TAG AAC TAG TGG ATC CCC CGG GCT CCT GGT CAG GCA GAC C 3′ |
| oAN50 | 5′ GGTCGACGATTACGCTACCGG 3′ |
| oAN130 | 5′ AGC CAT AGT GAC AAG AGT ATT ACC 3′ |
| oAN168 | 5′ GGC CCC TTG TCT CAG GTA GAG CCC GGG ACC CTG TCC AGG TAG GAC AC 3′ |
| oAN169 | 5′ GTG TCC TAC CTG GAC AGG GTC CCG GGC TCT ACC TGA GAC AAG G 3′ |
| oAN170 | 5′ GCT TAA TCT CCA GAT TTA TCT G 3′ |
| oAN193 | 5′ CTT TAA CTG CAG TTA ATT AAT GGT GAG CAA GGG CGA GG 3′ |
| oAN194 | 5′ GCA TGG ACG AGC TGT ACA AGT CTT CGA AAA CCT GGG ATG 3′ |
| oAN195 | 5′ CAT CCC AGG TTT TCG AAG ACT TGT ACA GCT CGT CCA TGC 3′ |
| oAN197 | 5′ CAA GCT TGC ATG CCT GCA GGT CGA CTC TAG AGG ATC GGG GAT CCA GCT TTT CCG TGA CTG GTT G 3′ |
| oAN198 | 5′ GGT AAC AGA ATC ACC CCA TCT GCA GGA ATT AAT TCG ATT GGC GCC ATG GTG AGC AAG GGC GAG GAG G 3′ |
| oAN199 | 5′ CCT CCT CGC CCT TGC TCA CCA TGG CGC CAA TCG AAT TAA TTC CTG CAG ATG GGG TGA TTC TGT TAC C 3′ |
| oAN200 | 5′ ACC TCT TGG TGC TGG TCG ACG GTA TCG ATA AGC TTG ATT GGC GCC CTT GTA CAG CTC GTC CAT GCC 3′ |
| oAN201 | 5′ GCA CGT AAG GCC TAC GTT CGA TGA CTT CTT CAC CCC GGT TTA TAG GTA GAA TTC CTG ATC TAA TCG ATA TCT ACC C 3′ |
| oAN202 | 5′ CGA ATT GGG TAC CGG GCC CCC CCT CGA GAA TTC GAC AGG TTA TCA GCA ACA ACA C 3′ |
| oAN204 | 5′ GAT GTC AAG CTT AAT TCC TGA TCT AAT CGA TAT C 3′ |
| oAN205 | 5′ CAA AAG CTG GAG CTC CAC CGC CCG GGC GGA TCC AGC TTT TCC GTG ACT GGT TG 3′ |
| oAN209 | 5′ CAA AAA TTA AAG CTT GAC ATC TCT AGA AGA TCC GAT TCG AGG TCT AAT AGA AAT TG 3′ |
| oAN210 | 5′ TCA AAA ATT AAA GCT TGA CCA AAC CTC TGG CGA AGA AGT CC 3′ |
| oAN211 | 5′ CGA ATT GGG TAC CCA GTA CCT CGA GCC TCT TCG CTA TTA CGC CA 3′ |
Plasmid pRP1400 was constructed by cutting plasmid pRP1085 (35) with BamHI and XbaI to add the in-frame cherry RFP (34) PCR product.
pRP1402, which expresses an RNA2, tagged with two MS2 binding sites in the 3′ untranslated region (UTR), was cloned in multiple steps. First, a SmaI site was introduced at position 2629 of RNA2. To introduce the SmaI site, two fragments of RNA2 were amplified from pB2NR3 (9) by primer extension using primers oAN50 and oAN60, and another fragment was amplified using primers oAN61 and oAN43. The two PCR products were gel purified and extended by mutually primed synthesis using primers oAN43 and oAN50. The final PCR product was cut with SphI and StuI and cloned into the SphI and StuI sites of pB2NR3 to create pB2NR3-SmaI. Then, an MscI/AfeI fragment of pSL-MS2-24 (6) containing a 24-MS2 binding module, was cloned into pB2NR3-SmaI cut with SmaI to create plasmid pB2NR3-MS2. Finally, the tagged RNA2 expression cassette cut from pB2NR3-MS2 using PvuII was cloned into the PvuII sites of pRS426 (10) to create pRP1402.
pB3RQ39-URA, a version of pB3RQ39 (18) with a URA3-selectable marker, was used for the construction of all RNA3 derivatives described below. pB3RQ39-URA was constructed by cloning the 3-kb EcoRI-SalI fragment from pB3RQ39 containing the RNA3 expression cassette into the EcoRI/SalI sites of pRS316 (10). pRP1403, which expresses RNA3 with a 24-MS2 tag inserted in the second open reading frame (coat protein), was constructed by inserting a blunt-ended EcoRV/NcoI fragment of pSL-MS2-6 (6) containing a 24-MS2 binding module into pB3RQ39-URA cut with StuI. Another MS2-tagged RNA3 containing the six MS2 binding sites in the 3′ UTR of RNA3 (plasmid pRP1405) was also constructed.
Plasmid pRP1405 was constructed in multiple stages. First, a SmaI restriction site was introduced into the 3′ UTR of RNA3 in pB4MK1 (28), which expresses the BMV subgenomic RNA4, which in effect is the 3′ half of RNA3. To introduce the SmaI site, a fragment of RNA4 was amplified from pB4MK1 by primer extension using primers oAN43 and oAN168, and another fragment was amplified using primers oAN169 and oAN170. The two PCR products were gel purified and extended by mutually primed synthesis using primers oAN43 and oAN170. The final PCR product was cut with KpnI and StuI and cloned into the KpnI and StuI sites of pB4MK1 to create pB4MK1-SmaI. Then, an MscI/AfeI fragment of pSL-MS2-6 (6) containing a six-MS2 binding module was cloned into pB4MK1-SmaI cut with SmaI to create plasmid 891. Finally, the tagged 3′ UTR of RNA3 was created by inserting the StuI/SphI fragment from plasmid 891 into pB3RQ39-URA, resulting in plasmid pRP1405. Tagged RNA3 from both pRP1403 and pRP1405 plasmids accumulated in Northern blots and in P bodies to similar levels (data not shown).
pRP1414 expressing a tagged version of RNA3 with the viral 5′ UTR replaced with the yeast GAL1 5′ UTR was cloned by cutting pMS114 (36) with EcoRI/MscI and cloning the resulting plasmid into the EcoRI/MscI sites of pRP1405. pRP1404, which expresses an RNA3 with the viral 3′ UTR replaced with a yeast MFA2 3′ UTR tagged with six MS2, was constructed by first amplifying a MFA2 3′ UTR tagged with six MS2 binding sites from pRP1083 (35) using primers oAN197 and oAN201, digesting it with StuI/SphI, and inserting it into pB3RQ39-URA cut with StuI/SphI. pRP1415 expressing a tagged version of RNA3 with the recruiting element (RE) deleted was cloned by cutting pMS46 (36) with EcoRI/MscI and cloning the resulting fragment into the EcoRI/MscI sites of pRP1405. pRP1406, which expresses an RNA3 with a yeast GAL1 5′ UTR, a deleted RE region, and an MFA2 3′ UTR tagged with six MS2 binding sites, was constructed in multiple stages.
First, the EcoRI/PflMI fragment from pB3MS114 (36) containing a replacement of the RNA3 5′ UTR with the yeast GAL1 leader was cloned into pRP1415 cut with EcoRI/PflMI to produce the intermediate plasmid 917. Second, the SphI/MscI fragment of pRP1404 containing the MFA2 3′ UTR tagged with six MS2 binding sites was cloned into the SphI/MscI sites of plasmid 917.
All partial deletions in the RE region of RNA3 were directly derived from the partial deletions produced by Sullivan and Ahlquist (36). To introduce the six partial RE deletions into a version of RNA3 tagged with six MS2, PflMI/MscI fragments from the plasmids described by Sullivan and Ahlquist (36) that contained the partial RE deletions were introduced into plasmid pRP1405 cut with PflMI/StuI to produce plasmids pRP1408 to -1413.
pRP1416 expressing a β-globin mRNA tagged with six MS2 binding sites was cloned by first amplifying the MFA2 3′ UTR tagged with six MS2 binding sites with primers oAN209 and oAN202 and cloning the resulting PCR product cut with HindIII/SacI into the HindIII/SacII sites of pRS426 (10) to produce the intermediate plasmid 931. Second, the β-globin expression cassette was amplified from pMS99 (36) with primers oAN204 and oAN205, and the resulting PCR product was cut with XhoI/XhoI and cloned into the XhoI/XhoI sites of plasmid 931 to produce pRP1416. pRP1417, expressing a version of the tagged β-globin mRNA that also contains the BMV RE, was cloned by amplifying the β-globin-RE expression cassette from pMS100 (36) with primers oAN210 and oAN202, cutting the resulting PCR product with XhoI/XhoI, and cloning it into the XhoI/XhoI sites of plasmid 931 to produce pRP1417.
The MS2-tagged β-globin containing the six partial RE deletions described by Sullivan and Ahlquist (36) was made by amplifying the β-globin expression cassettes containing the partial RE deletions from pMS100 (36) with primers oAN210 and oAN202, cutting the resulting PCR product with XhoI/XhoI, and cloning it into the XhoI/XhoI sites of plasmid 931 to produce pRP1418-pRP1423.
Coimmunoprecipitation analysis.
Strains were grown in SC supplemented with appropriate amino acids and 2% Gal at 30°C to an optical density at 600 nm of 0.6. All subsequent steps were carried out at 4°C. The cultures were centrifuged at 3,000 rpm, washed once in cold medium, and lysed with glass beads in lysis buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 2 mM MgCl2, 1 mM β-mercaptoethanol, 1× Complete mini-EDTA-free pellet [Roche], 0.1% NP-40). Immunoprecipitation was carried out using a Pierce ProFound HA tag IP/Co-IP Kit and protocol. Briefly, 200 μl of freshly prepared lysate was added to 6 μl of beads. The remainder of the lysate was stored at −80°C. Samples were rocked end over end for 2 h. The samples were then washed three times in 1× Tris-buffered saline with 0.1% Triton. The immunoprecipitates were eluted by boiling samples in buffer for 5 min at 100°C.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis were performed using standard methods. Monoclonal anti-protein 2a (05H05B02) was used at 1:1,000 for RdRp (29). Monoclonal anti-hemagglutinin (HA) (12CA5; Roche) was used at 1:5,000. Blots were developed using an ECL Western Blotting Detection Kit (GE Healthcare).
RESULTS
BMV RNAs accumulate in P bodies.
The functions of Lsm1-7p, Dhh1p, and Pat1p in BMV replication could be explained in two ways. First, the lsm1Δ, dhh1Δ, or pat1Δ lesions may alter the expression of a number of host mRNAs and thereby indirectly affect BMV function. Alternatively, BMV RNAs might interact directly with these proteins and accumulate in P bodies, which could facilitate a step in replication complex assembly. To test this possibility, we examined the subcellular locations of BMV RNAs. To visualize the subcellular locations of BMV RNAs, we inserted multiple copies of the 23-nucleotide bacteriophage MS2 coat protein-binding site into a nonfunctional part of the 3′ UTR of RNA2 and RNA3. Insertion of the MS2 binding sites in the RNA2 or RNA3 3′ UTR did not affect the level of RNA2 or RNA3 (Fig. 1A), indicating that these insertions did not alter the stability of RNA2 or RNA3. The subcellular location of each RNA was then monitored by coexpressing the tagged RNA2 and RNA3 with MS2 fused to green fluorescent protein (GFP). Viral RNA1 was omitted from this study, as it is the least characterized of the BMV transcripts and encodes the 1a protein, which recruits viral RNAs to a replication complex on the ER, potentially interfering with analysis of subcellular localization data. The subgenomic viral RNA4 was also omitted from the study, as stability of the MS2-tagged RNA4 was severely decreased compared to the wild-type RNA4 (data not shown).
FIG. 1.
BMV RNA2 and RNA3 accumulate in P bodies, and viral RNA expression increases the size and number of P bodies. (A) Insertion of MS2 sequences into BMV RNA2 or RNA3 does not affect the steady-state abundance of viral RNA. SCR1, loading control. (B) Wild-type strain yRP2065 expressing BMV RNA2 (pRP1402) or RNA3 (pRP1403), MS2-GFP (pRP1094), and Lsm1p-cherry RFP (pRP1400), or no virus control were examined for the accumulation of RNA2 or RNA3 in P bodies. BMV RNA2 and RNA3 steady-state levels were not affected by the addition of MS2 sequences. (C) Strains containing Dhh1p (yRP2081), Pat1p (yRP2082), or Lsm1p (yRP2083) fused to GFP, which can all accumulate in P bodies (35), were analyzed with or without RNA2 or RNA3 expression for the degree of accumulation of these proteins in P bodies.
An important result was that both RNA2 and RNA3 were observed in distinct cytoplasmic foci (Fig. 1B). These foci revealed a subcellular concentration of RNA2 and RNA3, since in the absence of their expression, MS2-GFP was diffused throughout the cell (Fig. 1B). Moreover, RNA2 and RNA3 foci colocalize with RFP-tagged versions of the P-body proteins Lsm1p and Dcp2p (Fig. 1B and data not shown). This indicates that RNA2 and RNA3 accumulate in P bodies.
Additional evidence that RNA2 and RNA3 accumulated in P bodies came from the observation that when RNA2 or RNA3 was expressed, Lsm1p-GFP, Dhh1p-GFP, and Pat1p-GFP were found in larger and more numerous foci (Fig. 1C). This is consistent with previous observations that RNA2 and RNA3 are poorly translated mRNAs in yeast (29) and that significant expression of poorly translating or nontranslating RNAs increases the visible pool of P bodies (5, 38).
cis-acting sequences target BMV RNAs to P bodies and correlate with elements required for replication.
The accumulation of BMV RNAs in P bodies and the requirement for P-body components in the recruitment of viral RNA to the replication complex suggested that this localization of RNA2 or RNA3 might play some role in replication complex assembly. A prediction of this hypothesis is that sequences required for targeting the RNAs to P bodies should be among the cis-acting signals important in viral RNA replication. To identify cis-acting sequences within BMV transcripts that target them to P bodies, we analyzed the accumulation of mutated RNA3 transcripts in P bodies. We utilized RNA3 for this study, since earlier experiments had examined the requirement for different regions of RNA3 for recruitment of RNA3 to a replication complex (4, 15, 33, 36).
Replacing the RNA3 5′ or 3′ UTRs with UTRs from host mRNAs still allowed accumulation in P bodies (Fig. 2A, Δ3′UTR and Δ5′UTR). However, P-body accumulation was abolished by deleting an intergenic segment comprising a template RE that is located just 3′ of the first open reading frame and is known to be required for recruiting RNA3 from translation to replication (36) (Fig. 2A, ΔRE). The absence of RNA foci in the ΔRE construct was not due to loss of the RNA, since this RNA was still efficiently expressed (data not shown). These data identify the RE region as being required for the accumulation of RNA3 in P bodies.
FIG. 2.
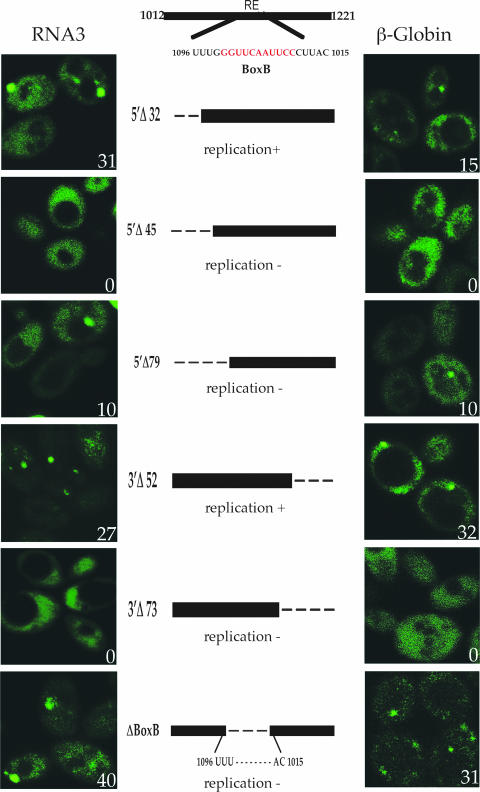
The cis-acting viral element RE is necessary and sufficient for targeting viral transcripts to P bodies. (A) Wild-type strain yRP2065 expressing full-length BMV RNA3 or various BMV RNA3 constructs lacking specific regions {from the top of the column to the bottom, pRP1403(RNA3-MS2), pRP1404 (Δ3′UTR/RNA3-MFA2), pRP1413 (Δ5′UTR/RNA3[GAL]), pRP1406 (RNA3 [GAL-RE-MFA2]), pRP1414 (ΔRE/RNA3[RE]), and MS2-GFP (pRP1094)}. (B) Wild-type strain yRP2065 expressing MS2-GFP (pRP1094) and the heterologous β-globin transcript with or without the RE {from the top of the column to the bottom, pRP1416 (β-globin[MFA2]) and pRP1415 (β-globin [RE-MFA2])}. The numbers located in the bottom right corners indicate the percentages of cells with foci out of 200 cells counted in three to five different experiments.
Previous results had shown that the RE was sufficient to target the heterologous β-globin mRNA for stabilization by viral protein 1a, which strongly correlates with RNA recruitment to the replication complex (33, 36). To test if the RE region was also sufficient for targeting heterologous RNAs to P bodies, we used MS2-GFP tagging to examine the subcellular location of a β-globin mRNA containing the 3′ UTR of MFA2 with MS2 sequences. We observed that the RE was sufficient to target the β-globin mRNA into P bodies (Fig. 2B). Thus, the RE is required for accumulation of RNA3 in P bodies and is sufficient to target a heterologous mRNA for accumulation in P bodies. Because the RE is also required for a step in the recruitment of RNA3 into replication complexes, these observations demonstrate a correlation between sequences required for recruitment of viral RNAs to replication and for accumulation of an RNA in a P body.
To examine this correlation further, we analyzed a series of deletions in the RNA3 intergenic region that were previously tested in both RNA3 and chimeric β-globin-RE transcripts to define the ∼190-nucleotide RE essential for recruitment of RNA3 to replication (4, 5, 36). We examined the effects of these deletions on the accumulation of both BMV RNA3 and β-globin mRNA in P bodies. We observed that the 5′Δ32 and 3′Δ52 deletions, which do not affect viral replication, still allowed RNA3 or β-globin-RE transcripts to accumulate in P bodies to an extent similar to that with the wild-type RNA (Fig. 3). In contrast, the 5′Δ45, 5′Δ79, and 3′Δ73 constructs, all of which prevent viral replication (36), showed no (5′Δ45 and 3′Δ73) or reduced (5′Δ79) P-body accumulation of RNA3 or β-globin-RE transcripts. Although these RNAs were absent from P bodies, they were still efficiently expressed in yeast (data not shown). Finally, we observed that RNA3 and β-globin-RE transcripts with a precise deletion of box B (ΔBoxB) still accumulated in P bodies (Fig. 3). Box B is an RE subsequence that is required for RNA recruitment into replication and duplicates exactly the invariant sequence and base pairing of tRNA TψC stem loops (4, 36). Taken together, these observations demonstrate that for five out of six deletion constructs there was a correlation between lesions that reduced or eliminated BMV RNA3 and β-globin mRNA accumulation in P bodies, and RNA recruitment to replication complexes. These observations suggest that accumulation of BMV RNAs in P bodies is important for their ability to form a replication complex. However, because the ΔBoxB RNA still accumulated in P bodies (Fig. 3) but was not recruited to replication complexes (36), these results also suggest that accumulation in a P body is not sufficient for recruitment into replication complexes (see Discussion).
FIG. 3.
Mutations in the RE that affect RNA replication prevent viral-transcript accumulation in P bodies. Wild-type strain yRP2065 expressing MS2-GFP (pRP1094) and either mutant forms of the RE in RNA3 [left column, pRP1408 (RNA3 Δ32-5′), pRP1409 (RNA3 Δ45-5′), pRP1410 (RNA3 Δ79-5′), pRP1411 (RNA3 Δ52-3′), pRP1412 (RNA3 Δ73-3′), pRP1407 (RNA3 bοx Β)] or β-globin [right column, pRP1418 (β-globin Δ32-5′), pRP1419 (β-globin Δ45-5′), pRP1420 (β-globin Δ79-5′), pRP1421(β-globin Δ52-3′), pRP1422 (β-globin Δ73-3′), pRP1417 (β-globin bοx Β)]. The numbers located in the bottom right corners indicate the percentages of cells with foci out of 200 cells counted in three to five different experiments.
The RdRp interacts with Lsm1p.
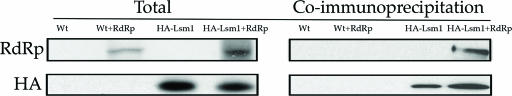
If P bodies play a role in the formation of viral replication complexes, then viral proteins involved in replication might be expected to interact with P-body components and/or accumulate in P bodies. To test this possibility, we first examined the interactions of the viral RdRp with the P-body component Lsm1p by coimmunoprecipitation. To do this experiment, we expressed Lsm1p tagged with the HA epitope at its carboxy terminus in strains also expressing the BMV RdRp. We then immunoprecipitated it with antisera against the HA epitope and probed the immunoprecipitates for RdRp using monoclonal anitsera specific for RdRp (40). We observed that the viral RdRp coimmunoprecipitated with Lsm1p (Fig. 4). In contrast, in strains expressing the RdRp without the HA-Lsm1p construct, no RdRp was observed in the HA immunoprecipitate (Fig. 4, lane Wt+RdRp). These observations demonstrate that Lsm1p and the BMV RdRp can physically interact in vivo, although whether the interaction is direct or requires additional components cannot be determined at this time.
FIG. 4.
Viral RdRp interacts with Lsm1p. Coimmunoprecipitation of RdRp (pRP1402) with HA-tagged Lsm1p (strain yRP1430). Epitope-tagged Lsm1p was immunoprecipitated with anti-HA antibody (12CA5 Roche), and the immunoprecipitate was analyzed by Western blotting for RdRp (29). Wt, wild type.
The interaction of Lsm1p and RdRp raised the possibility that the RdRp would accumulate in P bodies. To test this possibility, we created an RdRp-cherry RFP fusion protein and examined its subcellular distribution in cells expressing an Lsm1-GFP fusion protein. Similar to earlier reports (9), we observed that the RdRp was concentrated in cytoplasmic foci (Fig. 5). Moreover, the RdRp foci were often near or colocalized with P bodies (Fig. 5). However, since in some cases RdRp foci were distinct from Lsm1-GFP foci, this suggests either that RdRp can be found in other subcellular compartments or that there is heterogeneity in P-body composition, which has previously been described (3, 14). Nevertheless, the coimmunoprecipitation and partial colocalization of Lsm1p and RdRp in yeast cells indicate that RdRp interacts with P-body components and can accumulate in these structures.
FIG. 5.
Viral RdRp colocalizes with P bodies. Lsm1p-GFP (strain yRP2083) or Pat1p-GFP (strain yRP2082) expressing RdRp fused to cherry RFP (pRP1399) was analyzed by fluorescence microscopy.
DISCUSSION
P bodies facilitate the assembly of BMV replication complexes.
Previous work had shown that the Lsm1-7p complex, Pat1p, and Dhh1p, which are all components of P bodies, affect BMV genomic-RNA translation and the subsequent recruitment of genomic RNAs from translation to RNA replication (15, 26, 29). In this work, we presented multiple lines of evidence arguing that these proteins affect BMV replication, because accumulation of viral RNAs in P bodies is a significant step in the replication process. First, the replication substrates RNA2 and RNA3 accumulate in P bodies (Fig. 1). Second, there is a strong correlation between cis-acting elements required for RNA recruitment from translation to replication (22, 36) and for targeting RNAs to P bodies, with the only exception being the precise box B deletion (ΔBoxB) (Fig. 2 and 3). Third, the viral RdRp and the P-body component Lsm1p coimmunoprecipitate and show overlapping subcellular distributions (Fig. 4 and 5). These observations argue that accumulation of BMV RNAs in P bodies is an important step in the formation of replication complexes.
Previous work demonstrated that the actual BMV replication complexes are formed in invaginations of the ER (27). An unresolved issue is how these membrane-associated replication complexes are related to P bodies. Strikingly, at least a subpopulation of P bodies is associated with ER membranes in Drosophila oocytes (42), and multiple P-body components in yeast biochemically cofractionate with membranes (data not shown). These results suggest a possible sequential model for the formation of a BMV replication complex in which BMV genomic RNAs would first accumulate within ER-associated P bodies, along with viral replication proteins, followed by the formation of a replication complex facilitated by interactions between P-body components and membrane proteins.
BMV normally infects plant cells, and our analysis of BMV replication took place in yeast cells. However, plant cells contain P bodies with composition and function similar to those in yeast cells (17, 43). This suggests that P bodies are likely to be functionally significant in the replication of BMV in plant cells.
Our results also argue that accumulation of RNAs in P bodies is not sufficient for assembly into a replication complex. The critical observation is that RNAs lacking box B are still able to accumulate in P bodies (Fig. 3) but are unable to enter replication complexes (36). This suggests that, following accumulation of RNAs in P bodies, which would also contain host mRNAs, additional interactions with box B sequences would provide another level of specificity allowing only viral RNAs within P bodies to be selected for replication. One interesting possibility here is that the 1a protein, which is associated with ER membranes and has a DEAD box RNA helicase domain (40), might have direct interactions with box B and RdRp, thereby recruiting both the RNA and the RdRp to an ER-associated replication complex.
The use of P bodies to facilitate the assembly of RNA replication complexes could be mechanistically important, since it allows viruses to take advantage of a dynamic compartment of nontranslating mRNAs concentrated in discrete foci away from the translation machinery. This could allow replication complex assembly to be initiated without interference by elongating ribosomes. Indeed, the concentration of mRNAs and the absence of elongating ribosomes in P bodies make P bodies useful sites for other important transitions in viral life cycles. Consistent with that view, the Ty3 retrovirus-like element in yeast has been suggested to package virus-like particles in association with P bodies (5), and the antiviral proteins APOBEC3G and APOBEC3F, as well as the human immunodeficiency virus type 1 viral protein Vif, localize to P bodies in mammalian cells (41). Moreover, replication of hepatitis C virus is enhanced by the liver-specific microRNA, miR122 (23). In other studies, microRNAs have been shown to target mRNAs to P bodies in mammalian cells (25, 32). This suggests that targeting hepatitis C virus RNAs to P bodies may be important for replication. Taken together, these observations suggest that P bodies are critical sites for many viruses at critical transitions in their life cycles.
Acknowledgments
We thank members of the Parker laboratory for comments on the manuscript. We also thank the Molecular and Cellular Biology Image Processing Facility.
This work was supported by NIH grant GM45443 to R.P. and GM35072 to P.A. R.P. and P.A. are investigators of the Howard Hughes Medical Institute. H.R.L. and A.N. were supported by the WVU Research Corporation and WV Agricultural Forestry and Experiment Station.
Footnotes
Published ahead of print on 3 July 2007.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbee, S. A., P. S. Estes, A. M. Cziko, J. Hillebrand, R. A. Luedeman, J. M. Coller, N. Johnson, I. C. Howlett, C. Geng, R. Ueda, A. H. Brand, S. F. Newbury, J. E. Wilhelm, R. B. Levine, A. Nakamura, R. Parker, and M. Ramaswami. 2006. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52:997-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumstark, T., and P. Ahlquist. 2001. The brome mosaic virus RNA3 intergenic replication enhancer folds to mimic a tRNA TpsiC-stem loop and is modified in vivo. RNA 7:1652-1670. [PMC free article] [PubMed] [Google Scholar]
- 5.Beliakova-Bethell, N., C. Beckham, T. Giddings, M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertrand, E., P. Chartrand, M. Schaefer, S. Shenoy, R. Singer, and R. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2:437-445. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharyya, S. N., R. Habermacher, U. Martine, E. Closs, and W. Filipowicz. 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111-1124. [DOI] [PubMed] [Google Scholar]
- 8.Brengues, M., D. Teixeira, and R. Parker. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, J., A. O. Noueiry, and P. Ahlquist. 2001. Brome mosaic virus protein 1a recruits viral RNA2 to RNA replication through a 5′-proximal RNA2 signal. J. Virol. 75:3207-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christianson, T., R. Sikorski, M. Dante, J. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 11.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861-890. [DOI] [PubMed] [Google Scholar]
- 12.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decker, C. J., and R. Parker. 2006. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J. Cell Biol. 173:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diez, J., M. Ishikawa, M. Kaido, and P. Ahlquist. 2000. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 97:3913-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell. Biol. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 17.Goeres, D., J. Van Norman, W. Zhang, N. Fauver, M. Spencer, and L. Sieburth. 2007. Components of the Arabidopsis mRNA decapping complex are required for early seedling development. Plant Cell doi: 10.1105/tpc.106.0. 047621. [DOI] [PMC free article] [PubMed]
- 18.Holetz, F., A. Correa, A. Avila, C. Nakamura, M. Krieger, and S. Goldenberg. 2007. Evidence of P-body-like structures in Trypanosoma cruzi. Biochem. Biophys. Res. Commun. 356:1062-1067. [DOI] [PubMed] [Google Scholar]
- 19.Huh, W., J. Falvo, L. Gerke, A. Carroll, R. Howson, J. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 20.Ishikawa, M., M. Janda, and P. Ahlquist. 2000. The 3a cell-to-cell movement gene is dispensable for cell-to-cell transmission of brome mosaic virus RNA replicons in yeast but retained over 1045-fold amplification. J. Gen. Virol. 81:2307-2311. [DOI] [PubMed] [Google Scholar]
- 21.Janda, M., and P. Ahlquist. 1993. RNA-dependent replication, transcription, and persistence of brome mosaic virus RNA replicons in S. cerevisiae. Cell 72:961-970. [DOI] [PubMed] [Google Scholar]
- 22.Janda, M., and P. Ahlquist. 1998. Brome mosaic virus RNA replication protein 1a dramatically increases in vivo stability but not translation of viral genomic RNA3. Proc. Natl. Acad. Sci. USA 95:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jopling, C., M. Yi, A. Lancaster, S. Lemon, and P. Sarnow. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577-1581. [DOI] [PubMed] [Google Scholar]
- 24.Kushner, D. B., B. D. Lindenbach, V. Z. Grdzelishvilli, A. O. Noueiry, S. M. Paul, and P. Ahlquist. 2003. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA 100:15764-15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., M. Valencia-Sanchez, G. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mas, A., I. Alves-Rodrigues, A. Noueiry, P. Ahlquist, and J. Diez. 2006. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 80:246-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Noueiry, A. O., and P. Ahlquist. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77-98. [DOI] [PubMed] [Google Scholar]
- 28.Noueiry, A. O., J. Chen, and P. Ahlquist. 2000. A mutant allele of essential, general translation initiation factor DED1 selectively inhibits translation of a viral mRNA. Proc. Natl. Acad. Sci. USA 97:12985-12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noueiry, A. O., J. Diez, S. Falk, J. Chen, and P. Ahlquist. 2003. Yeast Lsm1p-7p/Pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell. Biol. 23:4094-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635-646. [DOI] [PubMed] [Google Scholar]
- 31.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 32.Pillai, R. 2005. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwartz M., J. Chen, M. Janda, M. Sullivan, J. den Boon, and P. Ahlquist. 2002. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9:505-514. [DOI] [PubMed] [Google Scholar]
- 34.Shaner, N., R. Campbell, P. Steinbach, B. Giepamans, A. Palmer, and R. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567-1572. [DOI] [PubMed] [Google Scholar]
- 35.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, M. L., and P. Ahlquist. 1999. A brome mosaic virus intergenic RNA3 replication signal functions with viral replication protein 1a to dramatically stabilize RNA in vivo. J. Virol. 73:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teixeira, D., and R. Parker. 2007. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 18:2274-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teixeira, D., U. Sheth, M. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tharun, S., W. He, A. Mayes, P. Lennertz, J. Beggs, and R. Parker. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404:515-518. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., W. Lee, T. Watanabe, M. Schwartz, M. Janda, and P. Ahlquist. 2005. Brome mosaic virus 1a nucleoside triphosphatase/helicase domain plays crucial roles in recruiting RNA replication templates. J. Virol. 79:13747-13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wichrolski, M. J., G. Robb, and T. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilhelm, J., M. Busczak, and S. Sayles. 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev. Cell. 9:675-685. [DOI] [PubMed] [Google Scholar]
- 43.Xu, J., J. Yang, Q. Niu, and N. Chua. 2006. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 18:3386-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]