Abstract
Human cytomegalovirus (HCMV) infections are prevalent in human populations and can cause serious diseases, especially in those with compromised or immature immune systems. The HCMV genome of 230 kb is among the largest of the herpesvirus genomes. Although the entire sequence of the laboratory-adapted AD169 strain of HCMV has been available for 18 years, the precise number of viral genes is still in question. We undertook an analysis of the HCMV transcriptome as an approach to enumerate and analyze the gene products of HCMV. Transcripts of HCMV-infected fibroblasts were isolated at different times after infection and used to generate cDNA libraries representing different temporal classes of viral genes. cDNA clones harboring viral sequences were selected and subjected to sequence analysis. Of the 604 clones analyzed, 45% were derived from genomic regions predicted to be noncoding. Additionally, at least 55% of the cDNA clones in this study were completely or partially antisense to known or predicted HCMV genes. The remarkable accumulation of antisense transcripts during infection suggests that currently available genomic maps based on open-reading-frame and other in silico analyses may drastically underestimate the true complexity of viral gene products. These findings also raise the possibility that aspects of both the HCMV life cycle and genome organization are influenced by antisense transcription. Correspondingly, virus-derived noncoding and antisense transcripts may shed light on HCMV pathogenesis and may represent a new class of targets for antiviral therapies.
Human cytomegalovirus (HCMV) is classified within the Betaherpesvirinae subfamily, and most humans are infected with HCMV during their lifetime. Like other herpesviruses, HCMV cannot be completely eliminated by the immune system and remains either as a low-level persistent infection or in a quiescent latent state for the lifetime of the infected person. Although infections in immunocompetent adults are usually benign, considerable morbidity is seen in congenitally infected infants and in persons with compromised immunity (reviewed in reference 63). HCMV infections are especially important in transplant patients, who may suffer graft loss, vascular disease, and other manifestations including hepatitis and pneumonitis (reviewed in 37, 63). An increasing number of studies have also raised the possibility that HCMV infections may be linked to some chronic diseases, including atherosclerosis, autoimmune diseases, and cancer (reviewed in reference 74). Unfortunately, no effective vaccine is currently available to prevent or ameliorate HCMV infections.
Progress in developing effective antiviral drugs and vaccine candidates will likely rely upon detailed knowledge of viral gene products and how they function in pathogenetic processes. For this reason, there is an intense interest in defining the gene products of HCMV and other herpesviruses. The HCMV genome of 230 kb is among the largest of the herpesvirus genomes. The genome is comprised of two unique regions, known as the unique long (UL) and unique short (US) regions, that are bounded by terminal (TRL or TRS) and internal (IRL and IRS) repeat regions. Although the entire sequence of the laboratory-adapted AD169 strain of HCMV was first available in 1989 (22), the precise number and nature of viral genes and gene products are still in question. After sequencing the HCMV genome, Chee and colleagues predicted approximately 200 open reading frames (ORFs) capable of coding for proteins (5, 22). It was subsequently discovered that the AD169 laboratory strain harbored a deletion of approximately 15 kb relative to clinical isolates. This region was predicted to encode 19 additional ORFs, suggesting that the HCMV genome encoded up to 220 genes (18). More recently, comparison of the HCMV genome with the chimpanzee cytomegalovirus genome led to a revised estimate for the protein-coding genes of AD169 to 145 (27). Likewise, application of an in silico approach based on the Bio-Dictionary gene finder algorithm to define the coding potential of HCMV supported elimination of 37 previously annotated ORFs (61). However, comparison of the AD169 genomic sequence to those of clinical isolates and use of proteomic experimental approaches predict that additional unannotated ORFs exist (62, 83).
Most studies of herpesvirus genomes have focused on protein-coding potentials of these viruses. However, rapid advances in understanding of the role of noncoding gene products and antisense (AS) transcripts are dramatically changing the paradigms applied to gene definitions, gene regulation, and gene functions (reviewed in references 51, 56, 60, and 93). In particular, the application of bioinformatics approaches to analyze expressed sequence databases has revealed that AS transcription is widespread in human and other genomes (reviewed in references 51 and 60).
Sense-antisense (S-AS) transcript pairs have been found in genomes from archaebacteria to humans (58, 79, 94), and a subset of S-AS pairs is recognized to be conserved across species (94). While some pioneering studies have estimated that between 1 and 15% of human or mouse genes were influenced by S-AS pairs (29, 47, 52, 73, 92), more recent estimates of up to 20 to 26% of human genes (24, 94) and 72% of mouse genes (45) suggest that AS-mediated gene regulation may be much more common than previously appreciated. Natural AS transcripts (NATs) are classified as cis or trans in nature. cis NATs are transcribed from opposite strands of the same genomic locus and are predicted to have longer and more perfect complementary sequences for S transcripts than trans NATs derived from separate loci. Regulatory functions of NATs are predicted to derive from double-stranded RNA-dependent and -independent mechanisms, including RNA editing, RNA interference, chromatin remodeling, transcriptional interference, and masking of RNA elements involved in splicing, localization, transport, and translation of RNAs (reviewed in references 51 and 60).
In this study, we examined the transcriptional products of HCMV during lytic infection of fibroblasts. Remarkably, of the 604 HCMV cDNA clones analyzed in this study, at least 45% were derived from genomic regions predicted to be noncoding. Of similar interest was our finding that 55% of the cDNA clones in this study were completely or partially AS to known or predicted HCMV genes. Moreover, cis NAT pairs were identified or predicted for 56 of the 191 genes currently annotated at the Los Alamos National Laboratory Sexually Transmitted Diseases Sequences Database (STD database) (now at the Oral Pathogens Sequences Database). We conclude that genomic maps based on ORF analyses and other in silico analyses may drastically underestimate the true complexity of viral gene products. In addition, the abundance of AS transcription in the HCMV viral genome raises the distinct possibility that AS-dependent gene regulatory mechanisms influence both viral gene expression and gene organization. These noncoding and AS transcripts may offer new insights into HCMV pathogenesis and may serve as novel targets for developing intervention strategies and treatments for HCMV-related diseases.
MATERIALS AND METHODS
Cells and virus.
Human foreskin fibroblasts (HFFs), telomerase-immortalized HFF cells (HFF-TEL, a kind gift of Tom Shenk [10]), MRC-5 human fibroblasts, and human umbilical vein endothelial cells (HUVECs) were used in this study. HFF and HFF-TEL cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, while MRC-5 cells were maintained in modified Eagle's medium supplemented with 10% fetal calf serum, 1.7 mM sodium bicarbonate, 1.4 mM sodium chloride, essential and nonessential amino acids, vitamins, and sodium pyruvate at manufacturer-recommended concentrations (Sigma-Aldrich, InVitrogen). All three cell lines were used between passages 2 and 10. HUVECs were isolated from vessels as previously described (71) and propagated in endothelial cell growth medium consisting of M-199 (GibcoBRL) supplemented with 20% fetal bovine serum (HyClone), 22.5 μg/ml bovine brain extract (BioWhittaker, Inc., Walkersville, MD), 12 U/ml sodium heparin (Sigma, St. Louis, MO), and 20 mM HEPES buffer. All growth surfaces for HUVECs were pretreated with human fibronectin (25 μg/ml; Upstate Biotechnology). Cells were passed weekly by brief trypsin digestion at a ratio of 1:4 and used in experiments at passages 5 to 7.
HCMV strain AD169 was obtained from ATCC and was propagated and titrated on MRC-5 cells by plaque assay (87). Cytomegalovirus strain VHL/E, originally isolated from duodenal biopsy material from a bone marrow transplant recipient (86), was propagated in HUVEC as detailed elsewhere (85) to preserve its natural endothelial cytopathogenicity.
Extraction of HCMV genomic DNA.
HCMV genomic DNA was extracted as described previously (81). Briefly, confluent HFF-TEL cells in two 175-cm2 flasks were exposed to 1 PFU per cell of AD169 or Towne strains. The cells were harvested at 72 h postinfection and collected by centrifugation. The cell pellets were resuspended in 5 ml of 150 mM NaCl, 10 mM Tris (pH 7.4), and 1.5 mM MgCl2. After incubation on ice, NP-40 was added to a final concentration of 0.1%. The lysate was centrifuged at 3,700 rpm for 20 min using a Beckman GS-6R centrifuge. The supernatant was collected and brought to a final concentration of 0.2% sodium dodecyl sulfate (SDS), 0.5 mM EDTA, and 50 mM β-mercaptoethanol. After incubation on ice and extraction with phenol-chloroform, the genomic DNA was precipitated with ethanol, resuspended in 1 ml of Tris-EDTA buffer, and treated with RNase (Sigma-Aldrich). The genomic DNA was further purified by centrifugation in a linear 5 to 20% (wt/vol) potassium acetate gradient at 40,000 rpm for 3.5 h at 20°C in a Beckman L7 Ultracentrifuge SW60 rotor. Following centrifugation, the DNA was collected, precipitated with ethanol, and resuspended in 50 μl distilled water. The purified genomic DNA was digested with MseI, followed by phenol-chloroform extraction and ethanol precipitation. The digested genomic DNA was finally resuspended in 50 μl sterile water.
Construction of HCMV AD169 cDNA libraries.
RNA was extracted from infected HFF cells cultured in 175-cm2 flasks under conditions that selected for immediate-early (IE), early (E), or late (L) viral transcripts. For all conditions, cells were exposed to 20 PFU per cell of the AD169 strain of HCMV. To select for IE transcripts, cells were treated with 100 μg/ml of cyclohexamide (Sigma-Aldrich) for 1 h prior to infection and throughout the 24-h infection period, when cells were harvested for RNA isolation. To select for E transcripts, 100 μM ganciclovir (Roche Pharma) was added to the medium after the infection period, and cells were harvested 72 h later. To select for L viral transcripts, untreated infected cells were harvested 72 h after infection. Prior to isolation of total RNA, cells from a small section of the flasks were scraped and collected. Cells were disrupted in SDS-polyacrylamide gel electrophoresis sample buffer (25 mM Tris-Cl, pH 6.8, 2.5% β-mercaptoethanol, 5% glycerol, and 0.5% SDS), boiled, and sonicated. Proteins were separated by denaturing gel electrophoresis and transferred to nitrocellulose (Amersham Bioscience). Efficacies of drug treatments were verified by immunoblot analyses for IE UL122/123 products, IE1/2 (Rumbaugh Goodwin no. 1203), the early UL55 product, glycoprotein B (gB) (Rumbaugh Goodwin no. 1201), and the L UL99 product, pp28 (Rumbaugh Goodwin no. 1207) (data not shown).
Unless otherwise stated, all extractions of total cellular RNA were performed using the TRIZOL reagent (Invitrogen), following the instructions of the manufacturer. Polyadenylated mRNA was isolated using an Oligotex kit (QIAGEN) according to the manufacturer's instruction. cDNA libraries were constructed using two cloning vectors, pAcCMV (96), derived from the pAcSG2 Baculovirus transfer vector (BD Biosciences), and pcDNA3.1(+) (Invitrogen). These vectors were modified by introducing recognition sites for two restriction enzymes, PacI and PmeI, that are absent in the AD169 genome. Specifically, sequences recognized by the PacI and PmeI enzymes were inserted between the StuI and KpnI sites of vector pAcCMV using the following oligonucleotides: 5′ CCTGTTTAAACCTAGGCGGCCGCTTAATTAAGGTAC and 5′ CTTAATTAAGCGGCCGCCTAGGTTTAAACAGG. The PmeI site at position 1007 in pcDNA3.1(+) was replaced with sequences specifying a PacI cutting site using a site-directed mutagenesis kit (Stratagene) and the following oligonucleotides: 5′ CTAGAGGGCCCGTTTAATTAAGCTGATCAGCCTCGACTG and 5′CAGTCGAGGCTGATCAGCTTAATTAAACGGGCCCTCTAG.
cDNA libraries were constructed by following the instruction manual for the SuperScript Plasmid System with Gateway Technology for cDNA Synthesis and Cloning (Invitrogen) with some minor modifications. Briefly, a poly(T)-tailed PacI primer-adapter was used for first-strand cDNA synthesis (5′GCGGCCGCTTAATTAACC(T)15). After second-strand synthesis, an EcoRI-PmeI adapter was added to the 5′ end of the cDNA that was generated from the following oligonucleotides: 5′AATTCAGGCCTGTTTAAACG and CGTTTAAACAGGCCTG. cDNA fragments were used in ligation reactions with modified pAcCMV or pcDNA3.1(+) vectors previously digested with the EcoRI and PacI restriction enzymes. Recombinant plasmids harboring cDNA sequences were transformed into XL1-Blue Supercompetent Escherichia coli cells (Stratagene).
cDNA library screening by colony hybridization.
Random transformed bacterial colonies were picked individually and transferred onto agarose grid plates (approximately 100 clones on each plate). Colonies on grid plates were transferred to Hybond-N+ nylon membranes (82 mm in diameter; Amersham Bioscience) and processed for hybridization as described by Hirsch (40). MseI-digested HCMV genomic DNA was labeled using the DIG High Prime DNA Labeling Detection Starter Kit II (Roche Applied Science). Probes were incubated with the membranes according to the manufacturer's instruction.
DNA sequencing and sequence analysis.
Bacterial colonies harboring HCMV-derived cDNA sequences were inoculated into 4 ml LB broth supplemented with 50 μg/ml ampicillin. Plasmid DNA was purified from overnight culture using QIAprep Spin Miniprep kits (QIAGEN). cDNA inserts were sequenced from the 5′ end using the T7 primer for pcDNA3.1(+) and a pAcCMV-specific primer (5′GGAGACGCCATCCACGCTGTTTTGACC) at the OSU Plant-Microbe Genomics Facility. In total, 870 clones were submitted for sequence analysis from the 5′ ends. Sequences were compared to the AD169 genome (GenBank accession no. NC_001347) using mega BLAST (95). Matched AD169 gene sequences were downloaded and aligned to corresponding cDNA clone sequences manually using BioEdit software (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) (39). Selected clones were subjected to a second round of sequence analysis using primers specific for the 3′ ends of the cDNA inserts for the pcDNA3.1(+) (5′GCACCTTCCAGGGTCAAGGAAG) and pAcCMV (5′GAGGTGCGTCTGGTGCAAAC) vectors. These primers failed to generate sequence from a subset of clones, presumably because of difficulties in reading through poly(A) tracts. Therefore, in some cases, 3′ ends were sequenced using standard poly(T) primers. Selected cDNA inserts were also sequenced using specifically designed internal primers. AD169 genomic positions (accession no. NC_001347) corresponding to the sequences of the cDNA clones were determined using the SPIDEY program for cDNA to genomic alignments (http://www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey/).
RT-PCR for detection of AS transcripts.
Confluent MRC-5 cells in six-well plastic tissue culture plates were exposed to 2 PFU per cell of the AD169 strain of HCMV. Cells were harvested at 24, 48, and 72 h after infection. Also, confluent HUVEC monolayers in six-well plastic tissue culture plates were inoculated with sonicated cell lysates containing VHL/E of HCMV (1 PFU/cell). To enhance infection efficiency of endothelial cells, plates were centrifuged at 300 × g for 30 min at room temperature. Cells were then incubated for an additional 30 min before removal of inoculum, followed by two washes with phosphate-buffered saline and addition of fresh medium. Cells were harvested at 24, 48, 72, 96, and 120 h after infection. Total RNA was isolated and treated with DNase I (Invitrogen). The reverse transcription-PCR (RT-PCR) analysis was performed using a ThermoScript kit (Invitrogen), following the manufacturer's instructions. cDNA synthesis was performed in the first step using total RNA and gene-specific primers at 55°C for 35 min (Table 1, reverse primers). In the second step, PCR was performed using primers specific for the gene of interest. A complete list of PCR primers is given in Table 1. Reactions were carried out at 94°C for 2 min, 30 cycles of 94°C for 30 s, 59°C for 45 s, and 72°C for 50 s, and final extension at 72°C for 10 min. RT-PCR products were analyzed by agarose gel (1% [wt/vol]) electrophoresis. No reverse transcriptase and no template controls were run in parallel.
TABLE 1.
Primers for RT-PCR and 5′ and 3′ RACE
| Primer name | Sequence (5′-3′) | Genomic positiona |
|---|---|---|
| AS-UL36-37-F | GGTGCCGCATTCCTGCTCCAGA | 49677-49698 |
| AS-UL36-37-R | CCTGCGTGTCTTGAACGAGAGTCG | 50566-50543 |
| AS-US30-31-F | GCTCCAAGAGCGACATGAGATCG | 223730-223708 |
| AS-US30-31-R | GGCGACGCCAAGTACGGTATTCGT | 223019-223042 |
| AS-UL61-62-F | CGAGGTTGGGTGTGGCCGGAA | 94987-95007 |
| AS-UL61-62-R | CGCTCCTAGGCTCTCGACGCCAT | 95612-95590 |
| AS-UL67-F | CCGTCACAGCCCTATGACGCGA | 97937-97958 |
| AS-UL67-R | GGTCGTGGTGACGATGATGATGT | 98720-98698 |
| AS-IGS-RL2-3-F | CCTCGCACTTCTTCGCTCTCA | 2939-2919-or 187459-187479 |
| AS-IGS-RL2-3-R | GGAGTGAAGAGGGTGAAGGCAA | 2140-2161 or 188258-188237 |
| AS-TRL8-9-F | CCGTTTCTCCTCAGCTGCCGT | 7791-7771-or 182607-182627 |
| AS-TRL8-9-R | CCACAGAGGGAGACGGGACGAAG | 7159-7181 or 183239-183217 |
| AS-UL27-28-F | GCTGCTGGGTTGGCAGTGGA | 34614-34633 |
| AS-UL27-28-R | GATGAGATCGTAGAGGGCGCGTG | 35289-35267 |
| AS-UL70-F | GCTGACGTGCGGTTCGTAGTGT | 101764-101785 |
| AS-UL70-R | CGAGCGCGACACAGAGGAGA | 102523-102506 |
| AS-UL87-88-F | GCTCGCGCACCGTACCGTCGT | 132216-132196 |
| AS-UL87-88-R | CACGGTCGAGAGAAGCGGCTGCA | 131608-131630 |
| AS-UL102-F | CGTGACTCGTTAAGCGTTGAGCC | 150083-150061 |
| AS-UL102-R | GGAGTGGTCATAACGGCGGTAGTG | 149395-149418 |
| AS-TRL3-F | CCAAGCCAGATTTCACGTCCATGAG | 3567-3543 or 186831-186855 |
| AS-TRL3-R | GCTTTGAGAGCGAAGAAGTGCGAG | 2915-2938 or 187483-187460 |
| AS-UL23-F | CGTTTTCGTGGCAGCGTGTCA | 27954-27974 |
| AS-UL23-R | GAAGGACGGGATGCACAGCGT | 28566-28546 |
| AS-UL24-F | CGTTGTGGCCGCTGAAACGTC | 28985-29005 |
| AS-UL24-R | CGTGTTGTCGCCGGATCAGCA | 29601-29581 |
| AS-UL25-F | CGAACGGCTCTGGTTGTTGACC | 30979-30958 |
| AS-UL25-R | GGTGGCGACGATAACAGCAGCAG | 30467-30489 |
| AS-UL47-F | CGTAGGGCATGAGCGTGCAG | 61984-61965 |
| AS-UL47-R | CGGAGGCGTTAAATACCGTGAGC | 61388-61410 |
| AS-UL88-F | GCTTTCCGGCCAACATGCCCAG | 133032-133011 |
| AS-UL88-R | GGTGGTGGAATGCGGTCAGTTG | 132425-132446 |
| gB E1 | TCCAACACCCACAGTACCCGT | 83769-83749 |
| gB E2 | CGGAAACGATGGTGTAGTTCG | 83502-83522 |
| GAPDH-F | CCATGGGGAAGGTGAAGGTCGGAGC | |
| GAPDH-R | GGTGGTGCAGGCATTGCTGATG | |
| pL8212-5RACEsp1 | GCTCTAGACCTTATCCCCATCGTCGTG | 29138-29119 |
| pL8212-5RACEsp2 | GATCTAGACGACCGCTACATCCGGGTG | 29081-29061 |
| pL5312-5RACEsp1 | GCTCTAGATGAGGCACACAGCGGTCTT C | 29555-29577 |
| pL5312-5RACEsp2 | CGTTCACGAAGCGACCCACGAAG | 29606-29628 |
| PL222-5RACEsp1 | GATCTAGACTTCGTCCTCTTCGGGCAC G | 149904-149924 |
| PL222-5RACEsp2 | CGTCTAGAACGCTTAACGGTCACG | 150066-150083 |
| pE104114-5RACEsp1 | GATCTAGAGGGTCCGGGCTTTATGCG | 49426-49407 |
| pE104114-5RACEsp2 | GCTCTAGAAGGACTTTCTGCGGAACG | 49316-49297 |
| pE104114-3RACEsp1 | CCGAATTCCCGTAGGTAAAAACCCAC | 50904-50924 |
Nucleotide positions correspond to the AD169 genome sequence under GenBank accession no. NC_001347.
Northern blots.
Confluent MRC-5 cells in six-well plastic tissue culture plates were exposed to 2 PFU per cell of the AD169 strain of HCMV. Cells were harvested at 24, 48, and 72 h after infection. Total RNA was isolated and subjected to denaturing agarose gel (1% [wt/vol]) electrophoresis in the presence of formaldehyde. Nucleic acids were transferred to positively charged nylon membranes (Millipore) using a Posi-Blot 30-30 pressure blotter (Stratagene). All probes were labeled using a DIG Northern starter kit (Roche Applied Science) according to the manufacturer's instructions.
Plasmids pE10422 and pE104114, harboring cDNAs with gene sequences in S and AS orientation, were used to generate probes complementary to AS and S transcripts, respectively, of UL36 (see Table S1 in the supplemental material for a description of cDNA clones). Plasmids pE10422 and pE104114 were linearized with BssHII and NdeI, respectively. RNA probes were generated using T7 polymerase. The AS probe corresponds to nucleotides 49791 to 49577 and 49473 to 48961, while the S probe corresponds to nucleotides 49135 to 50065, of the AD169 genome. Plasmids pL5312 and pL8212, harboring cDNAs with gene sequences in S and AS orientation, were used to generate probes complementary to AS and S transcripts, respectively, of UL24. Plasmids pL5312 and pL8212 were linearized with SalI and NcoI, respectively. RNA probes were generated using T7 polymerase. The AS probe corresponds to nucleotides 29806 to 29136, while the S probe corresponds to nucleotides 28949 to 29362, of the AD169 genome.
Plasmid pL222 carrying the AS UL102 sequence was digested with EcoRI and XbaI (corresponding to nucleotides 150184 to 149378 of AD169), and this fragment was inserted into pBluescript II KS+ (Stratagene). The plasmid was linearized with EcoRI to generate a probe complementary to the AS transcripts of UL102 using the T7 promoter or linearized with XbaI to generate a probe complementary to the S transcripts using the T3 promoter.
Plasmid pE1033, carrying the AS RL5 and S RL4 sequences, was digested with BamHI and SalI (corresponding to nucleotides 4555 to 3958 or 185843 to 186440 of AD169) and inserted into pBluescript II KS+. This plasmid was linearized with BamHI to generate a probe complementary to the AS RL4 transcripts using the T3 promoter. Because the S-specific probe generated from this entire fragment exhibited nonspecific binding, this plasmid was linearized with AvaII to generate a probe complementary to the S transcripts of β2.7 using the T7 promoter (corresponding to nucleotides 4555 to 4399 or 185843 to 185999 of AD169).
Plasmid pE10335, carrying the AS UL61 and AS UL62 sequences, was digested with EcoRI and XhoI (corresponding to nucleotides 94467 to 94735 of AD169), and this fragment was inserted into pBluescript II KS+. This plasmid was linearized with EcoRI to generate a probe complementary to the AS UL61 and UL62 transcripts using the T3 promoter. Because the S-specific probe generated from the T7 promoter exhibited nonspecific binding, a second S-specific probe was generated by linearizing plasmid pE10335 with MluI (corresponding to nucleotides 94467 to 95174 of AD169) and using the T7 promoter.
RACE.
Rapid amplification of cDNA ends (RACE) was performed to determine 5′ and 3′ ends of the AS clones of UL36 (E) and the 5′ ends of AS clones for UL24 (E) and UL102 (L). Total RNA was isolated from MRC-5 cells in six-well plastic tissue culture plates exposed to 2 PFU per cell of the AD169 strain of HCMV at 48 h (for UL36 RACE) or 72 h (for UL24 and UL102 RACE) after infection. RNA was treated with DNase I (Roche Applied Science). 5′ or 3′ cDNA ends were amplified using the 5′/3′ RACE kit (Roche Applied Science), following the manufacturer's instructions. The products of the RACE reactions were inserted into a TOPO TA vector (Invitrogen) and sequenced at the OSU Plant-Microbe Genomics Facility. Primers used for RACE experiments are listed in Table 1.
RESULTS
Construction and analysis of HCMV cDNA libraries.
Three cDNA libraries were generated from primary human fibroblasts infected with HCMV. We chose to use AD169 HCMV for these studies because it is the most extensively characterized with respect to analyses of viral gene products. HCMV, like other herpesviruses, is known to express genes in a temporal and regulated cascade. The most clearly recognized temporal classes are IE, E, and L. In order to make predictions regarding temporal relationships of viral transcripts, RNA was isolated under conditions that selected for expression of IE, E, or L genes as described in Materials and Methods. We employed positive selection using viral genomic DNA as a probe to isolate cDNA clones harboring viral gene sequences. A total of 870 bacterial colonies that were positive by this screening method were individually expanded. Plasmid DNA isolated from each culture was subjected to DNA sequence analysis, and usable sequence data were obtained for 618 clones. After excluding 14 clones (2.3%) that were recombinants of viral gene sequences or viral and cellular gene sequences, 604 clones were subjected to analysis (109 from the IE library, 180 from the E library, and 315 from the L library). A complete list of the cDNA clones and their positions relative to genomic DNA is listed in Table S1 in the supplemental material. We assigned these 604 clones to 184 transcript groups based on the similarity of cDNA sequences.
Many indicators suggest that these libraries accurately reflect temporal regulation, splicing, and abundance of viral gene products. cDNA clones isolated include S sequences overlapping 125 of the 191 unique genes currently annotated for the AD169 strain of HCMV in the STD database, including full-length sequences for 92 annotated genes. Examples of correct temporal expression and splicing of viral genes in these libraries were supported by clones representing the UL123 and UL4 genes. The major transcriptional activator (IE72) encoded by the UL123 gene is known to be expressed during IE times (78). At least 15 cDNA clones isolated from the IE library were full-length and fully spliced transcripts capable of coding for the IE72 protein. Also, transcripts with short and long 5′ untranslated regions (UTRs) for the UL4 gene have been characterized to accumulate at early and late times after infection, respectively (1, 15, 20). We identified 14 transcripts harboring longer 5′ UTR regions, all of which were derived from the L library. We also identified 15 clones that had shorter 5′ UTRs, the majority of which (9) were found in the E library. In addition, studies from other laboratories suggest that the most abundant transcripts found in infected cells overlap the TRL4/IRL4 genes (TRL/IRL region genes are henceforth referred to as RL) (35), and this was reflected in our libraries. Indeed, 169 clones in our E and L libraries harbored RL4 gene sequences, 141 of which were classified within the same transcript group (see Table S1, group 148, in the supplemental material). Taken together, these and other indicators suggest that our libraries reasonably reflect the range and abundance of HCMV transcriptional products.
A more detailed comparison of the temporal profiles of transcripts isolated in this study relative to microarray and Northern studies from other laboratories is shown in Table S2 in the supplemental material. This table also lists the clones characterized in this study, organized by gene name, and provides references for other transcript mapping studies performed for HCMV. The primary conclusion from this analysis is that the tentative temporal class assignments made in this study are largely congruent with temporal class assignments made using Northern and microarray analyses. The major difference is that we found many more genes represented in the transcripts of the IE class relative to findings with other methods. Specifically, we identified 45 genes with at least 1 clone isolated in the IE library. For many of these genes, the majority of clones were isolated at E or L times with only one or a few clones isolated at IE times. There are two non-mutually exclusive explanations to account for these unexpected transcripts found in the IE library: (i) they represent tegument-associated viral transcripts delivered by the virion rather than newly synthesized viral transcripts; and (ii) these IE transcripts reflect leaky control of E and L gene expression in the IE period in HCMV-infected fibroblasts. Although the latter possibility could be due to incomplete blockade of protein synthesis by cycloheximide treatment, the efficacy of drug treatments used to generate the cDNA libraries was verified by measuring viral protein accumulation (data not shown). It is worthy of note that while the cDNA library approach employed in this study is subject to different biases relative to other methods (see Discussion and reference 19), it is free of bias introduced by employing gene-specific probes. Thus, it is possible that a cDNA library approach offers a specific advantage relative to Northern and microarray approaches in capturing minor transcripts available at various temporal phases of infection.
Another unexpected feature of our libraries was the prevalence of transcripts overlapping genes predicted to be noncoding genes. While we expected transcripts overlapping the RL4 gene to be abundant (35), we also isolated numerous cDNA clones from other repeat region genes. We found that 230 of the 604 clones analyzed mapped to the RL2-RL9 region. We also found 29 clones derived from the UL61 to UL68 gene region and 13 clones derived from the UL106 to UL111 gene region, also recently revised as likely to be noncoding regions (27). Altogether, we obtained 274 clones (45% of the total) overlapping annotated genes predicted to be noncoding.
AS transcription in the HCMV transcriptome.
One of the most striking features of our transcriptome study was the prevalence of transcripts in AS orientation to known or predicted genes. For this analysis, we compared our cDNA sequences to the genomic map of the AD169 strain (GenBank accession no. NC_001347) of HCMV and the STD database. This map includes up-to-date revisions in annotation proposed by Davison and colleagues, including annotation of those genes revised as noncoding (27).
Of the 604 sequences we analyzed, 257 represented one or more genes strictly in the S orientation (Table 2). Remarkably, 347 sequences were partially or completely AS to genes annotated on the STD database map, representing 57% of the cDNA clones isolated in our libraries. Because experimental evidence verifying the existence of gene products derived from a number of viral genes is lacking, we considered the possibility that the only products derived from a subset of these genes are those we identified in our library and that our calculation for the number of AS transcripts could be overestimated. When we excluded those genes for which we could find no evidence in our libraries or in the literature for a product derived from the S orientation of the gene (orphaned AS transcripts; see Table 5), we estimated that 271 clones (45%) represented transcriptional products strictly in one orientation with respect to gene sequences (designated the S orientation) and 333 clones (55%) were completely or partially in AS orientation. The AS sequences fell into two categories: a minority (54 clones) overlapped one or more genes strictly in AS orientation, whereas 279 clones overlapped more than one gene with sequences in both S and AS orientations. ORF analysis of AS clones predicts that these are predominantly noncoding (see Table S1 in the supplemental material). When these clones are included in the calculations for coding and noncoding transcripts, we estimate that up to 49.5% of the clones isolated in this study are noncoding in nature.
TABLE 2.
Orientation of cDNA clones
| Classification | No. (%b) of cDNA clones relative to STD database gene map | No. (%b) of cDNA clones, revised based on exptl evidencea |
|---|---|---|
| cDNA clones in only S orientation | 257 (43) | 271 (45) |
| cDNA clones completely or partially in AS orientation | 347 (57) | 333 (55) |
| cDNA clones in only AS orientationc | 51 (8) | 54 (9) |
| cDNA clones with both AS and S orientationsd | 296 (49) | 279 (46) |
Revised analysis based on excluding those AS clones for which we found no experimental evidence for an S transcript.
Relative to total number of clones in the cDNA library.
Clones spanning one or more genes only in AS orientation.
Orientation of gene regions for clones spanning two or more genes.
TABLE 5.
Orphaned AS transcripts
| Group | Gene | No. of AS clones | Library |
|---|---|---|---|
| 1 | UL28 | 1 | L |
| 2 | UL79 | 2 | L |
| 3 | UL90 | 1 | L |
| 4 | US1 | 3 | IE, E |
| 5 | US4 | 2 | L |
| 6 | US5 | 2 | L |
| 7 | US31 | 1 | E |
| 8 | US35 | 4 | IE, E |
| 9 | J1S | 1 | L |
A complete list of genes for which we identified S-AS pairs is shown in Table 3. We obtained direct evidence for the existence of S-AS transcript pairs derived from 38 known or predicted viral genes. We also predict that S-AS pairs exist for 18 additional annotated genes listed in Table 4. In these cases, we identified AS but not S transcripts overlapping the indicated gene sequences. Considering that evidence for S products derived from these genes was published by other laboratories, we predict S-AS pairs also exist for these genes. As mentioned earlier, we also identified AS transcripts derived from gene sequences of nine genes but could find no evidence of a corresponding S product in virus-infected cells or in the literature. We designated this group as orphaned AS transcripts (Table 5). Together our analysis indicates that cis natural S-AS pairs are generated during infection for at least 56 of the 191 known or predicted unique genes annotated at the STD database for the AD169 strain of HCMV.
TABLE 3.
Genes for which cis natural S-AS pairs were identified and their properties
| Pair | Genea | No. of S clones | Library | No. of AS clones | Library | Predicted temporal association | Predicted classification of AS pairsb |
|---|---|---|---|---|---|---|---|
| 1 | UL12 | 1 | E | 2 | L | Discordant | Divergent |
| 2 | UL13 | 6 | IE, L | 1 | E | Discordant | Divergent, full overlap |
| 3 | UL21 | 3 | IE | 2 | IE | Concordant | Divergent |
| 4 | UL22A | 2 | IE | 3 | IE | Concordant | Intronic |
| 5 | UL23 | 3 | IE, L | 1 | L | Discordant | Divergent |
| 6 | UL24 | 2 | L | 3 | L | Concordant | Divergent, convergent |
| 7 | UL25 | 5 | L | 1 | IE | Discordant | Divergent, intronic |
| 8 | UL29 | 3 | IE | 1 | L | Discordant | Divergent |
| 9 | UL36 | 3 | IE, E | 1 | E | Concordant | Divergent, full overlap |
| 10 | UL37 | 5 | IE, L | 1 | E | Discordant | Convergent |
| 11 | UL52 | 6 | E, L | 1 | L | Concordant | Divergent |
| 12 | UL61 | 2 | L | 10 | E, L | Concordant | Divergent |
| 13 | UL62 | 3 | L | 12 | E, L | Concordant | Convergent, divergent, full overlap |
| 14 | UL63 | 19 | E, L | 3 | L | Concordant | Convergent, divergent, full Overlap |
| 15 | UL64 | 4 | L | 21 | E, L | Concordant | Convergent, divergent, full overlap |
| 16 | UL65 | 20 | E, L | 3 | L | Concordant | Convergent, divergent, full overlap |
| 17 | UL66 | 1 | L | 18 | E, L | Concordant | Divergent, full overlap |
| 18 | UL67 | 1 | L | 17 | E, L | Concordant | Divergent, full overlap |
| 19 | UL70 | 1 | L | 4 | IE, L | Concordant | Divergent, full overlap |
| 20 | UL72 | 1 | L | 3 | E, L | Concordant | Divergent, full overlap |
| 21 | UL73 | 3 | E, L | 1 | L | Concordant | Full overlap |
| 22 | UL74 | 1 | L | 3 | E, L | Concordant | Full overlap, intronic |
| 23 | UL88 | 1 | L | 2 | L | Concordant | Divergent |
| 24 | UL89 | 1 | L | 2 | L | Concordant | Divergent, convergent |
| 25 | UL92 | 1 | L | 1 | L | Concordant | Divergent, intronic |
| 26 | UL102 | 2 | L | 2 | L | Concordant | Convergent, divergent, |
| 27 | UL108 | 2 | IE, L | 1 | IE | Concordant | Convergent |
| 28 | UL111 | 6 | IE, L | 2 | IE, L | Concordant | Convergent, full overlap |
| 29 | UL111A | 1 | L | 2 | L | Concordant | Full overlap |
| 30 | UL112 | 5 | IE, L | 1 | L | Concordant | Divergent |
| 31 | UL115 | 4 | IE, L | 2 | L | Concordant | Divergent, intronic, full overlap |
| 32 | UL116 | 2 | IE | 2 | L | Discordant | Divergent, intronic, full overlap |
| 33 | RL2 | 6 | E, L | 15 | E, L | Concordant | Convergent, divergent, full overlap |
| 34 | RL3 | 7 | E, L | 189 | E, L | Concordant | Convergent, divergent, full overlap |
| 35 | RL4 | 167 | E, L | 4 | E, L | Concordant | Convergent |
| 36 | TRS1 | 1 | IE, E, L | 1 | L | Concordant | Full overlap |
| 37 | US32 | 1 | L | 1 | L | Concordant | Divergent |
| 38 | US33 | 1 | L | 4 | IE, E, L | Concordant | Convergent, divergent, intronic |
Bold indicates sequence analysis confirmed a genuine poly(A) tail of the AS member of the pair.
Italicized “full overlap” designations are tentative because the 3′ ends were not sequenced.
TABLE 4.
Genes with predicted S-AS pairs
| Predicted pair | Genea | No. of AS clones | Library | Reference for S gene product |
|---|---|---|---|---|
| 1 | UL2 | 3 | E, L | 19 |
| 2 | UL3 | 32 | IE, E, L | 19 |
| 3 | UL11 | 1 | E | 41 |
| 4 | UL27 | 1 | L | 67 |
| 5 | UL30 | 1 | L | 34 |
| 6 | UL39 | 1 | L | 34 |
| 7 | UL47 | 1 | IE | 44 |
| 8 | UL48 | 4 | E, L | 4 |
| 9 | UL68 | 19 | E, L | 34 |
| 10 | UL87 | 1 | L | 34 |
| 11 | UL107 | 2 | IE, L | 11, 64 |
| 12 | RL5 | 150 | E, L | 34 |
| 13 | RL8 | 15 | IE, E, L | 19 |
| 14 | RL9 | 3 | E, L | 19 |
| 15 | RL10 | 1 | L | 75 |
| 16 | RL11 | 2 | L | 2, 54 |
| 17 | US29 | 3 | E, L | 19 |
| 18 | US30 | 2 | E, L | 19 |
Bold indicates sequence analysis confirmed a genuine poly(A) tail of the AS member of the pair.
The abundance of the S and AS transcripts forming pairs fell into two groups. For most genes, we isolated between one and six clones representing S-oriented transcripts and one and six clones representing AS-oriented transcripts. There were two dramatic exceptions to this finding: the gene region from UL61 to UL67 and the gene region from RL2 to RL5. cDNA clones representing transcripts from these two gene regions were among the most abundant in our libraries, and this was reflected in the number of S-AS pairs. For example, we found seven clones overlapping RL3 gene sequences in the S orientation and 189 clones overlapping RL3 gene sequences in the AS orientation. One notable feature of both classes of genes was dominance of either S or AS transcripts of a pair. We observed that clones in one orientation outnumbered those in opposite orientations between 1.5- and 41-fold. In fact, an inverse relationship (at least 2:1) in abundances of S and AS transcripts was observed for 30 of the 38 S-AS pairs identified in this study.
Based upon the library in which the clones were isolated, we made predictions regarding the temporal association of S and AS transcripts derived from the same gene. Accordingly, 7 S-AS pairs were discordant relative to the library in which they were identified, whereas most (28) were concordant inasmuch as they were isolated from the same library. Keeping in mind that the E library is expected to contain both IE and E transcripts and the L library could contain transcripts expressed at IE, E, or L temporal classes, together these findings suggest that the majority of S-AS pairs are concordantly and inversely expressed during infection.
We also classified S-AS pairs with respect to the nature of the complementary overlapping sequences (Table 3). We used combinations of classification schemes proposed by others (45, 94) divided into one of four categories: full overlap, intronic, convergent, and divergent. Full overlap was defined as one gene sequence being completely contained within the gene sequence of the other member of the pair. Intronic was defined as one gene sequence starting within the intron of the other member of the pair and ending beyond the start of its pair. Divergent was defined as S-AS pairs exhibiting overlap in their 5′ regions in a head-to-head manner. Finally, convergent was defined as S-AS pairs exhibiting overlap in their 3′ regions in a tail-to-tail manner. Each potential pair was assigned to only one category, and the order of stringency was full overlap, intronic, divergent, and convergent. Using this scheme, we identified S-AS pairs that fell into each of these groups. Although the least abundant class, intronic pairs were observed for S-AS pairs overlapping seven genes. Convergent overlap was common, with 1 or more S-AS pairs for 14 genes falling into this class. Pairs exhibiting full overlap were also abundant. We found 1 or more S-AS pairs overlapping 19 genes in this class. However, it should be noted that in the absence of 3′ end sequence for all of the cDNA clones, it is possible that the number of genes with S-AS pairs exhibiting full overlap is currently overestimated. Finally, we found that 29 of 38 genes included 1 or more S-AS pairs that were divergent in their overlap. Also, of those genes with multiple S-AS pairs, the divergent class was typically most abundant. Thus, while we observe a diversity of S-AS pair classes derived from HCMV genes, pairs with divergent or head-to-head overlap were most common.
Finally, we classified S-AS pairs according to functional class of known or predicted gene products (Table 6). Several S-AS pairs overlap genes known to be involved in DNA replication and packaging, including those genes (UL70 and UL102) that encode the components of the HCMV helicase-primase complex. We also found S-AS pairs for the viral inhibitors of apoptosis encoded by UL36 and UL37 genes and the recently described noncoding β2.7 transcript overlapping the RL4 gene (69). Additionally, we identified S-AS pairs that overlap genes encoding tegument proteins, glycoproteins, and proteins involved in subversion of immune responses (UL111A and RL11) (2, 54) or cellular antiviral defense mechanisms (TRS1 and IRS1) (17). Finally, we identified S-AS pairs for at least 32 genes of unknown function, most of which are predicted to be noncoding. This aside, these findings indicate that there is not a clear bias of S-AS pairs for genes of specific functional classes and that S-AS pairs exist for both coding and noncoding genes.
TABLE 6.
Functional classes of genes with verified or predicted cis natural S-AS pairs
| Functional class | Genes for which S-AS pairs were identifieda |
|---|---|
| DNA replication/packaging | UL27, UL52, UL70, UL72, UL89, UL102, UL112 |
| Gene expression | IRS1, TRS1 |
| Antiapoptotic | UL36, UL37, RL4 |
| Glycoproteins | UL11, UL13, UL22A, UL73, UL74, UL115, UL116, RL10, RL11 |
| Subversion of immune or antiviral defense | UL111A, IRS1/TRS1, RL11 |
| Tegument proteins | UL23, UL24, UL25, UL47, UL48, UL88 |
| Unknown | UL2, UL3, UL11, UL12, UL13, UL21, UL27, UL29, UL30, UL39, UL61, UL62, UL63, UL64, UL65, UL66, UL67, UL68, UL87, UL92, UL107, UL108, UL111, RL2, RL3, RL5, RL8, RL9, RL10, US29, US32, US33 |
Bold indicates sequence analysis confirmed a genuine poly(A) tail of the AS member of the pair.
Verification of HCMV AS transcripts.
At least two features of a subset of AS clones suggest they cannot represent artifacts of our infection conditions or library construction. First, at least 68 of the clones harboring AS sequences possess genuine poly(A) tails (see Table S3 in the supplemental material). Because not all clones were sequenced from the 3′ ends, it is likely additional clones with genuine poly(A) tails will be identified. These 68 clones constitute at least 1 AS member for 25 of the 38 genes with S-AS pairs identified in this study (Table 3, genes identified in bold) and 13 of 18 genes with predicted S-AS pairs (Table 4). AS clones with genuine poly(A) tails were also found in each of the functional classes listed in Table 6. A second feature is exemplified by the S-AS pair overlapping the UL36 gene. In this case, we identified three S clones, all of which represented mature, spliced transcripts lacking the UL36 intron sequences (see Table S1, transcript group 35, in the supplemental material). The AS clone overlapping the UL36 gene region (transcript group 36) contains this intron sequence, indicating that it could not represent aberrant cloning of the S cDNA product.
Our analysis of cis NATs from the HCMV genome relied upon isolation of virally derived transcripts from human foreskin-derived fibroblasts. If this is a truly robust phenomenon, we predict that cis NATs will be observed in other infected cell types and in cells infected with other strains of HCMV. To test this, we used RT-PCR to identify AS transcripts in lung-derived human fibroblasts infected with the laboratory-adapted strain AD169 of HCMV and endothelial cells infected with the VHL/E clinical strain of HCMV. We designed primers specific for AS transcripts from both coding and noncoding viral genes. We also utilized primers specific for S-oriented viral UL55 transcripts coding for gB or cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts. For specificity controls, RNA isolated from virus-infected and mock-infected cells was analyzed in the presence and in the absence of reverse transcriptase.
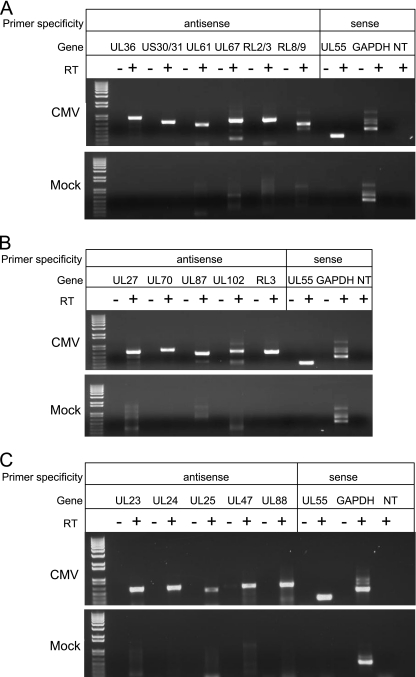
In the first set of experiments, RT-PCR was used to amplify AS transcripts identified in the E and L libraries using RNA isolated from AD169-infected MRC-5 fibroblasts at 48 and 72 h after infection, respectively. As shown in Fig. 1A, we specifically amplified portions of AS transcripts found in the early library derived from genes known or predicted to be protein coding (UL36 and US30/31) and genes predicted to be noncoding (UL61, UL67, RL2/3, and RL8/9). Similarly, we specifically amplified products of expected sizes for AS transcripts derived from genes known to be coding (UL27, UL70, UL87, and UL102) and predicted to be noncoding (RL3) from RNA isolated at 72 h after infection (Fig. 1B). Finally, we specifically amplified products of expected sizes for AS transcripts derived from genes specifying tegument proteins (UL23, UL24, UL25, UL47, and UL88) (Fig. 1C). Except for the cellular GADPH, we could not amplify these transcripts from mock-infected cells. The absence of bands of predicted sizes in reactions conducted without reverse transcriptase ensures that these results do not reflect contamination of viral DNA in our reactions. We conclude from these experiments that virally derived AS transcripts are generated during lytic HCMV infections of fibroblast cells and that these AS transcripts do not reflect DNA contamination during the cDNA library construction.
FIG. 1.
Verification of virally derived AS transcripts. Digital images of agarose gels used to separate PCR products specific for virally derived AS and S transcripts. Confluent MRC-5 cells in six-well tissue culture plates were exposed to 2 PFU per cell of the AD169 strain of HCMV (top panels) or were mock infected (bottom panels). (A) RT-PCR of AS transcripts identified in the E library using total RNA isolated at 48 h after infection. (B) RT-PCR of AS transcripts identified in the L library using total RNA isolated at 72 h after infection. (C) RT-PCR of AS transcripts specific to tegument genes using total RNA isolated at 24 h after infection (UL47) or 72 h after infection (UL23, UL24, UL25, and UL85). Total RNA was isolated and subjected to RT-PCR using primers specific to virally derived AS transcripts (Table 1). Primers specific for the S transcripts of viral UL55 (gB) or cellular GAPDH were included as positive controls. No-reverse-transcriptase and no-template (NT) controls were run in parallel. At 48 h, the no-template control included primers specific for AS US30/31. At 72 h, the no-template control included primers specific for AS UL87/88. In panel C, the no-template control included primers specific for AS UL23. RT-PCR products were separated by agarose gel (1% [wt/vol]) electrophoresis, and bands were visualized by exposure to UV light.
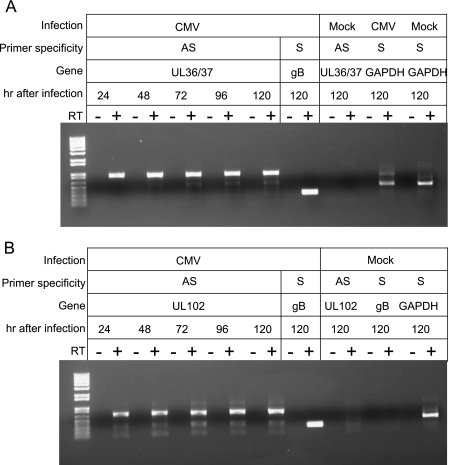
In the second set of experiments, we analyzed two specific AS transcripts derived from the UL36/37 gene region and the UL102 gene in primary endothelial cells infected with the VHL/E strain of HCMV. Because lytic replication in endothelial cells infected with VHL/E proceeds much more slowly than in fibroblasts infected with laboratory-adapted strains, we performed RT-PCR over a 120-h time course (Fig. 2). Specific AS transcripts were amplified at each time point examined for these two gene regions. We conclude that generation of cis NATs is a common feature of infection by both laboratory-adapted and clinical isolates of HCMV and that cis NATs occur during infection of clinically relevant cell types. Also, while cycloheximide and ganciclovir were used to generate the IE and E libraries, all of these experiments were performed in the absence of any drug treatment. This rules out the possibility that these AS transcripts represent aberrant transcriptional products that accumulate in cells exposed to protein synthesis or viral polymerase inhibitors.
FIG. 2.
Verification of AS transcripts in endothelial cells infected with a clinical strain of HCMV. Digital image of an agarose gel used to separate PCR products specific for virally derived AS and S transcripts. Confluent HUVEC monolayers in six-well tissue culture plates were inoculated with the VHL/E strain of HCMV (1 PFU/cell). Cells were harvested at 24, 48, 72, 96, and 120 h postinfection or were mock infected for 120 h. Total RNA was isolated and subjected to RT-PCR using primers specific to virally derived AS transcripts for UL36/37 (A) or UL102 (B). Primers specific for the S transcripts of viral UL55 (gB) or cellular GAPDH were included as positive controls. No-reverse-transcriptase and no-template controls were run in parallel. RT-PCR products were separated by agarose gel (1% [wt/vol]) electrophoresis, and bands were visualized by exposure to UV light.
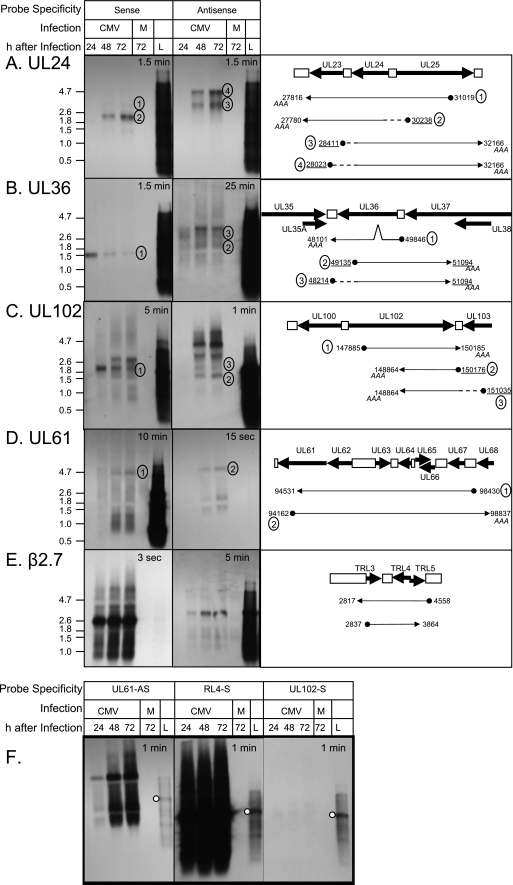
To more carefully analyze the relationship between accumulation of S-AS pairs, we performed Northern analyses using RNA from AD169-infected MRC-5 cells at various times after infection with probes specific to either S or AS products of the UL24, UL36, UL102, UL61, and RL4/5 genes (Fig. 3). We identified two S and three AS clones overlapping the UL24 gene in our libraries. By using a probe specific to the S transcripts of UL24, we identified a major band between the 1.8- and 2.6-kb markers beginning at 24 h after infection and increasing in abundance through 72 h after infection and a larger, weaker band just over 3 kb expressed with the same temporal profile (Fig. 3A). These bands correspond to predicted sizes of clones pIE811 and pL5312, described in Table S1 in the supplemental material. RACE was performed to verify the 5′ ends of pL5312. We identified genomic positions of 30238 and 30029 as initiation positions for transcripts that are 432 and 223 bp larger than clone pL5312. Using a probe specific for the AS transcript of UL24, we identified two major bands between 3.0 and 4.0 kb at 48 and 72 h after infection. The largest AS UL24 clone isolated was 3.2 kb (pL8212). By RACE analysis, we identified two transcript initiation sites, at genomic positions 28411 and 28023, predicting transcripts of 3,755 and 4,143 bp in this region, that correspond well with the two bands observed by Northern blotting. Together these findings confirm the presence of cis NATs derived from the UL24 gene in infected cells and validate our prediction of concordant expression and our finding of similar abundance of the UL24 cis NATs.
FIG. 3.
Northern blot analysis of S and AS transcripts. Film images of S and AS transcripts analyzed by Northern blotting and schematic depictions of select S and AS transcripts. Confluent MRC-5 cells in six-well tissue culture plates were exposed to 2 PFU per cell of the AD169 strain of HCMV or were mock infected (M). Cells were harvested at 24, 48, and 72 h after infection. Total RNA was isolated, subjected to denaturing agarose gel electrophoresis, and transferred to nylon membranes. Prelabeled RNA molecular mass markers were loaded for each group (L). Membranes were incubated with probes specific for the S and AS transcripts of UL24, 3 μg RNA/lane (A); UL36, 3 μg RNA/lane (B); UL102, 4 μg RNA/lane (C and F); UL61, 5 μg RNA/lane (D and F); or RL4, 4 μg RNA/lane (E and F) as described in Materials and Methods. Exposure times are indicated at the top of each panel. A schematic of transcripts represented by select cDNA clones isolated in our library relative to the genome is shown in the right panels. Gene regions and intergenic regions are depicted by thick arrows and white boxes, respectively. Transcripts cloned in this study are represented as thin arrows below the gene regions. 5′ ends of transcripts are depicted with filled circles. Clones in which we identified the poly(A) tails are indicated (AAA). The genomic positions of the 5′ and 3′ ends of the clones in the libraries are shown. Underlined genomic positions are those verified by RACE. Dashed lines represent presumptive transcript sequences based on RACE analysis. Tentative assignment of bands corresponding to clones identified in this study is indicated with circled numbers. In panel F, the relative abundances of transcripts overlapping the RL4 and UL61 gene regions are compared to those of transcripts derived from the UL102 gene region. The white circle indicates the position of the 2.6-kb marker band. No images were altered.
We identified three S clones and one AS clone overlapping the UL36 gene. As shown in Fig. 3B, we identified a band of the expected size (1.6 kb) for the spliced S product of the UL36 gene (80a) that corresponds to the clones listed in transcript group 35 in Table S1 in the supplemental material. Also as expected, we could identify this band throughout the 72-h infection period, though it was most abundant at 24 h after infection. In our E library, we isolated one clone (pE104114) of approximately 1.9 kb that was AS to UL36. When we used a probe specific for the AS transcript, we identified only a weak band of 1.9 kb at 48 and 72 h after infection. We also observed a larger, more abundant band at 48 and 72 h. RACE was performed in an attempt to identify the boundaries of this larger transcript. In this analysis we confirmed the boundaries of the AS clone pE104114 and provided evidence for a transcript initiating at genomic position 48214, predicting a 2,880-bp transcript AS to UL36. This may represent the larger, more abundant AS transcript identified on the blot. However, because of the limitation in resolving the bands in this size range, we cannot exclude the possibility that this represents the faint band observed between the two major bands on this blot. In either case, we predict that there exists an additional AS transcript derived from the UL36 gene. This analysis also confirms our prediction of concordant and inverse expression for the UL36-derived cis NATs.
Similar to that observed for the UL36 AS transcripts, we found that expression of the S UL102 transcript precedes expression of the AS UL102 transcripts (Fig. 3C). We identified a 2.3-kb S transcript in our library represented by clone pL2211. An additional larger S transcript is also observed by Northern analysis, consistent with that reported previously (73a). RACE analysis confirmed the 5′ end of AS clone pL222 and identified another, larger AS transcript of 2.1 kb initiating at genomic position 151035. Additional, even larger bands were observed by Northern analysis that have yet to be characterized. We predicted that cis NATs of UL102 should be expressed with similar abundance and concordantly. While concordant expression was confirmed, the difference in exposure times required to visualize these transcripts indicates inverse accumulation of the S-AS transcripts.
Finally, we used a Northern blot approach to ascertain whether the abundance of clones in our libraries overlapping the RL4/5 and UL61/UL62 genes reasonably reflects the abundance of these transcripts in infected cells (Fig. 3D, E, and F). First we set out to verify the existence of S and AS clones from these gene regions. Clone pL537 was the longest S clone overlapping UL61 in our library, predicting a band of 3.9 kb, which corresponds well with the largest band observed on this Northern blot (Fig. 3D). Numerous clones overlapped UL61 in the AS orientation, the longest of which are listed in transcript group 59 (see Table S1 in the supplemental material), which predict a band of 4.6 kb. These clones correspond well with the largest band identified with the AS-specific probe for UL61 (Fig. 3D). The smaller bands identified by both S and AS probes specific for UL61 may represent distinct smaller transcripts or degradation products of the larger transcripts isolated in this study. The major 2.7-kb transcript overlapping the RL4/5 genes has been described previously (35) and represented the major band identified by the S-specific probe (Fig. 3E). However, most or possibly all of the clones overlapping the RL4 genes isolated in this study appear to be initiated from genomically encoded poly(A) tracts and thus do not represent the full-length 2.7-kb transcript (represented by clone pE103121, depicted in the right panel of Fig. 3E). We identified a band of approximately 3 kb using a probe that would recognize transcripts that are in AS orientation relative to the 2.7-kb transcript. Although the precise boundaries of the AS RL4/5 transcripts (represented by clone pE103210 in the right panel) as well as the UL61-derived transcripts have yet to be verified, the different exposures times required to visualize the S and AS transcripts clearly demonstrate inverse expression of the UL61-derived AS transcripts relative to the UL61-derived S transcripts and inverse expression of RL4-derived S transcripts relative to the RL4-derived AS transcripts. Finally, we compared the abundance of clones overlapping the UL61/UL62 and RL4/5 gene regions to that derived from the UL102 gene region (Fig. 3F). In our libraries, we isolated two S UL102 clones, five times as many AS UL61 clones, and 85 times as many S RL4 clones. The abundance of these clones in our libraries correlated with the signal intensities of transcripts that bound the S-specific RL4 probe and the AS-specific UL61 probe relative to the S-specific UL102 probe. Taken together, these experiments prove the existence of cis NATs derived from coding (UL24, UL36, and UL102) and predicted noncoding (UL61 and RL4) genes. Furthermore, these studies indicate that the representation of S and AS transcripts in our libraries reasonably reflects the abundance and temporal expression patterns of S and AS transcripts generated in HCMV-infected fibroblasts.
DISCUSSION
The HCMV transcriptome.
In this study, we isolated and characterized transcripts generated during lytic infection of HCMV-infected fibroblasts. Selecting for cDNA clones representing viral genes and obtaining sequence data on each isolated clone constituted a de facto analysis of the HCMV transcriptome. This analysis revealed two novel findings. First, there was an unexpectedly high percentage of clones derived from genomic regions currently considered to be noncoding (27). While we expected a high prevalence of RL4 transcripts based on reports from other studies (35, 43, 57), additional noncoding transcripts were also represented at high frequencies, especially those from the UL61-UL67 gene region. Altogether, 45% of the clones isolated in this study were derived from gene regions predicted to be noncoding (27), suggesting further studies of the function and regulation of these gene regions are needed. Second, we identified a striking number of transcripts, predominantly AS, that were not predicted by the STD database annotation of the HCMV genome. We therefore conclude that genomic maps based on ORF analyses and comparisons to related viral genomes drastically underestimate the true complexity of viral gene products.
The discovery of widespread AS transcription also bears upon the use of viral gene arrays for transcriptome analyses and recombinant viruses for gene function studies. Specifically, these findings suggest that tiling arrays using overlapping probes from both DNA strands rather than probes based upon ORF predictions will be required to capture an accurate representation of viral transcripts. Additionally, these findings raise the possibility that AS as well as S gene products could be affected when recombinant viruses harboring mutations or deletions are generated.
There are several factors that could influence the composition of this library that are relevant to the interpretation of these data. First, because highly abundant transcripts are expected to be preferentially represented, we infer that transcripts from genes not represented in our library are of relatively low abundance. While abundance of these transcripts is likely a key factor, we cannot exclude the possibility that artifactual pressures might also influence library composition. For example, it is possible that there were selective advantages for isolating transcripts from gene regions with long or repetitive tracts of adenosines, such as the RL gene region. These transcripts may have been preferentially enriched during the purification of polyadenylated RNAs prior to cDNA library construction. Similarly, genomically encoded poly(A) tracts may have served as primer binding sites during the reverse transcription step of the cDNA library construction. While many cDNA clones appear to have genuine poly(A) tracts, we also found a number of clones, especially from predicted noncoding regions, in which the 3′ end sequence corresponded to a genomically encoded poly(A) tract. Therefore, further studies will be necessary to define precise ends of a subset of transcripts, as well as their relative abundance in infected cells. Despite these caveats, Northern analyses of transcripts derived from single-copy and repeat-region genes suggest that composition of our library reasonably reflects the abundance and temporal expression patterns of transcripts in infected fibroblasts.
AS transcripts of the HCMV genome.
Individual AS transcripts have been described for many herpesviruses, including the betaherpesviruses, (6, 48, 82), the gammaherpesviruses, (65, 66, 72), and especially the alphaherpesviruses, (8, 9, 13, 16, 21, 25, 26, 42, 49, 50, 53, 68, 84, 88, 89, 91). In fact, Roizman and colleagues predicted that genes in AS orientation to known herpesvirus genes could be common (16). However, to our knowledge, our study is the first to systematically document cis NATs derived from a herpesvirus. We found that at least 55% of the clones analyzed in this study contain sequences in AS orientation overlapping 56 known or predicted genes. These figures are dramatic inasmuch as they suggest that more than half of all virally derived transcripts harbor AS sequences. Nevertheless, three factors suggest that our study may have underestimated AS transcription in HCMV. First, our libraries do not contain all of the S transcripts expressed by HCMV, and thus, it is likely that they do not contain all of the AS transcripts that accumulate during infection. Indeed, at least two previously reported AS transcripts overlapping the HCMV UL82 and UL123 genes were not isolated in our libraries (6, 48). Second, we selected for polyadenylated transcripts during construction of our libraries, and studies suggest that a large fraction of AS transcripts are poly(A) negative (46). Finally, we classified only cis NATs identified in our libraries. Considering that the HCMV genome includes numerous repeat elements, it is likely that trans-derived NATs are also generated during infection.
Relative to eukaryotic genomes, herpesvirus genomes are small and densely populated with genes. In fact, many genes are directly adjacent to one another, and a number of viral genes overlap each other, often in opposite orientations. These characteristics suggest that the occurrence of S-AS pairs may only reflect the dense organization of viral genes and the relative paucity of intergenic regions relative to larger genomes. Nevertheless, S-AS pairs derived as a consequence of this gene organization may be functionally relevant with respect to regulation of viral genes. This raises the interesting possibility that regulatory consequences of S-AS pairs may constitute one evolutionary pressure influencing function and orientation of adjacent viral genes.
Given these properties of the HCMV genome, it seems reasonable to predict that higher proportions of viral genes would be associated with S-AS pairs than would be the case for genes of eukaryotic genomes. In this study, we found that at least 29% of the currently annotated 191 HCMV genes are associated with S-AS pairs. While this figure is similar to estimates of 22 to 26% (24, 94) of human genes associated with S-AS pairs, it is lower than the estimate of 72% of mouse genes (45) predicted to be influenced by S-AS pairs. These preliminary findings suggest that the proportion of HCMV genes involved in the generation of S-AS pairs is similar to that observed for the human genome, despite the striking differences in sizes and structures of these genomes. Another common feature between mammalian and viral S-AS pairs is the nature of the complementary sequences. Our analyses indicate that most S-AS pairs overlap either in 5′ or 3′ regions, with intronic organization as the least frequent class of S-AS pairs, and this is similar to reports by other groups for the human genome (52, 92, 94). One prediction from this type of overlap is that the potential regulatory consequences of these S-AS pairs relate to function, accessibility, or processing of the UTRs of viral transcripts (80). Since posttranscriptional processing of HCMV viral transcripts has been described previously (7, 14, 30-33, 55, 76, 90), it will be of interest to determine whether such events are influenced by AS transcripts. Also, in mammalian genomes, S-AS pairs appear to be overrepresented among genes involved in genomic imprinting (45), metabolic, catalytic, and cell organization functions (94), and translational regulation (24). However, our findings indicate that there is not a clear functional bias among HCMV genes for which S-AS pairs were identified.
A key question raised by our findings is whether the virally derived S-AS pairs have regulatory consequences during lytic or latent viral infections. AS transcripts can impact viral gene expression though multiple mechanisms, such as influencing splicing, editing, stability, localization, and translation of transcripts (51, 60). In addition, double-stranded intermediates generated from direct interaction of S-AS pairs may lead to gene regulation by RNA silencing or chromatin remodeling (51, 60). S-AS pairs involved in regulatory functions are predicted to be expressed concordantly and to accumulate in an inverse manner (23). Indeed, coexpressed and inversely expressed S-AS pairs not only are more frequent in the human genome than would be expected by chance but are also evolutionarily conserved (80). Our results predict that the majority of HCMV S-AS pairs are expressed with just such a profile, and we provide experimental evidence for concordant expression and inverse accumulation for S-AS pairs derived from the UL36, UL102, UL61/UL62, and RL4/5 gene regions.
Another possibility is that AS transcripts may serve as primary transcripts for microRNAs (miRNA), as was recently suggested for the AS latency-associated transcripts of herpes simplex virus and Marek's disease virus (12, 38). Several studies have identified putative or validated miRNAs of HCMV (28, 36, 64). Interestingly, a number of miRNAs that have been predicted or validated are derived from the AS strand of viral genes, including the UL31, UL53, UL70, UL102, UL114, UL150, and US29 genes (36, 64). In support of this possibility, we identified AS transcripts from several of these genes, including UL70, UL102, and US29. While further studies are required to establish regulatory roles for these AS transcripts, we predict that these AS transcripts may dramatically alter our understanding of viral gene regulation during both lytic and latent infections.
To summarize, the remarkable accumulation of noncoding and AS transcripts during infection suggests that currently available genomic maps based on ORF and other in silico analyses may drastically underestimate the true complexity of viral gene products. These findings also raise the possibility that aspects of both the HCMV life cycle and genome organization are influenced by AS transcription. These noncoding and AS transcripts may offer new insights into HCMV pathogenesis and may serve as novel targets for developing intervention strategies and treatments for HCMV-related diseases.
Supplementary Material
Acknowledgments
We gratefully acknowledge the technical efforts of Pete Zimmerman, RuthAnn Weimer, Abbey Wright, and Mary Sivulich in cDNA library construction.
This work was supported by grant AI51411-03 from the National Institutes of Health.
Footnotes
Published ahead of print on 8 August 2007.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Alderete, J. P., S. J. Child, and A. P. Geballe. 2001. Abundant early expression of gpUL4 from a human cytomegalovirus mutant lacking a repressive upstream open reading frame. J. Virol. 75:7188-7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atalay, R., A. Zimmermann, M. Wagner, E. Borst, C. Benz, M. Messerle, and H. Hengel. 2002. Identification and expression of human cytomegalovirus transcription units coding for two distinct Fcγ receptor homologs. J. Virol. 76:8596-8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awasthi, S., J. A. Isler, and J. C. Alwine.2004. Analysis of splice variants of the immediate-early 1 region of human cytomegalovirus. J. Virol. 78:8191-8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bankier, A. T., S. Beck, R. Bohni, C. M. Brown, R. Cerny, M. S. Chee, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, et al. 1991. The DNA sequence of the human cytomegalovirus genome. DNA Seq. 2:1-12. [DOI] [PubMed] [Google Scholar]
- 6.Bego, M., J. Maciejewski, S. Khaiboullina, G. Pari, and S. St Jeor. 2005. Characterization of an antisense transcript spanning the UL81-82 locus of human cytomegalovirus. J. Virol. 79:11022-11034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergamini, G., M. Reschke, M. C. Battista, M. C. Boccuni, F. Campanini, A. Ripalti, and M. P. Landini. 1998. The major open reading frame of the beta2.7 transcript of human cytomegalovirus: in vitro expression of a protein posttranscriptionally regulated by the 5′ region. J. Virol. 72:8425-8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohenzky, R. A., M. Lagunoff, B. Roizman, E. K. Wagner, and S. Silverstein. 1995. Two overlapping transcription units which extend across the L-S junction of herpes simplex virus type 1. J. Virol. 69:2889-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borchers, K., U. Wolfinger, B. Lawrenz, A. Schellenbach, and H. Ludwig. 1997. Equine herpesvirus 4 DNA in trigeminal ganglia of naturally infected horses detected by direct in situ PCR. J. Gen. Virol. 78:1109-1114. [DOI] [PubMed] [Google Scholar]
- 10.Bresnahan, W. A., G. E. Hultman, and T. Shenk. 2000. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J. Virol. 74:10816-10818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 12.Burnside, J., E. Bernberg, A. Anderson, C. Lu, B. C. Meyers, P. J. Green, N. Jain, G. Isaacs, and R. W. Morgan. 2006. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J. Virol. 80:8778-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantello, J. L., A. S. Anderson, and R. W. Morgan. 1994. Identification of latency-associated transcripts that map antisense to the ICP4 homolog gene of Marek's disease virus. J. Virol. 68:6280-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao, J., and A. P. Geballe. 1996. Coding sequence-dependent ribosomal arrest at termination of translation. Mol. Cell. Biol. 16:603-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao, J., and A. P. Geballe. 1998. Ribosomal release without peptidyl tRNA hydrolysis at translation termination in a eukaryotic system. RNA 4:181-188. [PMC free article] [PubMed] [Google Scholar]
- 16.Carter, K. L., P. L. Ward, and B. Roizman. 1996. Characterization of the products of the U(L)43 gene of herpes simplex virus 1: potential implications for regulation of gene expression by antisense transcription. J. Virol. 70:7663-7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cassady, K. A. 2005. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J. Virol. 79:8707-8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, C. P., C. L. Malone, and M. F. Stinski. 1989. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J. Virol. 63:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang, Y. E., L. Menotti, F. Filatov, G. Campadelli-Fiume, and B. Roizman. 1998. UL27.5 is a novel gamma2 gene antisense to the herpes simplex virus 1 gene encoding glycoprotein B. J. Virol. 72:6056-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 23.Chen, J., M. Sun, L. D. Hurst, G. G. Carmichael, and J. D. Rowley. 2005. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 21:326-329. [DOI] [PubMed] [Google Scholar]
- 24.Chen, J., M. Sun, W. J. Kent, X. Huang, H. Xie, W. Wang, G. Zhou, R. Z. Shi, and J. D. Rowley. 2004. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res. 32:4812-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chesters, P. M., R. Allsop, A. Purewal, and N. Edington. 1997. Detection of latency-associated transcripts of equid herpesvirus 1 in equine leukocytes but not in trigeminal ganglia. J. Virol. 71:3437-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung, A. K. 1990. The BamHI J fragment (0.706 to 0.737 map units) of pseudorabies virus is transcriptionally active during viral replication. J. Virol. 64:977-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 28.Dunn, W., P. Trang, Q. Zhong, E. Yang, C. van Belle, and F. Liu. 2005. Human cytomegalovirus expresses novel microRNAs during productive viral infection. Cell Microbiol. 7:1684-1695. [DOI] [PubMed] [Google Scholar]
- 29.Fahey, M., T. F. Moore, and D. G. Higgins. 2002. Overlapping antisense transcription in the human genome. Comput. Funct. Genomics 3:244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geballe, A. P., F. S. Leach, and E. S. Mocarski. 1986. Regulation of cytomegalovirus late gene expression: gamma genes are controlled by posttranscriptional events. J. Virol. 57:864-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geballe, A. P., and E. S. Mocarski. 1988. Translational control of cytomegalovirus gene expression is mediated by upstream AUG codons. J. Virol. 62:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geballe, A. P., R. R. Spaete, and E. S. Mocarski. 1986. A cis-acting element within the 5′ leader of a cytomegalovirus beta transcript determines kinetic class. Cell 46:865-872. [DOI] [PubMed] [Google Scholar]
- 33.Goins, W. F., and M. F. Stinski. 1986. Expression of a human cytomegalovirus late gene is posttranscriptionally regulated by a 3′-end-processing event occurring exclusively late after infection. Mol. Cell. Biol. 6:4202-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodrum, F. D., C. T. Jordan, K. High, and T. Shenk. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc. Natl. Acad. Sci. USA 99:16255-16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenaway, P. J., and G. W. Wilkinson. 1987. Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res. 7:17-31. [DOI] [PubMed] [Google Scholar]
- 36.Grey, F., A. Antoniewicz, E. Allen, J. Saugstad, A. McShea, J. C. Carrington, and J. Nelson. 2005. Identification and characterization of human cytomegalovirus-encoded microRNAs. J. Virol. 79:12095-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths, P. D., A. V. Cope, A. F. Hassan-Walker, and V. C. Emery. 1999. Diagnostic approaches to cytomegalovirus infection in bone marrow and organ transplantation. Transpl. Infect. Dis. 1:179-186. [DOI] [PubMed] [Google Scholar]
- 38.Gupta, A., J. J. Gartner, P. Sethupathy, A. G. Hatzigeorgiou, and N. W. Fraser. 2006. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature 442:82-85. [DOI] [PubMed] [Google Scholar]
- 39.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 40.Hirsch, P. R. 2004. Detection of microbial DNA sequences by colony hybridization, p.345-356. In G. A. Kowalchuk, F. J. de Bruijn, I. M. Head, A. D. Akkermans, and J. D. van Elsas, (ed.), Molecular microbial ecology manual, 2nd ed., vol. 2.0.9. Kluwer Academic, Dordrecht, The Netherlands.
- 41.Hitomi, S., H. Kozuka-Hata, Z. Chen, S. Sugano, N. Yamaguchi, and S. Watanabe. 1997. Human cytomegalovirus open reading frame UL11 encodes a highly polymorphic protein expressed on the infected cell surface. Arch. Virol. 142: 1407-1427. [DOI] [PubMed] [Google Scholar]
- 42.Holden, V. R., R. N. Harty, R. R. Yalamanchili, and D. J. O'Callaghan. 1992. The IR3 gene of equine herpesvirus type 1: a unique gene regulated by sequences within the intron of the immediate-early gene. DNA Seq. 3:143-152. [DOI] [PubMed] [Google Scholar]
- 43.Hutchinson, N. I., R. T. Sondermeyer, and M. J. Tocci. 1986. Organization and expression of the major genes from the long inverted repeat of the human cytomegalovirus genome. Virology 155:160-171. [DOI] [PubMed] [Google Scholar]
- 44.Hyun, J. J., H. S. Park, K. H. Kim, and H. J. Kim. 1999. Analysis of transcripts expressed from the UL47 gene of human cytomegalovirus. Arch. Pharm. Res. 22:542-548. [DOI] [PubMed] [Google Scholar]
- 45.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564-1566. [DOI] [PubMed] [Google Scholar]
- 46.Kiyosawa, H., N. Mise, S. Iwase, Y. Hayashizaki, and K. Abe. 2005. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 15:463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiyosawa, H., I. Yamanaka, N. Osato, S. Kondo, and Y. Hayashizaki. 2003. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 13:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krause, P. R., K. D. Croen, S. E. Straus, and J. M. Ostrove. 1988. Detection and preliminary characterization of herpes simplex virus type 1 transcripts in latently infected human trigeminal ganglia. J. Virol. 62:4819-4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lagunoff, M., and B. Roizman. 1994. Expression of a herpes simplex virus 1 open reading frame antisense to the gamma(1)34.5 gene and transcribed by an RNA 3′ coterminal with the unspliced latency-associated transcript. J. Virol. 68:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavorgna, G., D. Dahary, B. Lehner, R. Sorek, C. M. Sanderson, and G. Casari. 2004. In search of antisense. Trends Biochem. Sci. 29:88-94. [DOI] [PubMed] [Google Scholar]
- 52.Lehner, B., G. Williams, R. D. Campbell, and C. M. Sanderson. 2002. Antisense transcripts in the human genome. Trends Genet. 18:63-65. [DOI] [PubMed] [Google Scholar]
- 53.Li, D. S., J. Pastorek, V. Zelnik, G. D. Smith, and L. J. Ross. 1994. Identification of novel transcripts complementary to the Marek's disease virus homologue of the ICP4 gene of herpes simplex virus. J. Gen. Virol. 75:1713-1722. [DOI] [PubMed] [Google Scholar]
- 54.Lilley, B. N., H. L. Ploegh, and R. S. Tirabassi. 2001. Human cytomegalovirus open reading frame TRL11/IRL11 encodes an immunoglobulin G Fc-binding protein. J. Virol. 75:11218-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez, J., and S. C. St Jeor. 1986. Molecular cloning and analysis of three cDNA clones homologous to human cytomegalovirus RNAs present during late infection. J. Virol. 60:531-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mattick, J. S., and I. V. Makunin. 2006. Non-coding RNA. Hum. Mol. Genet. 15(Spec. no. 1):R17-R29. [DOI] [PubMed] [Google Scholar]
- 57.McDonough, S. H., and D. H. Spector. 1983. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology 125:31-46. [DOI] [PubMed] [Google Scholar]
- 58.Merino, E., P. Balbas, J. L. Puente, and F. Bolivar. 1994. Antisense overlapping open reading frames in genes from bacteria to humans. Nucleic Acids Res. 22:1903-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mocarski, E. S., L. Pereira, and N. Michael. 1985. Precise localization of genes on large animal virus genomes: use of lambda gt11 and monoclonal antibodies to map the gene for a cytomegalovirus protein family. Proc. Natl. Acad. Sci. USA 82:1266-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Munroe, S. H., and J. Zhu. 2006. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol. Life Sci. 63:2102-2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy, E., I. Rigoutsos, T. Shibuya, and T. E. Shenk. 2003. Reevaluation of human cytomegalovirus coding potential. Proc. Natl. Acad. Sci. USA 100:13585-13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy, E., D. Yu, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pass, R. 2001. Cytomegalovirus,vol. 2. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 64.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 65.Prang, N., H. Wolf, and F. Schwarzmann. 1995. Epstein-Barr virus lytic replication is controlled by posttranscriptional negative regulation of BZLF1. J. Virol. 69:2644-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prang, N., H. Wolf, and F. Schwarzmann. 1999. Latency of Epstein-Barr virus is stabilized by antisense-mediated control of the viral immediate-early gene BZLF-1. J. Med. Virol. 59:512-519. [DOI] [PubMed] [Google Scholar]
- 67.Prichard, M. N., D. C. Quenelle, D. J. Bidanset, G. Komazin, S. Chou, J. C. Drach, and E. R. Kern. 2006. Human cytomegalovirus UL27 is not required for viral replication in human tissue implanted in SCID mice. Virol. J. 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Randall, G., M. Lagunoff, and B. Roizman. 2000. Herpes simplex virus 1 open reading frames O and P are not necessary for establishment of latent infection in mice. J. Virol. 74:9019-9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reeves, M. B., A. A. Davies, B. P. McSharry, G. W. Wilkinson, and J. H. Sinclair. 2007. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316:1345-1348. [DOI] [PubMed] [Google Scholar]
- 70.Reference deleted.
- 71.Sedmak, D. D., W. H. Roberts, R. E. Stephens, W. J. Buesching, L. A. Morgan, D. H. Davis, and W. J. Waldman.1990. Inability of cytomegalovirus infection of cultured endothelial cells to induce HLA class II antigen expression. Transplantation 49:458-462. [DOI] [PubMed] [Google Scholar]
- 72.Segouffin, C., H. Gruffat, and A. Sergeant. 1996. Repression by RAZ of Epstein-Barr virus bZIP transcription factor EB1 is dimerization independent. J. Gen. Virol. 77:1529-1536. [DOI] [PubMed] [Google Scholar]
- 73.Shendure, J., and G. M. Church. 2002. Computational discovery of sense-antisense transcription in the human and mouse genomes. Genome Biol. 3:RESEARCH0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73a.Smith, J. A., and G. S. Pari. 1995. Human cytomegalovirus UL102 gene. J. Virol. 69:1734-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soderberg-Naucler, C. 2006. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J. Intern. Med. 259:219-246. [DOI] [PubMed] [Google Scholar]
- 75.Spaderna, S., H. Blessing, E. Bogner, W. Britt, and M. Mach. 2002. Identification of glycoprotein gpTRL10 as a structural component of human cytomegalovirus. J. Virol. 76:1450-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stamminger, T., E. Puchtler, and B. Fleckenstein. 1991. Discordant expression of the immediate-early 1 and 2 gene regions of human cytomegalovirus at early times after infection involves posttranscriptional processing events. J. Virol. 65:2273-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reference deleted.
- 78.Stenberg, R. M., D. R. Thomsen, and M. F. Stinski.1984. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 49:190-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stolt, P., and W. Zillig. 1993. Antisense RNA mediates transcriptional processing in an archaebacterium, indicating a novel kind of RNase activity. Mol. Microbiol. 7:875-882. [DOI] [PubMed] [Google Scholar]
- 80.Sun, M., L. D. Hurst, G. G. Carmichael, and J. Chen. 2005. Evidence for a preferential targeting of 3′-UTRs by cis-encoded natural antisense transcripts. Nucleic Acids Res. 33:5533-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80a.Tenney, D. J., and A. M. Colberg-Poley. 1991. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology 182:199-210. [DOI] [PubMed] [Google Scholar]
- 81.Tognon, M., E. Cassai, A. Rotola, and B. Roizman. 1983. The heterogenous regions in herpes simplex virus 1 DNA. Microbiologica 6:191-198. [PubMed] [Google Scholar]
- 82.van Cleef, K. W., M. J. Blok, K. G. Savelkouls, G. E. Grauls, C. A. Bruggeman, and C. Vink. 2005. Identification and characterization of two antisense transcripts from the major immediate early region of rat cytomegalovirus. Arch. Virol. 150:2593-2599. [DOI] [PubMed] [Google Scholar]
- 83.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Voss, J. H., and B. Roizman. 1988. Properties of two 5′-coterminal RNAs transcribed part way and across the S component origin of DNA synthesis of the herpes simplex virus 1 genome. Proc. Natl. Acad. Sci. USA 85:8454-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Waldman, W. J., P. W. Adams, C. G. Orosz, and D. D. Sedmak. 1992. T lymphocyte activation by cytomegalovirus-infected, allogeneic cultured human endothelial cells. Transplantation 54:887-896. [DOI] [PubMed] [Google Scholar]
- 86.Waldman, W. J., W. H. Roberts, D. H. Davis, M. V. Williams, D. D. Sedmak, and R. E. Stephens. 1991. Preservation of natural endothelial cytopathogenicity of cytomegalovirus by propagation in endothelial cells. Arch. Virol. 117:143-164. [DOI] [PubMed] [Google Scholar]
- 87.Wentworth, B. B., and L. French. 1970. Plaque assay of cytomegalovirus strains of human origin. Proc. Soc. Exp. Biol Med. 135:253-258. [DOI] [PubMed] [Google Scholar]
- 88.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wirth, U. V., B. Vogt, and M. Schwyzer. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wright, D. A., and D. H. Spector. 1989. Posttranscriptional regulation of a class of human cytomegalovirus phosphoproteins encoded by an early transcription unit. J. Virol. 63:3117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamaguchi, T., S. L. Kaplan, P. Wakenell, and K. A. Schat. 2000. Transactivation of latent Marek's disease herpesvirus genes in QT35, a quail fibroblast cell line, by herpesvirus of turkeys. J. Virol. 74:10176-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yelin, R., D. Dahary, R. Sorek, E. Y. Levanon, O. Goldstein, A. Shoshan, A. Diber, S. Biton, Y. Tamir, R. Khosravi, S. Nemzer, E. Pinner, S. Walach, J. Bernstein, K. Savitsky, and G. Rotman. 2003. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 21:379-386. [DOI] [PubMed] [Google Scholar]
- 93.Zamore, P. D., and B. Haley. 2005. Ribo-gnome: the big world of small RNAs. Science 309:1519-1524. [DOI] [PubMed] [Google Scholar]
- 94.Zhang, Y., X. S. Liu, Q. R. Liu, and L. Wei. 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res. 34:3465-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang, Z., S. Schwartz, L. Wagner, and W. Miller. 2000. Megablast: a greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7:203-214. [DOI] [PubMed] [Google Scholar]
- 96.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.