Abstract
Lethal mutagenesis is the mechanism of action of ribavirin against poliovirus (PV) and numerous other RNA viruses. However, there is still considerable debate regarding the mechanism of action of ribavirin against a variety of RNA viruses. Here we show by using T7 RNA polymerase-mediated production of PV genomic RNA, PV polymerase-catalyzed primer extension, and cell-free PV synthesis that a pyrimidine ribonucleoside triphosphate analogue (rPTP) with ambiguous base-pairing capacity is an efficient mutagen of the PV genome. The in vitro incorporation properties of rPTP are superior to ribavirin triphosphate. We observed a log-linear relationship between virus titer reduction and the number of rPMP molecules incorporated. A PV genome encoding a high-fidelity polymerase was more sensitive to rPMP incorporation, consistent with diminished mutational robustness of high-fidelity PV. The nucleoside (rP) did not exhibit antiviral activity in cell culture, owing to the inability of rP to be converted to rPMP by cellular nucleotide kinases. rP was also a poor substrate for herpes simplex virus thymidine kinase. The block to nucleoside phosphorylation could be bypassed by treatment with the P nucleobase, which exhibited both antiviral activity and mutagenesis, presumably a reflection of rP nucleotide formation by a nucleotide salvage pathway. These studies provide additional support for lethal mutagenesis as an antiviral strategy, suggest that rPMP prodrugs may be highly efficacious antiviral agents, and provide a new tool to determine the sensitivity of RNA virus genomes to mutagenesis as well as interrogation of the impact of mutational load on the population dynamics of these viruses.
Viruses with RNA genomes are the causative agents of numerous infectious diseases of clinical relevance, and some are considered significant threats as agents of bioterrorism. With the exception of drugs targeting human immunodeficiency virus, few therapeutics exist for the treatment of RNA virus infection. Thus, the development of broad-spectrum antiviral treatments for RNA virus infections remains a crucial medical research goal.
In the past several years, a number of studies have appeared that support the use of lethal mutagenesis as a general antiviral strategy (reviewed in reference 14). RNA viruses replicate with a high error frequency due to the lack of proofreading activity in the virus-encoded polymerases (11, 50). The resulting heterogeneous virus population has often been termed a quasispecies (12). Quasispecies theory predicts the existence of an error threshold, an upper limit to the mutation frequency beyond which genome viability is severely compromised. In one study, poliovirus (PV) viability was reduced 99.3% when the mutation frequency was increased only 9.7-fold beyond the normal level (6). Thus, lethal mutagenesis is an antiviral strategy which aims to increase the mutation rate of RNA viruses beyond the threshold where virus viability can be maintained.
The antiviral nucleoside analogue ribavirin has been demonstrated to act via lethal mutagenesis in vitro against a variety of RNA viruses, including poliovirus (7), hepatitis C virus (28, 60), GB virus B (27), Hantaan virus (48), food-and-mouth disease virus (1), and West Nile virus (9). Furthermore, other antiviral nucleoside and nucleobase analogues have been proposed to act via lethal mutagenesis, most notably 5-hydroxy-2′-deoxycytidine against human immunodeficiency virus (30), 5-azacytidine against foot-and-mouth disease virus (49), and 5-fluorouracil against poliovirus and vesicular stomatitis virus (24), lymphocytic choriomeningitis virus (16, 46), and foot-and-mouth disease virus (41, 42, 49).
However, there is still debate regarding the capacity for ribavirin to act as a lethal mutagen, particularly whether direct incorporation of a mutagen into viral RNA (vRNA) is sufficient for antiviral activity. Ribavirin has been shown to act via a number of different mechanisms, all of which may contribute to its antiviral activity (reviewed in reference 15).
In addition, the applicability of quasispecies theory, error catastrophe, and lethal mutagenesis concepts to viral evolution has been controversial (10, 25, 26, 38). An important theoretical advance was made by Schuster and Swetina in proposing “survival of the flattest” (47). Using digital organisms, Wilke et al. demonstrated that for populations evolving at high mutation rates, high-fitness organisms could be outcompeted by lower-fitness organisms if the latter existed in a “flatter” region of the fitness landscape (59). That is, populations subjected to high mutation rates will evolve to exist in “flat” regions of the fitness landscape where the density of neutral mutations is high. This has become known as “mutational robustness” (54).
Whereas lethal mutagenesis and “survival of the flattest” have been described theoretically and in digital models (58, 59), there has been little advance in these theories from an in vitro or in vivo perspective. The interplay between mutational load, polymerase fidelity, and mutational robustness is largely unexplored. One approach to evaluating these theories in live virus populations is by manipulating population error frequencies through the use of mutagenic nucleoside analogues.
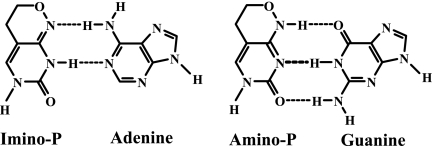
We have previously investigated the use of “universal” bases with minimal hydrogen bonding potential as lethal mutagens of PV (17, 19). Here we investigate the antiviral and mutagenic potential of a nucleoside analogue with the capacity to stably base pair with two natural nucleobases. The ribonucleoside analogue 6-(β-d-ribofuranosyl)-3,4-dihydro-8H-pyrimido[4,5-c][1,2]oxazin-7-one (hereafter referred to as rP) (Fig. 1) is a pyrimidine analogue with degenerate hydrogen-bonding properties (51). Tautomerization of the nucleobase (named P) allows for two distinct configurations of hydrogen bond donors and acceptors, permitting stable base pairing with either adenine or guanine (Fig. 1).
FIG. 1.
P base pairs with adenine and guanine. P exists as two tautomers: imino-P and amino-P. The former hydrogen bonds with adenine; the latter hydrogen bonds with guanine. The imino-P/amino-P ratio is approximately 11:1 (20).
Previous work with the ribonucleoside analogue (rP) and related 2′-deoxyribonucleoside analogue (dP) has established that they can be utilized as substrates by multiple RNA and DNA polymerases (22, 35, 52), and incorporation of rP into a biologically active RNA molecule had little effect on its structure or activity (35). Furthermore, dP was shown to induce the predicted transition mutations in Escherichia coli after phosphorylation through the bacterial thymidine kinase (TK) pathway (40). dP has also been shown to induce C-to-U and U-to-C transition mutations at a high rate in an in vitro retroviral replication model (36). Templates containing rPMP were also able to direct incorporation of either G or A by human immunodeficiency virus and avian myeloblastosis virus reverse transcriptases (52). Accordingly, rP is considered to be an ambiguous pyrimidine nucleoside analogue with high potential to induce mutagenesis if utilized as a substrate by RNA virus polymerases during genome replication. Herein, we evaluated the abilities of rP and rPTP to function as lethal mutagens of poliovirus.
We find that incorporation of rP into PV genomes synthesized in vitro results in substantial loss of RNA specific infectivity, indicating that incorporation of a mutagenic nucleotide into viral RNA is sufficient to reduce virus fitness. Furthermore, rPTP is a highly efficient and promiscuous substrate for the PV-encoded RNA-dependent RNA polymerase (RNAP) 3Dpol. Unfortunately, rP does not have antiviral activity in cell culture, due to insufficient accumulation of the active triphosphate, although treatment with the nucleobase (P) demonstrated mild antiviral activity and mutagenesis. In addition, a poliovirus variant with a high-fidelity polymerase (G64S) demonstrated increased sensitivity to direct incorporation of this nucleotide into transcribed genomes. Direct incorporation of rP into viral RNA genomes can therefore be used to elucidate the effect of mutation frequency on the infectivity of viral genomes and dynamics of the resulting virus population independent of the error rate of the cognate polymerase or the ability of the polymerase to incorporate the analogue.
MATERIALS AND METHODS
Cells and viruses.
HeLa S3 cells (obtained from ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM)/F-12 medium supplemented with 2% dialyzed fetal bovine serum, 50 U/ml penicillin, and 50 μg/ml streptomycin (Invitrogen). Cells were maintained at 37°C in a 5% CO2 atmosphere. Antiviral activity assays were performed as previously described (19). A guanidine resistance assay was performed as previously described (7). Statistical analysis was performed using Prism 4 for Windows (GraphPad Software, Inc.). PV-WT and PV-G64S virus stocks were generated by transfecting HeLa cells with full-length genomic RNA transcribed using T7 RNA polymerase from a plasmid containing the viral cDNA (pMoRA or pMoRA-G64S) as previously described (21, 56). Recovered virus was passaged a minimum of five times in HeLa cells to allow for diversification of the virus population from the initial cDNA sequence.
Expression and purification of T7 RNAP.
T7 RNAP was purified as previously described for PV 3Dpol (13) with the following modifications: (i) ammonium sulfate was added to 40% saturation; (ii) the phosphocellulose column was eluted using a linear gradient (six column volumes) from 50 mM to 700 mM NaCl in buffer A; (iii) the Q-Sepharose column was loaded and washed and the protein eluted using a linear gradient (six column volumes) from 50 mM to 400 mM NaCl in buffer A. Protein-containing fractions were pooled as before. Additional steps to concentrate the protein were not necessary.
Incorporation of RTP by T7 RNA polymerase.
Incorporation by T7 RNAP opposite templating bases cytosine or thymine in assembled enzyme-primer/template complexes was examined for ribavirin triphosphate (RTP) and the correct natural nucleotide (GTP opposite cytosine, ATP opposite thymine). Assay conditions were modified from those described in reference 53. The RNA primer sequence was 5′-UUUUGCCGCGCC-3′. The DNA template sequence providing templating C (underlined) was 5′-GGGAATGTACGGCGCGGC-3′. The DNA template sequence providing templating T (underlined) was 5′-GGGAATGCATGGCGCGGC-3′. RNA primer was 5′-end labeled with [γ-32P]ATP by using T4 polynucleotide kinase according to the manufacturer's protocol (New England BioLabs). Annealing of 32P-end-labeled RNA primer with DNA template was accomplished by heating to 90°C for 1 min followed by cooling to 10°C at a rate of 5°C per min. T7 RNAP was allowed to preassemble with the primer/template duplex for 10 min at room temperature immediately prior to reactions. Reactions were at 30°C for 30 or 180 s in 50 mM HEPES pH 7.5, 10 mM 2-mercaptoethanol, and 5 mM MgCl2 with 2 μM T7 RNAP, 1 μM primer/template duplex, and 0.5 mM RTP or natural nucleotide. Reactions were initiated by addition of nucleotide and terminated by quenching in 100 mM EDTA-90% formamide gel loading dye. Reaction products were separated by denaturing polyacrylamide gel electrophoresis (PAGE), and gels were visualized using a Typhoon 8600 variable mode imager (Molecular Dynamics) as previously described (13).
In vitro transcription by T7 RNA polymerase.
A plasmid containing the PV genomic cDNA under the control of a T7 promoter (pMoRA) (21) was linearized via digestion with the restriction enzyme ApaI (New England BioLabs). The transcription reaction mixture contained 350 mM HEPES (pH 7.5), 32 mM magnesium acetate, 40 mM dithiothreitol, 2 mM spermidine, 7 mM each nucleoside triphosphate (ATP, CTP, GTP, and UTP), 0.5 μg linearized plasmid DNA, and 0.5 μg purified T7 RNA polymerase in a final volume of 0.02 ml. rPTP was added at various concentrations with an equimolar amount of magnesium acetate. The mixture was incubated for 3 h at 37°C followed by a 2-min centrifugation at 14,000 × g. The pellet was discarded, and 2 U of RQ1 DNase (Promega) was added to the supernatant, followed by incubation at 37°C for an additional 30 min to digest template DNA. RNA was precipitated by the addition of 0.05 ml of 7.5 M lithium chloride with 50 mM EDTA. This mixture was frozen on dry ice for 15 min and centrifuged at 14,000 × g for 30 min at 4°C. The supernatant was discarded, and the RNA pellet was washed twice with 70% ethanol, air dried, and resuspended in deionized water. Quality of full-length genomic RNA was verified by agarose gel, and RNA was quantitated by fluorescence in the presence of ethidium bromide by comparing to a known RNA standard.
Digestion of T7-transcribed RNA to component nucleosides.
RNA digestion was performed essentially as previously published (5). Briefly, RNA suspended in ∼20 μl deionized water (approximately 2 μg/μl) was denatured at 100°C for 3 min and then rapidly chilled in an ice water slush. A 1/10 volume of 0.1 M ammonium acetate (pH 5.3) was added, followed by addition of 2 U nuclease P1 (MP Biochemicals) and incubation for 2 h at 45°C. A 1/10 volume of 1 M ammonium bicarbonate (pH 7.8) was then added to the digest, followed by 1 U shrimp alkaline phosphatase (USB Corporation) and further incubation for 1 h at 37°C. The solution was heated to 95°C for 10 min to inactivate enzymes prior to high-performance liquid chromatography (HPLC) separation.
HPLC separation and detection method.
HPLC separations were performed on a Hewlett Packard (Agilent) 1100 series instrument equipped with an in-line solvent degasser and diode array detector. RNA digests were separated on an Aquasil C18 column (4.6 by 250 mm, 5 μm; Keystone Scientific Inc., Thermo Electron Corp.) and eluted with the following linear gradient (1-ml/min flow rate): 1 to 95% acetonitrile in 100 mM monobasic potassium phosphate buffer (KH2PO4, pH 6.0) over 15 min. The phosphate buffer was prepared by dissolving KH2PO4 in distilled and deionized water (ddH2O) at 100 mM and adjusting the pH to 6.0 by addition of 10% KOH (aqueous). Acetonitrile employed in the separation was HPLC grade. The elution of ribonucleosides (cytidine, uridine, adenosine, and guanosine) was detected at 254 nm; elution of rP was detected at 295 nm. Injection volumes for treatments ranged from 7.5 μl to 15 μl; the injector port was fitted with a 20-μl injection loop. Peak areas were obtained by standard integration of the appropriate peak by the ChemStation for LC 3D software (rev. A.09.03; Agilent Technologies).
Generation of standard stocks and fitting of data points.
rP was dissolved in ddH2O at a 2.5 mM concentration, and serial dilutions were prepared. Ribonucleoside standards were prepared by serial dilution of 25 mM stocks of cytidine and uridine (in ddH2O) and guanosine and adenosine (in dimethyl sulfoxide). Individual analyses of a range of ribonucleoside concentrations were performed for each standard, and the resulting peak areas were plotted against the quantity of material injected (in nmoles). Linear regression analysis of the tabulated calibration lines found an R2 of >0.99 for each of the standards.
Identification of peaks and tabulation of the abundance of P in transcribed RNA.
The identities of nucleosides from the RNA digestion were assigned by doping the transcription mixture with known standards and observing an increase in the area of the corresponding peak (compared with nondoped analyses). The relative abundance of each nucleoside in the transcriptional mix was determined by fitting the area of the separated peak to the corresponding standard line formula. The percentage of rP in each RNA digestion was tabulated by dividing the nanomoles of rP by the total nanomoles of nucleosides in the sample. Each digestion was analyzed in triplicate, with the mean representing the average of these individual analyses. As a control to ensure that residual rPTP was adequately washed away from transcribed RNA, a control experiment was performed in which transcription was performed with only the four natural nucleotides. After digestion of the DNA template and immediately prior to lithium chloride precipitation, rPTP was added at the highest concentration used for transcription. This RNA was precipitated, washed, and digested, and the nucleosides were separated by HPLC as described above. No rP was detected in this control experiment, indicating that unincorporated nucleotides were adequately removed from the RNA solution prior to digestion and separation (data not shown).
Transfection and infectious center assays.
An infectious center assay to determine the specific infectivity of transcribed PV genomic RNA was performed as previously described (6). HeLa S3 cells were detached via treatment with trypsin-EDTA (Invitrogen), washed with 1× phosphate-buffered saline (PBS), and resuspended in 1× PBS at 3 × 106 cells/ml. The cell suspension (0.4 ml) was mixed with 0 to 10 μg T7-transcribed RNA and transferred to an electroporation cuvette (0.2-mm gap; VWR International). Electroporation was performed using a Gene Pulser (Bio-Rad) set at 500 μF and 130 V. Electroporated cells were serially diluted in 10-fold increments in PBS, and 0.1 ml of each dilution and the undiluted electroporated cell suspension was plated onto a subconfluent HeLa S3 monolayer plated the previous day at 5 × 105 cells/well in six-well plates containing 1.5 ml of growth medium. Cells were incubated for 1 h at 37°C to allow for attachment. Wells were aspirated, and the monolayers were covered with 3 ml of DMEM/F-12 with 2% dialyzed fetal bovine serum and 1× penicillin-streptomycin supplemented with 1% low-melting-point agarose (American Bioanalytical). Plates were incubated for 3 days at 37°C before removal of the agar plug and staining with 0.1% crystal violet solution in 20% ethanol for visualization of plaques. For the experiments shown in Fig. 4B, below, data were normalized such that for each individual experiment the number of plaques resulting from RNA transcribed in the absence of rPTP was set to 100.
FIG. 4.
rPMP incorporation into PV genomic RNA results in a dose-dependent decrease in specific infectivity. (A) HeLa S3 cells were transfected with various concentrations of in vitro-transcribed RNA and serially diluted on HeLa S3 monolayers. Resulting plaques increased linearly up to ∼5 μg. (B) PV genomic RNA was transcribed in vitro in the presence of various concentrations of rPTP, and infectivity was determined in an infectious center assay. The number of rPMP incorporations per genome is plotted on the x axis, as determined by extrapolating the data in Fig. 3B for transcription in the presence of various amounts of rPTP. Specific infectivity was normalized such that for each experiment the number of plaques resulting from RNA transcribed in the absence of rPTP was set to 100. The mean and standard deviation of at least three independent samples are shown for each data point.
For the experiment comparing wild-type and G64S PV, RNA was transfected as described above. Following electroporation, 0.2 ml of cell suspension was added to 0.8 ml of growth medium and incubated in a 37°C water bath for 6 h. The cell suspension was then subjected to three freeze-thaw cycles for cell lysis and centrifugation for 2 min at 14,000 × g to remove cellular debris. The supernatant titer was determined on HeLa cell monolayers for 3 days followed by staining with crystal violet solution.
Nucleotide incorporation by PV 3Dpol in vitro.
PV 3Dpol was expressed and purified as previously described (13). Extension assays utilizing symmetrical primer/template substrates (s/s) were performed as described elsewhere (2). s/s RNAs were synthesized by Dharmacon, Inc., and 5′-end labeled with [γ-32P]ATP by using T4 polynucleotide kinase according to the manufacturer's protocol (New England BioLabs). Annealing of 32P-end-labeled RNA primer to form the duplex substrate was accomplished by heating to 90°C for 1 min followed by cooling to 10°C at a rate of 5°C per min. 3Dpol was incubated with the appropriate s/s duplex for 90 s at 30°C to allow formation of preinitiation enzyme-RNA complexes. Extension reactions were initiated by the addition of nucleotide and incubated at 30°C for the indicated times. The initiated reaction mixture contained 1 μM 3Dpol, 1 μM s/s (0.5 μM duplex), 50 mM HEPES (pH 7.5), 10 mM 2-mercaptoethanol, 5 mM MgCl2, 60 μM ZnCl2, and nucleoside triphosphate as indicated for each experiment. Reactions were quenched by addition of EDTA (pH 8.0) to 50 mM. Polymerase was diluted immediately prior to use in 50 mM HEPES (pH 7.5), 10 mM 2-mercaptoethanol, 60 μM ZnCl2, and 20% glycerol. For all experiments, 100 μM nonradiolabeled s/s “trap” was added along with initiating nucleotide to prevent reinitiation of dissociated enzyme. Product was added to an equal volume of loading buffer (90% formamide, 0.025% bromphenol blue, and 0.025% xylene cyanol) and heated to 65°C prior to loading on a denaturing polyacrylamide gel containing 23% acrylamide, 1× TBE (89 mM Tris base, 89 mM boric acid, and 2 mM EDTA), and 7 M urea. Electrophoresis was performed in 1× TBE at 80 W for ∼2 h. Products were visualized using a Typhoon 8600 variable mode imager (Molecular Dynamics) as previously described (13). Quantitation was performed using ImageQuant software (Molecular Dynamics) and fit by nonlinear regression using KaleidaGraph 3.5 software (Synergy Software, Reading, PA).
Thymidine kinase assay.
Purified recombinant herpes simplex virus type 1 (HSV-1) TK and ganciclovir (GCV) were a gift of Richard R. Drake (Eastern Virginia Medical School). Nucleoside substrates were assayed as described previously (23). The reaction mixture consisted of 0.01 to 1 mM nucleoside, 20 mM potassium phosphate (pH 7.6), 1 mM dithiothreitol, 0.160 μM ATP, 0.106 μM [α-32P]ATP (MP Biochemicals), 5 mM MgCl2, 25 mM NaF, 40 mM KCl, and 0.5 mg/ml bovine serum albumin. The mixture was incubated for 5 min at 37°C prior to addition of purified HSV-1 TK to a final concentration of 3.85 μg/ml. The mixture was incubated at 37°C for 5 min and then quenched by addition of EDTA (pH 8.0) to 80 mM. One μl of the quenched reaction mixture was spotted on polyethyleneimine-cellulose F thin-layer chromatography plates (EMD Chemicals) and resolved with 300 mM potassium phosphate, pH 7.0. The thin-layer chromatography plate was exposed to a phosphor screen (Molecular Dynamics) for 30 min and visualized using a Typhoon 8600 variable mode imager (Molecular Dynamics). Quantitation and curve fitting were performed using ImageQuant 5.0 (Molecular Dynamics) and KaliedaGraph 3.5 (Synergy Software).
Selection of a HeLa S3 cell line expressing HSV-1 TK.
A mammalian expression plasmid containing the HSV-1 TK gene (pLTKEN) was provided by Richard R. Drake (Eastern Virginia Medical School) (32). HeLa S3 cells were transfected with pLTKEN via lipofection with DMRIE-C reagent (Invitrogen) according to the manufacturer's protocol. Stably transfected cells were selected in the presence of 0.4 mg/ml G418 sulfate (Invitrogen). After 2 weeks of selection, 12 colonies were isolated, expanded, and screened for sensitivity to 25 μM ganciclovir. Five clones demonstrating robust growth and sensitivity to ganciclovir were then screened for HSV-1 TK expression by lysing cells (4 × 107 cells/ml) in 1× cell culture lysis reagent (Promega). Total protein concentration was determined by the Bradford assay (Bio-Rad Laboratories), and 15 μg of protein from each sample was separated via sodium dodecyl sulfate (SDS)-PAGE using a 10% acrylamide gel followed by transfer to nitrocellulose. HSV-1 TK was detected via Western blotting using rabbit HSV-1 TK polyclonal antiserum (provided by Margaret E. Black, Washington State University) and goat anti-rabbit immunoglobulin G-horseradish peroxidase. The blot was developed using Amersham ECL Western blotting detection reagents and exposed to X-ray film. The clone showing the highest expression of HSV-1 TK (hereafter referred to as HeLa-TK) was expanded and used exclusively for further experiments.
Nucleotide extraction from HeLa and HeLa-TK cells.
Nucleotide extraction was modified from a previously published procedure (45). A total of 7.5 × 106 HeLa S3 or HeLa-TK cells were plated in a 100-mm dish 15 to 18 h before treatment. Cells were treated with 2.5 μg/ml actinomycin D (Sigma) for 15 min at 37°C, and then rP was added to the medium to a final concentration of 2 mM. Cells were incubated for 3 h at 37°C. After incubation, the medium was aspirated, plates were washed with 5 ml PBS, and 1 ml prewarmed trypsin-EDTA solution (Invitrogen) was added to each plate. Cells were incubated for 5 min at 37°C to facilitate detachment, after which cells were collected, pelleted, and resuspended in 0.05 ml ice-cold 0.6 M trichloroacetic acid (Sigma). The cell suspension was incubated on ice for 10 min and then centrifuged at 14,000 × g for 2 min at 4°C. The supernatant was collected and extracted with an equal volume of ice-cold 0.5 M trioctylamine in 1,1,2-trichlorotrifluoroethane (Sigma). Samples were then vortexed for 30 s and centrifuged for 30 s at 14,000 × g and 4°C. The upper (aqueous) layer was removed and analyzed on a Hewlett Packard 1100 series instrument equipped with an Aquasil C18 analytical column (4.6 by 250 mm, 5 μm; Keystone Scientific Inc., Thermo Electron Corp.) running the following mobile phase (flow rate, 1 ml/min): gradient of 1% to 15% CH3CN in 100 mM potassium phosphate buffer (KH2PO4, pH 6.0) over 20 min, followed by 15 to 80% CH3CN in 100 mM KH2PO4 buffer over 10 min.
Cells and viruses for cell-free translation and replication.
HeLa R19 cell monolayers and suspension cultures of HeLa S3 cells were maintained in DMEM supplemented with 10% fetal bovine calf serum. Poliovirus for vRNA preparation was amplified on HeLa R19 cells as described previously (31). The infectivity of virus stocks was determined by plaque assays on HeLa R19 monolayers, as described previously (31).
Preparation of poliovirus vRNA.
Virus stocks were grown and purified by CsCl gradient centrifugation (31). Viral RNA was isolated from the purified virus stocks with a 1:1 mixture of phenol and chloroform. The purified RNA was precipitated by the addition of two volumes of ethanol.
Preparation of HeLa cytoplasmic extracts.
HeLa S10 extracts were prepared as previously described (8, 33) except for the following modifications: (i) packed cells from 2 liters of HeLa S10 were resuspended in 1.0 volume (relative to packed cell volume) of hypotonic buffer; (ii) the final extracts were not dialyzed.
Translation-RNA replication reactions with HeLa cell extracts and plaque assays.
Viral RNA was translated at 34°C in the presence of unlabeled methionine, 200 μM each of CTP, GTP, and UTP, and 1 mM ATP in a total volume of 25 μl (8, 33). After incubation for 12 to 15 h, the samples were diluted with phosphate-buffered saline and were added to HeLa cell monolayers. Virus titers were determined by plaque assay, as described previously (31). A 1 mM concentration of rPTP was added to the reaction mixture as indicated.
Translation-RNA replication reactions with HeLa cell extracts and in vitro translation.
Translation reaction mixtures (25 μl) containing 8.8 μCi of Trans35S-label (ICN Biochemicals) and vRNA were incubated for 12 h at 34°C (8). A 1 mM concentration of rPTP was added to the reaction mixture as indicated. The samples were analyzed by electrophoresis on SDS-12% polyacrylamide gels, followed by autoradiography.
Translation-RNA replication reactions with HeLa cell extracts and luciferase assay.
The P/L replicon (wild type) (29) was used to measure the luciferase activity in translation-RNA replication reactions. The P/L replicon was linearized with DraI (New England BioLabs) prior to transcription by T7 RNA polymerase. The transcript RNA was purified by phenol-chloroform extraction and ethanol precipitation. After the P/L replicon was translated for 12 h at 34°C in the presence of unlabeled methionine, 200 μM each of CTP, GTP, and UTP and 1 mM ATP in a total volume of 25 μl (8, 33, 34), the total reaction mixture was transferred to a tube. A 100-μl aliquot of luciferase assay reagent (Promega) was mixed with 25 μl of lysate, and the firefly luciferase activity was measured in an Optocomp I luminometer (MGM Instruments, Inc.). Puromycin (Sigma) was added to the reaction mixture as indicated.
RESULTS
Incorporation of ribavirin triphosphate by T7 RNA polymerase.
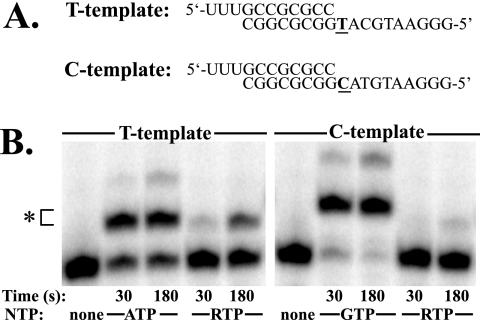
Lethal mutagenesis as the mechanism of action of ribavirin could be supported by synthesizing PV genomic RNA containing a known number of ribavirin monophosphate (RMP) substitutions and evaluating the phenotype of virus populations produced from these RNAs. The synthesis of P is described in supplemental material which can be found at (http://bmb.psu.edu/faculty/cameron/lab/JVI2007Supplement.html). We investigated the ability of T7 RNAP to utilize RTP as a substrate by using a primer-extension assay. As shown in Fig. 2, RTP was a poor substrate for T7 RNAP compared with the correct nucleotides when either C or T was used as the first templating base. Thus, poor incorporation of RTP by T7 RNA polymerase precludes its incorporation into PV genomic RNA in vitro at a frequency sufficient to reduce genome infectivity.
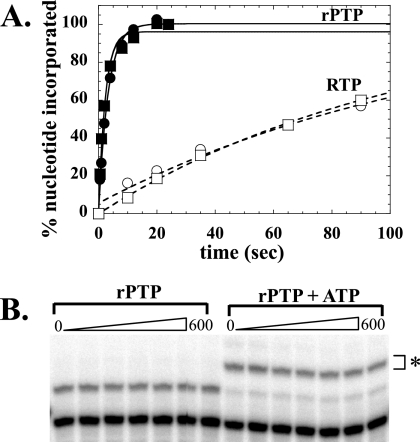
FIG. 2.
RTP is not an efficient substrate for T7 RNAP. (A) Substrates used for the T7 RNAP extension assay are illustrated with DNA template (top strand) and RNA primer (bottom strand). The templating nucleotide is indicated in bold. (B) Primer extension by T7 RNAP is shown after 30 and 180 seconds when either RTP or the correct nucleotide was provided as substrate. RMP was incorporated inefficiently compared to the correct (natural) substrate. The asterisk indicates an 11-mer extended RNA substrate.
Incorporation of rP into PV genomic RNA in vitro.
The nucleotide analogue rPTP has been established as an ambiguous substrate for T7 RNAP (35). To determine the effect of rP incorporation into PV genomic RNA, rPTP was added along with all four naturally occurring nucleotides during in vitro transcription of the PV genome by T7 RNAP. No significant reduction in the RNA yield by T7 transcription was observed when rPTP was added at up to 20% of the total nucleotide concentration, indicating that rPTP does not cause inhibition of RNA synthesis or premature termination when utilized as a substrate for T7 RNAP under these conditions (Fig. 3C).
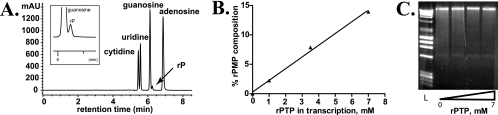
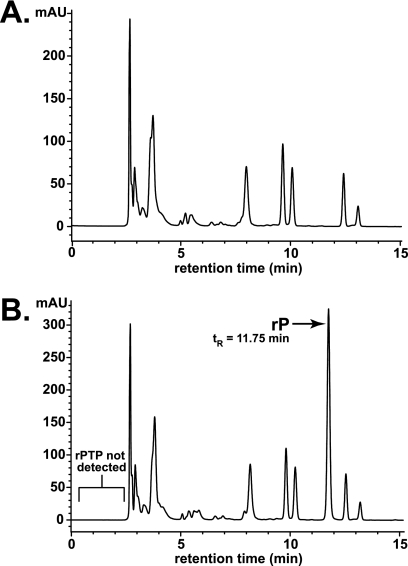
FIG. 3.
rPMP is incorporated into genomic RNA during transcription with T7 RNAP. (A) HPLC separation of RNA transcribed in the presence of rPTP. RNA was digested to component nucleosides as described in the text. rP was observed along with the four natural nucleosides. (B) rPMP composition of RNA (as determined by HPLC of digested RNA) is plotted against the concentration of rPTP present in the in vitro transcription reaction mixture. The PV genome is 7.5 kb in length. (C) RNA transcribed in the presence of increasing concentrations of rPTP was run on a 1% agarose gel and stained with ethidium bromide. Differences in RNA quality or length were not observed when rPTP was added to the transcription reaction mixture at up to 20% of the total nucleotide pool. L, DNA ladder.
To demonstrate incorporation of rPMP into transcribed RNA, the RNA was precipitated by lithium chloride followed by washing with 70% ethanol to remove residual nucleotides. The RNA was then digested to its nucleoside constituents using nuclease P1 and shrimp alkaline phosphatase. Nucleoside products were separated via reversed-phase HPLC and detected by UV absorbance. A peak corresponding to rP could be seen for RNA transcribed in the presence of PTP (Fig. 3A). The identity of each peak was verified by doping the RNA digest with standards of rP and all four naturally occurring nucleosides (data not shown).
Quantitation of the rP peak in relation to the four naturally occurring nucleoside peaks provided a measure of the incorporation of rPMP into RNA by T7 RNAP when various concentrations of rPTP were added to the in vitro transcription reaction mixture. The rP content of RNA increased linearly with increasing concentrations of rPTP available during transcription (Fig. 3B). When rPTP was added equimolar to each of the four natural nucleotides (7 mM), 15% of the digested RNA product consisted of rP.
Specific infectivity of PV genomic RNA containing rP.
The infectivity of PV genomic RNA containing rP was measured by infectious center assay as previously described (6). PV genomic RNA was transcribed as described above and transfected into HeLa S3 cells by electroporation. Serial dilutions of transfected cells were then added to subconfluent HeLa S3 monolayers and incubated under an agar overlay for 3 days. Replication-competent RNA genomes launch an authentic PV infection upon transfection, resulting in virus spread and a plaque in the cell monolayer. This allows for quantitation of infectious RNA genomes.
To determine the concentrations of RNA that would result in a linear response for plaque formation, various amounts of T7 RNAP-transcribed PV genomic RNA were transfected into HeLa cells (Fig. 4A). The number of productively infected cells (as measured by subsequent plaque formation) was found to be linear when up to 5 μg of RNA was transfected. Three μg was used for all subsequent experiments.
PV genomic RNA was transcribed by T7 RNAP in the presence of various concentrations of rPTP. When this RNA was transfected into HeLa S3 cells, an rPTP-dependent decrease in RNA infectivity was observed (Fig. 4B). Specific infectivity was defined as the number of plaques observed per μg of RNA per ml of transfected cells. Specific infectivity was normalized such that the number of plaques resulting from transfection with RNA containing no rPMP was set to 100. Data described above for the amount of rP in RNA based on transcription conditions (Fig. 3B) were extrapolated to estimate the number of rPMP incorporations per genome required to provide a given reduction in genome specific activity. Approximately 20 molecules of rPTP incorporated per genome (∼0.3% nucleotide composition) were sufficient for a 10-fold reduction in specific infectivity. Thus, the presence of rP in viral genomes has a dramatic effect on the ability of those genomes to launch productive infections.
Incorporation of rP into RNA by PV 3Dpol.
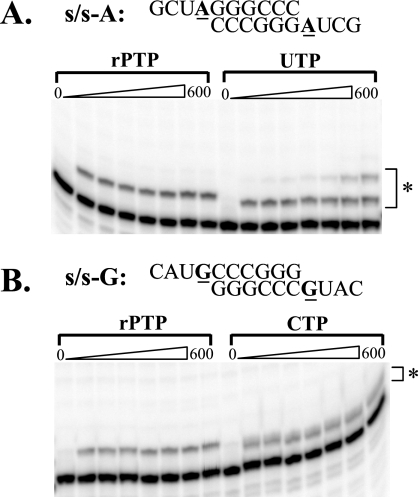
To confirm that rP acts as an ambiguous nucleoside during poliovirus replication, the ability of the PV RNA-dependent RNA polymerase (3Dpol) to utilize rPTP as a substrate was determined using an in vitro primer-extension assay. Symmetrical primer/template substrates (s/s) were utilized as previously described (2). Purified PV 3Dpol was mixed with RNA duplex primer/template for 90 s to allow for the formation of stable elongation complexes, at which time nucleotide substrate was added. The reaction was stopped at various times post-nucleotide addition, and product was separated by denaturing PAGE (Fig. 5). rPMP was efficiently incorporated into RNA by 3Dpol when the templating base was either adenosine or guanosine. As indicated by the kpol/Kdapp, rPMP (from the rPTP substrate) was incorporated into RNA by PV 3Dpol approximately 100-fold more efficiently than incorporation of the known mutagen ribavirin (Table 1). Furthermore, incorporation was equally efficient opposite either adenosine or guanosine, confirming the promiscuous base-pairing properties of rP (Fig. 6A). Incorporation was not detected when cytidine or uridine was utilized as the templating base (data not shown).
FIG. 5.
PV polymerase incorporates rPMP opposite both adenine and guanine in RNA. Either s/s-A (A) or s/s-G (B) was employed. Utilization of rPTP was compared directly to utilization of the correct nucleotide: UTP (A) or CTP (B). Polymerase-s/s complexes were assembled and nucleotide added to a final concentration of 500 μM. Product formed after 15, 30, 45, 60, 120, 300, and 600 s was separated from substrate by denaturing PAGE and visualized by phosphorimaging. Extended RNA product is indicated by the asterisks.
TABLE 1.
Kinetic and thermodynamic constants for PV 3Dpol-catalyzed nucleotide incorporation
| Primer and templatea | Nucleotide | Kd(app) (μM) | kpol (s−1) | kpol/Kd(app) (s−1 μM−1) |
|---|---|---|---|---|
| s/s-A | ||||
| GCUAGGGCCC | UTPb | 98 ± 2 | 266 ± 2 | 2.7 |
| CCCGGGAUCG | rPTP | 134 ± 48 | 0.47 ± 0.05 | 0.0035 |
| s/s-C | ||||
| GAUCGGGCCC | GTPb | 3.8 ± 0.7 | 57 ± 3 | 15 |
| CCCGGGCUAG | RTPc | 430 ± 79 | 0.019 ± 0.002 | 0.000044 |
| s/s-G | ||||
| CAUGCCCGGG | CTPb | 19 ± 3 | 157 ± 8 | 8.2 |
| GGGCCCGUAC | rPTP | 132 ± 29 | 0.59 ± 0.05 | 0.0045 |
| s/s-U | ||||
| CGAUGGGCCC | ATPb | 134 ± 18 | 86.7 ± 3.7 | 0.86 |
| CCCGGGUACG | RTPc | 496 ± 21 | 0.014 ± 0.001 | 0.000028 |
FIG. 6.
PV 3Dpol utilizes rPTP more efficiently than ribavirin triphosphate. (A) Poliovirus polymerase 3Dpol was incubated with s/s for 120 s prior to initiating the reaction by addition of the appropriate nucleotide to a final concentration of 500 μM. The solid and dashed lines represent fits of the data to a single exponential with kobs of 0.30 ± 0.02 s−1 and 0.44 ± 0.05 s−1 for rPMP incorporation opposite A (•) and G (▪) and kobs of 0.008 ± 0.001 s−1 and 0.009 ± 0.004 s−1 for rRMP incorporation across U (○) and C (□), respectively. (B) A chain termination experiment was performed with s/s-A as described in the legend for Fig. 6A. Incorporation was monitored in the presence of rPTP alone (left panel) or in the presence of rPTP and ATP, the next correct nucleotide (right panel). Quantitative incorporation of AMP was observed (the n + 2 product is indicated by the asterisk).
Next, the ability of PV 3Dpol to further extend RNA having rPMP at the terminal 3′ position was evaluated. An extension reaction using s/s-A was performed as described above, but with ATP added to the reaction mixture along with rPTP. AMP should be incorporated in the second templated position immediately after incorporation of rPMP, resulting in quantitative extension of the 10-mer s/s-A substrate to a 12-mer product (Fig. 6B). AMP was found to be incorporated quantitatively as the +2 nucleotide, indicating that incorporation of rPMP does not result in “chain termination” by preventing further extension at the 3′ end of the RNA. These experiments suggest that rPTP can be templated efficiently by either cytidine or uridine during poliovirus RNA replication and that incorporation of rPMP should not result in premature termination of the nascent RNA.
Treatment of HeLa S3 cells with rP during PV infection.
In light of the observation that rPMP incorporation causes a reduction in specific infectivity of PV genomic RNA, the antiviral properties of rP were evaluated against PV in cell culture. HeLa S3 cells were pretreated with rP for 1 h and infected with PV at a multiplicity of infection of 0.01 in order to maximize the cumulative effects of replication in the presence of a mutagen. PV infection was allowed to proceed in the presence of rP until all cells demonstrated cytopathic effect, at which time virus was collected from the medium and the titer was determined. Treatment of cells with up to 2 mM rP failed to reduce the time required to reach cytopathic effect and did not cause a reduction in virus titer (data not shown). Furthermore, analysis of cellular nucleotide pools (as described below) failed to detect any phosphorylated rP metabolites in extracts of HeLa cells treated with 2 mM rP for 3 h (data not shown).
rP is a substrate for HSV-1 thymidine kinase.
The failure of rP to exhibit antiviral activity could potentially be due to inefficient phosphorylation of the nucleoside by cellular nucleoside and nucleotide kinases, which would prevent accumulation of the nucleoside triphosphate necessary for incorporation into RNA. The rate-limiting step for intracellular accumulation of a nucleoside triphosphate in cells is often the initial phosphorylation that results in production of the nucleoside monophosphate (55). TK from HSV-1 is a nucleoside kinase with broad substrate specificity (44). We examined the ability of HSV-1 TK to phosphorylate rP in vitro using purified HSV-1 TK. For comparison, the natural substrate thymidine (T) and the antiherpesvirus nucleoside analogue GCV were evaluated in parallel. Steady-state kinetics produced the kinetic constants in Table 2. The kcat/Km for rP was reduced 10-fold relative to that for T and 3-fold relative to that for GCV, primarily due to an increase in the Km. Nonetheless, HSV-1 TK is able to phosphorylate rP in vitro with an efficiency comparable to known in vivo nucleoside substrates.
TABLE 2.
Steady-state kinetic constants of HSV-1 thymidine kinasea
| Nucleoside or analogue | Km (μM) | kcat (10−3 s−1) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| T | 1.7 ± 1.1 | 3.5 ± 0.3 | 2,100 |
| GCV | 7.8 ± 2.9 | 4.0 ± 0.3 | 510 |
| rP | 29 ± 9.1 | 5.5 ± 0.6 | 190 |
Values are means ± standard deviations.
We reasoned that expression of HSV-1 TK in HeLa cells may be sufficient for activation of rP, resulting in antiviral activity due to lethal mutagenesis. To this end, we introduced an HSV-1 TK expression plasmid conferring neomycin resistance (32) into HeLa cells and selected stably transfected cells by addition of 400 μg/ml G418 sulfate to the culture medium. Colonies were screened for sensitivity to GCV, and then a Western blot assay was performed to assess the degree of expression of HSV-1 TK (data not shown). The colony with the most favorable expression, referred to as HeLa-TK, was expanded and used for subsequent experiments.
Determination of intracellular phosphorylation states of rP in HeLa-TK cells.
To investigate the intracellular metabolism of rP, HeLa-TK cells were treated with 2 mM rP for 3 h, followed by nucleotide extraction and HPLC analysis of nucleotide pools as previously described (45). Cells were treated with actinomycin D for 15 min prior to addition of rP to the medium in order to prevent incorporation of phosphorylated rP metabolites into cellular RNA. Phosphorylated forms of rP were not detected by this method (Fig. 7). However, the unmodified nucleoside was readily detected, indicating that the absence of phosphorylated metabolites was not due solely to exclusion of the nucleoside from the cytoplasmic compartment. Furthermore, treatment with rP did not reduce the titer of PV produced by infected HeLa-TK cells (data not shown).
FIG. 7.
rPTP is not detected in HeLa-TK cells treated with rP. (A) Separation of extracts from untreated HeLa-TK cells (100-μl injection). (B) Separation of extracts from HeLa-TK cells that were treated with 2 mM rP for 3 h (100-μl injection). rP (retention time [tR], 11.8 min) was clearly observed in treated extracts. No new peaks with rPTP retention (tR, 2.2 min) or characteristic UV trace were observed. The absorbance wavelength for all traces was 295 nm.
P exhibits mild antiviral activity and mutagenesis of PV.
Treatment of HeLa cells with rP did not result in detectable levels of rPTP in cells and did not exhibit any observable antiviral effect or increased mutagenesis of PV genomes. This may be due to the inability of rP to be phosphorylated through the cellular de novo nucleotide metabolism pathways. However, administration of the nucleobase (P) might allow metabolism via the cellular nucleotide salvage enzymes, resulting in accumulation of the active triphosphate to sufficient intracellular levels. To explore this possibility, we treated HeLa S3 cells with P at concentrations of up to 2 mM. No overt cellular toxicity was observed by visual inspection through 3 days of continuous exposure to the nucleobase at these concentrations (data not shown).
P was tested for antiviral activity against PV under one-step growth conditions. HeLa S3 cells were pretreated with P for 1 hour prior to infection with PV (MOI, 5). Fresh P in medium was then added prior to incubation for 6 h at 37°C, followed by collection of cell-associated virus and determination of virus titer. A mild decrease in titer was observed, approximately twofold with treatment with 0.5 mM or higher (Fig. 8A).
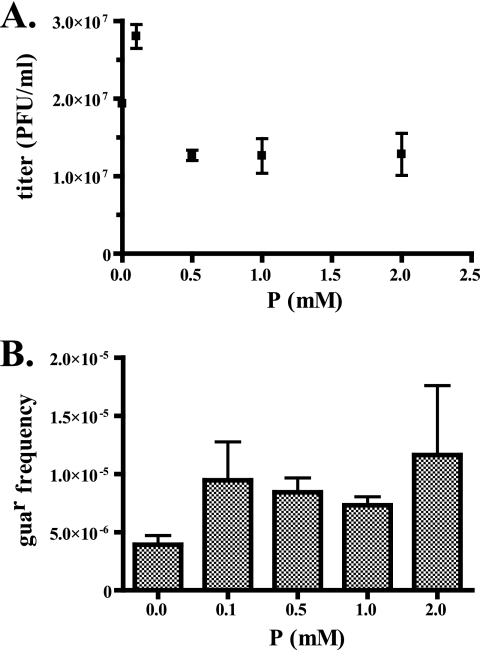
FIG. 8.
P exhibits mild antiviral and mutagenic properties. (A) HeLa cells were pretreated for 1 hour with P and then infected with PV (MOI, 5), followed by a 6-h incubation at 37°C. Cell-associated virus was recovered by freeze-thaw, and the titer was determined. Experiments were performed in triplicate. (B) A guanidine resistance assay was performed in triplicate as described in the text.
To probe the mechanism responsible for this antiviral activity, the mutation frequencies of the resulting virus populations were estimated using a previously established assay for guanidine resistance (7). Poliovirus replication is completely inhibited in the presence of 3 mM guanidine hydrochloride, but a single transition mutation is sufficient to restore viability. Hence, measurement of the frequency of guanidine-resistant PV variants can be used as an estimate of mutation frequency. When PV, grown in the presence of various concentrations of P, was assayed in this manner, a mild increase (twofold) in guanidine-resistant variants was detected (Fig. 8B). However, this increase was not statistically significant when analyzed by analysis of variance (P > 0.05) (37).
Antiviral activity of rPTP in a PV cell-free replication system.
To examine the effects of rPTP on poliovirus infection, we utilized a well-established cell-free replication system employing HeLa cell extracts (33). When programmed with PV genomic RNA, translation, replication, and production of infectious virus particles can be observed in these extracts. Addition of 1 mM rPTP to the cell-free translation reaction mixture resulted in no reduction in translation of replicon RNA as monitored by luciferase production, whereas addition of the translation inhibitor puromycin caused a complete block of luciferase production (Fig. 9A). Furthermore, no detectable effect on the processing of the PV polyprotein was observed in the presence of rPTP (Fig. 9B). However, infectious virus production in the presence of rPTP was reduced approximately 80-fold (Fig. 9C). Because the presence of rPTP itself does not inhibit cell-free translation of the input RNA, the decreased infectivity of virus synthesized in cell extracts containing rPTP reflects a posttranslational effect.
FIG. 9.
Cell-free translation of PV genomic RNA is unaffected by the presence of rPTP. (A) Cell-free reactions were programmed with PV replicon RNA containing luciferase in place of the capsid-coding sequence. Translation was measured after 12 h via a luciferase activity assay. (B) Cell-free reactions were programmed with PV vRNA. After 12 h of translation, products were separated via SDS-PAGE. (C) Extracts were programmed with PV vRNA, and viable virus produced was quantitated by plaque assay.
rPTP substitution causes a replication defect in the poliovirus subgenomic replicon.
To determine whether incorporated rPMP can affect virus replication, a poliovirus subgenomic replicon, wherein the capsid coding region is replaced by a luciferase reporter (21), was transcribed in the presence of rPTP as previously described (18). Reporter activity was monitored following transfection into HeLa S3 cells. When RNA containing approximately 16.5 substitutions per genome was transfected, translation of input RNA was reduced approximately 50% as determined by incubation in the presence of 3 mM guanidine hydrochloride, a poliovirus replication inhibitor (Fig. 10A). Translation was further diminished as additional rPMP substitutions were made (Fig. 10B). While this may reflect translational effects due to erroneous codon recognition of rPMP-containing RNA, it also likely reflects at least some loss of functionality due to mutation of the luciferase coding region induced by rPMP incorporation.
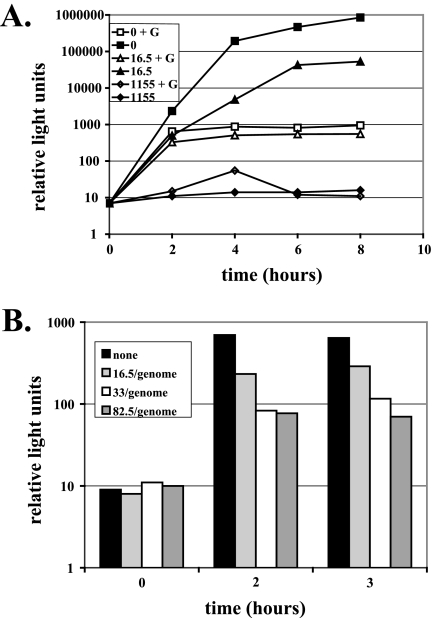
FIG. 10.
The PV replicon containing rPMP shows a reduction in active luciferase reporter activity. The PV replicon contains the luciferase reporter in place of the capsid-coding sequence. Replicons were transcribed under conditions previously shown to result in 0, 16.5, 33, or 82.5 rPMP substitutions per RNA molecule. (A) Replicon-transfected cells were incubated in the presence (open symbols) or absence (closed symbols) of 3 mM guanidine hydrochloride for 8 h. (B) Replicon-transfected cells were incubated for up to 3 h in the presence of 3 mM guanidine hydrochloride. Activity of the luciferase reporter was reduced by a maximum of 10-fold.
rPMP-substituted RNA was transfected into HeLa cells and monitored for reporter activity over 8 h (Fig. 10A). Genomes containing an average of 16.5 substitutions per genome exhibited diminished replication kinetics and an approximately 15-fold reduction in reporter activity at 8 h. Genomes containing an average of 1,155 substitutions per genome exhibited no reporter activity in the presence or absence of 3 mM guanidine hydrochloride.
A high-fidelity PV variant has increased sensitivity to rPTP incorporation.
The preceding experiments suggest that incorporation of rPMP is deleterious to the PV genome through increased mutagenesis of the virus genomic sequence. A high-fidelity PV (G64S) variant was previously identified as resistant to ribavirin treatment due to a twofold increase in fidelity of the virus-encoded RNA-dependent RNA polymerase (4, 43). If decreased infectivity of rPTP-containing genomes were due to increased mutagenesis, a high-fidelity variant would be less likely to “correct” those mutations during subsequent rounds of replication, resulting in an increased rate of fixation of deleterious mutations. Therefore, the high-fidelity G64S variant should be more sensitive to mutations introduced into the genome by T7 RNAP-mediated transcription.
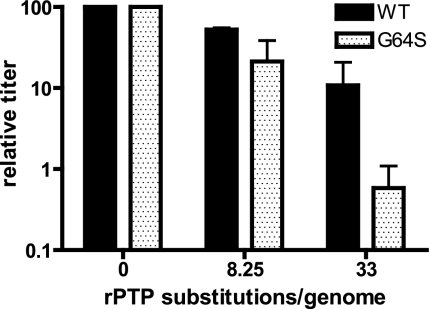
We tested this hypothesis by transcribing both wild-type and high-fidelity (G64S) PV genomes in the presence of various concentrations of rPTP. These RNA genomes were transfected into HeLa cells via electroporation, and the transfected cells were added to growth medium and incubated for 6 h at 37°C to allow for virus amplification over one replicative cycle. Resultant virus population titers were determined on HeLa monolayers. The G64S variant showed increased sensitivity to rPTP genomic substitution (Fig. 11). At approximately 33 substitutions per genome, the G64S virus titer was reduced 20-fold compared to wild-type virus under the same conditions.
FIG. 11.
rPMP substitution in genomic RNA is more deleterious to a high-fidelity PV variant (G64S). Cells were transfected by T7 RNAP-transcribed RNA containing a known number of rPMP substitutions, followed by incubation at 37°C for 6 h. Cell-associated virus was collected by freeze-thaw, and the titer was determined. Titers were normalized such that the titer of virus produced by RNA with no rPMP substitutions was set to 100. The titer resulting from transfection of wild-type (WT) genomes containing no PTP was 2.3 ×106 PFU/ml; for G64S, the titer was 6.6 ×106 PFU/ml.
DISCUSSION
Lethal mutagenesis as an antiviral strategy remains controversial, primarily due to the fact that mutagenesis alone has not been directly demonstrated to be sufficient for antiviral activity in vitro or in vivo. Known nucleoside analogues, including ribavirin, generally have multiple pathways through which they can exert antiviral activity, including polymerase inhibition and perturbation of cellular nucleotide pools through interaction with the enzymes of nucleotide metabolism. In addition, ribavirin and other purine analogues must compete with relatively large intracellular purine pools to act as effective mutagens. Here we have demonstrated that mutagenesis of viral genomic RNA itself is sufficient for substantial reductions in virus infectivity. While the nucleoside analogue evaluated herein did not exhibit antiviral activity in cell culture, we have directly demonstrated the application of lethal mutagenesis as an antiviral strategy, as well as the potential of nucleobase analogues with tautomeric constants near unity to act as lethal mutagens when used as substrates for RNA-dependent RNA polymerases. Further research in the area of tautomeric nucleobases should lead to analogues that are effective in vivo.
Previous work has shown that rP and dP can be incorporated promiscuously by a number of RNA and DNA polymerases, respectively. We demonstrated that rP is also recognized as an ambiguous pyrimidine analogue by a virus-encoded RNA-dependent RNA polymerase (PV 3Dpol). Crotty and colleagues observed a 3.2-fold reduction in RNA specific infectivity when ribavirin-treated poliovirus genomic RNA was found to have 1.9 mutations per genome through sequencing of capsid coding regions (6). Here, we show approximately 16 incorporations of rPMP are required for an equivalent reduction in RNA specific infectivity. This apparent discrepancy may be reconciled by noting that every incorporation event will not result in a mutation. Furthermore, the tautomerization of P (KT of approximately 11) favors the imino form (mimicking uridine) (20). Accordingly, T7 RNA polymerase demonstrates a marked preference for recognition of rPTP as a uridine analogue (35). Mutational assays in E. coli (40) and an in vitro retroviral replication model (36) both showed a bias towards C-to-U (or T) mutations over U- (or T)-to-C by nearly a factor of 2, consistent with the imino form of P as the most thermodynamically favorable conformation.
The loss of RNA infectivity when rPMP is incorporated into RNA may be due to a number of factors. P is an ambiguously hydrogen-bonding base, and its incorporation into genomic RNA should result in transition mutations. Thus, increased mutation of the coding sequence caused by genomic incorporation of rPMP may elicit loss of protein function, including the generation of dominant negative mutations. Secondly, the unconventional structure of rP may preclude its recognition as part of a codon during RNA translation. The presence of unnatural bases may also affect the stability of the RNA genome, resulting in more rapid degradation of the RNA in the cellular milieu. This can impact the infectivity of the genome if the effective lifetime of the molecule is too short to allow a productive infection to be established. Finally, the presence of P may affect the ability of the RNA to form secondary and tertiary structures required for translation and virus replication. Thus, mutation may affect not only the primary (protein coding) sequence of the virus but also the higher-order structures that are known to play essential roles during the virus life cycle as well as in genome stability.
Inefficient phosphorylation of rP by cellular nucleotide and nucleoside kinases likely explains the lack of antiviral activity of this nucleoside in cell culture. Experiments performed with standards of the nucleoside indicated a lower limit of detection of approximately 1 nmol under the HPLC conditions employed (data not shown). Based on the extraction conditions, this would allow detection of intracellular P metabolites present at approximately 20 μM or higher. Our failure to detect any metabolites other than the ribonucleoside indicate that, even if rP were recognized as a substrate for intracellular kinases, phosphorylated forms would not accumulate to high levels. Additionally, the ability to detect the nucleoside (rP) at substantial levels indicates that the lack of activity is likely not due to degradation or metabolism of the nucleoside, or failure of the nucleoside to accumulate intracellularly. While our studies with the nucleobase (P) did demonstrate antiviral activity and mutagenesis (albeit statistically insignificant), this does not necessarily mean that rPTP accumulates intracellularly after treatment with the nucleobase. The nucleobase itself could have other activities which caused the observed mutagenic effect, such as perturbation of cellular nucleotide pools through interaction with the nucleotide biosynthetic enzymes of the cell.
The failure to detect intracellular phosphorylated metabolites of rP highlights an important complication of nucleoside-based antiviral therapy, as relatively high levels of metabolically downstream nucleotides may need to be achieved for sufficient antiviral activity. Mutagenic nucleoside analogues are prodrugs in that they must be activated by cellular enzymes (nucleoside and nucleotide kinases) in order for the active (triphosphorylated) form to accumulate. The specificity of cellular nucleoside, nucleotidyl, and nucleoside diphosphate kinases is therefore a crucial factor in the activity of nucleoside-based therapeutics. A pronucleotide approach could potentially be utilized in delivering rPMP (or even rPTP) to cells, protecting the charged phosphates to allow cellular entry and liberation of the phosphorylated form in the cytoplasm by cellular metabolic enzymes (reviewed in reference 57). However, it is still unknown if available rPMP in the cytoplasm of mammalian cells will lead to accumulation of the triphosphorylated form (rPTP) and observable antiviral activity. Given the high capacity for mutagenesis exhibited by P, a prodrug of rP containing masked 5′ phosphates has the potential for significant antiviral activity.
Lethal mutagenesis was proposed as an antiviral strategy almost 15 years ago. Despite this, previous attempts to design novel mutagenic nucleotide analogues that are capable of inducing mutation in RNA viruses have had only limited success (17-19, 39). In this study, we have demonstrated that rPTP is an efficient and ambiguous substrate of a viral RNA-dependent RNA polymerase. rPTP was incorporated 100-fold more efficiently than the known mutagenic nucleoside ribavirin in vitro. P therefore represents an important lead compound for the development of clinically useful antiviral therapies based on lethal mutagenesis of RNA virus genomes.
Direct mutagenesis of viral RNA through T7-mediated transcription as described herein also holds potential for unraveling the sensitivity of RNA viruses to mutagenesis outside of the context of the virus-encoded polymerase. A number of positive-sense RNA viruses have genomes that can be manipulated in vitro to examine the effects of mutational load on population fitness. As such, this study introduces a potential methodology to screen for viruses that may exhibit enhanced sensitivity to lethal mutagens due to low mutational robustness. It may also be used as a tool to understand the interplay between polymerase fidelity, infectivity, genome conservation, and virus evolution.
Acknowledgments
Financial support was provided by the National Institutes of Health (AI054776 to C.E.C. and B.R.P.; AI015122 to D.F. and A.V.P), the MRC Development Gap Fund (financial assistance to K.T.), and the American Heart Association (Established Investigator award 0340028N to C.E.C. and predoctoral fellowships to D.A.H. and J.P.E.).
We thank Christian Castro (Penn State University) for purification of CVB3 3Dpol and Jamie Arnold (Penn State University) for expression and purification of T7 RNA polymerase. We thank Richard R. Drake (Eastern Virginia Medical School) for the gift of HSV-1 TK expression plasmids and purified enzyme. We thank Margaret E. Black (Washington State University) for rabbit HSV-1 TK polyclonal antiserum. We thank Kirk U. Knowlton (University of California, San Diego) for the gift of CVB3 cDNA.
Footnotes
Published ahead of print on 8 August 2007.
REFERENCES
- 1.Airaksinen, A., N. Pariente, L. Menendez-Arias, and E. Domingo. 2003. Curing of foot-and-mouth disease virus from persistently infected cells by ribavirin involves enhanced mutagenesis. Virology 311:339-349. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, J. J., and C. E. Cameron. 2000. Poliovirus RNA-dependent RNA polymerase (3Dpol). Assembly of stable, elongation-competent complexes by using a symmetrical primer-template substrate (sym/sub). J. Biol. Chem. 275:5329-5336. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, J. J., and C. E. Cameron. 2004. Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+. Biochemistry 43:5126-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, J. J., M. Vignuzzi, J. K. Stone, R. Andino, and C. E. Cameron. 2005. Remote site control of an active site fidelity checkpoint in a viral RNA-dependent RNA polymerase. J. Biol. Chem. 280:25706-25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crain, P. F. 1990. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol. 193:782-790. [DOI] [PubMed] [Google Scholar]
- 6.Crotty, S., C. E. Cameron, and R. Andino. 2001. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 98:6895-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 8.Cuconati, A., A. Molla, and E. Wimmer. 1998. Brefeldin A inhibits cell-free, de novo synthesis of poliovirus. J. Virol. 72:6456-6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day, C. W., D. F. Smee, J. G. Julander, V. F. Yamshchikov, R. W. Sidwell, and J. D. Morrey. 2005. Error-prone replication of West Nile virus caused by ribavirin. Antivir. Res. 67:38-45. [DOI] [PubMed] [Google Scholar]
- 10.Domingo, E. 2002. Quasispecies theory in virology. J. Virol. 76:463-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Domingo, E., C. Escarmis, N. Sevilla, A. Moya, S. F. Elena, J. Quer, I. S. Novella, and J. J. Holland. 1996. Basic concepts in RNA virus evolution. FASEB J. 10:859-864. [DOI] [PubMed] [Google Scholar]
- 12.Domingo, E., E. Martinez-Salas, F. Sobrino, J. C. de la Torre, A. Portela, J. Ortin, C. Lopez-Galindez, P. Perez-Brena, N. Villanueva, R. Najera, et al. 1985. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance—a review. Gene 40:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Gohara, D. W., C. S. Ha, S. Kumar, B. Ghosh, J. J. Arnold, T. J. Wisniewski, and C. E. Cameron. 1999. Production of “authentic” poliovirus RNA-dependent RNA polymerase (3Dpol) by ubiquitin-protease-mediated cleavage in Escherichia coli. Protein Expr. Purif. 17:128-138. [DOI] [PubMed] [Google Scholar]
- 14.Graci, J. D., and C. E. Cameron. 2004. Challenges for the development of ribonucleoside analogues as inducers of error catastrophe. Antivir. Chem. Chemother. 15:1-13. [DOI] [PubMed] [Google Scholar]
- 15.Graci, J. D., and C. E. Cameron. 2006. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 16:37-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grande-Perez, A., E. Lazaro, P. Lowenstein, E. Domingo, and S. C. Manrubia. 2005. Suppression of viral infectivity through lethal defection. Proc. Natl. Acad. Sci. USA 102:4448-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harki, D. A., J. D. Graci, J. P. Edathil, C. Castro, C. E. Cameron, and B. R. Peterson. 2007. Synthesis of a universal 5-nitroindole ribonucleotide and incorporation into RNA by a viral RNA-dependent RNA polymerase. ChemBioChem 8:1359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harki, D. A., J. D. Graci, J. E. Galarraga, W. J. Chain, C. E. Cameron, and B. R. Peterson. 2006. Synthesis and antiviral activity of 5-substituted cytidine analogues: identification of a potent inhibitor of viral RNA-dependent RNA polymerases. J. Med. Chem. 49:6166-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harki, D. A., J. D. Graci, V. S. Korneeva, S. K. Ghosh, Z. Hong, C. E. Cameron, and B. R. Peterson. 2002. Synthesis and antiviral evaluation of a mutagenic and non-hydrogen bonding ribonucleoside analogue: 1-beta-D-ribofuranosyl-3-nitropyrrole. Biochemistry 41:9026-9033. [DOI] [PubMed] [Google Scholar]
- 20.Harris, V. H., C. L. Smith, W. Jonathan Cummins, A. L. Hamilton, H. Adams, M. Dickman, D. P. Hornby, and D. M. Williams. 2003. The effect of tautomeric constant on the specificity of nucleotide incorporation during DNA replication: support for the rare tautomer hypothesis of substitution mutagenesis. J. Mol. Biol. 326:1389-1401. [DOI] [PubMed] [Google Scholar]
- 21.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill, F., D. Loakes, and D. M. Brown. 1998. Polymerase recognition of synthetic oligodeoxyribonucleotides incorporating degenerate pyrimidine and purine bases. Proc. Natl. Acad. Sci. USA 95:4258-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinds, T. A., C. Compadre, B. K. Hurlburt, and R. R. Drake. 2000. Conservative mutations of glutamine-125 in herpes simplex virus type 1 thymidine kinase result in a ganciclovir kinase with minimal deoxypyrimidine kinase activities. Biochemistry 39:4105-4111. [DOI] [PubMed] [Google Scholar]
- 24.Holland, J. J., E. Domingo, J. C. de la Torre, and D. A. Steinhauer. 1990. Mutation frequencies at defined single codon sites in vesicular stomatitis virus and poliovirus can be increased only slightly by chemical mutagenesis. J. Virol. 64:3960-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holmes, E. C., and A. Moya. 2002. Is the quasispecies concept relevant to RNA viruses? J. Virol. 76:460-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins, G. M., M. Worobey, C. H. Woelk, and E. C. Holmes. 2001. Evidence for the non-quasispecies evolution of RNA viruses. Mol. Biol. Evol. 18:987-994. [DOI] [PubMed] [Google Scholar]
- 27.Lanford, R. E., D. Chavez, B. Guerra, J. Y. Lau, Z. Hong, K. M. Brasky, and B. Beames. 2001. Ribavirin induces error-prone replication of GB virus B in primary tamarin hepatocytes. J. Virol. 75:8074-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lanford, R. E., B. Guerra, H. Lee, D. R. Averett, B. Pfeiffer, D. Chavez, L. Notvall, and C. Bigger. 2003. Antiviral effect and virus-host interactions in response to alpha interferon, gamma interferon, poly(I)-poly(C), tumor necrosis factor alpha, and ribavirin in hepatitis C virus subgenomic replicons. J. Virol. 77:1092-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, X., H. H. Lu, S. Mueller, and E. Wimmer. 2001. The C-terminal residues of poliovirus proteinase 2Apro are critical for viral RNA replication but not for cis- or trans-proteolytic cleavage. J. Gen. Virol. 82:397-408. [DOI] [PubMed] [Google Scholar]
- 30.Loeb, L. A., J. M. Essigmann, F. Kazazi, J. Zhang, K. D. Rose, and J. I. Mullins. 1999. Lethal mutagenesis of HIV with mutagenic nucleoside analogs. Proc. Natl. Acad. Sci. USA 96:1492-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, H. H., C. F. Yang, A. D. Murdin, M. H. Klein, J. J. Harber, O. M. Kew, and E. Wimmer. 1994. Mouse neurovirulence determinants of poliovirus type 1 strain LS—a map to the coding regions of capsid protein VP1 and proteinase 2Apro. J. Virol. 68:7507-7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMasters, R. A., R. L. Saylors, K. E. Jones, M. E. Hendrix, M. P. Moyer, and R. R. Drake. 1998. Lack of bystander killing in herpes simplex virus thymidine kinase-transduced colon cell lines due to deficient connexin 43 gap junction formation. Hum. Gene Ther. 9:2253-2261. [DOI] [PubMed] [Google Scholar]
- 33.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 34.Molla, A., A. V. Paul, and E. Wimmer. 1993. Effects of temperature and lipophilic agents on poliovirus formation and RNA synthesis in a cell-free system. J. Virol. 67:5932-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriyama, K., K. Negishi, M. S. Briggs, C. L. Smith, F. Hill, M. J. Churcher, D. M. Brown, and D. Loakes. 1998. Synthesis and RNA polymerase incorporation of the degenerate ribonucleotide analogue rPTP. Nucleic Acids Res. 26:2105-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriyama, K., C. Otsuka, D. Loakes, and K. Negishi. 2001. Highly efficient random mutagenesis in transcription-reverse-transcription cycles by a hydrogen bond ambivalent nucleoside 5′-triphosphate analogue: potential candidates for a selective anti-retroviral therapy. Nucleosides Nucleotides Nucleic Acids 20:1473-1483. [DOI] [PubMed] [Google Scholar]
- 37.Motulsky, H. 1995. Intuitive biostatistics. Oxford University Press, New York, NY.
- 38.Moya, A., S. F. Elena, A. Bracho, R. Miralles, and E. Barrio. 2000. The evolution of RNA viruses: a population genetics view. Proc. Natl. Acad. Sci. USA 97:6967-6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami, E., A. Basavapathruni, W. D. Bradley, and K. S. Anderson. 2005. Mechanism of action of a novel viral mutagenic covert nucleotide: molecular interactions with HIV-1 reverse transcriptase and host cell DNA polymerases. Antivir. Res. 67:10-17. [DOI] [PubMed] [Google Scholar]
- 40.Negishi, K., D. M. Williams, Y. Inoue, K. Moriyama, D. M. Brown, and H. Hayatsu. 1997. The mechanism of mutation induction by a hydrogen bond ambivalent, bicyclic N4-oxy-2′-deoxycytidine in Escherichia coli. Nucleic Acids Res. 25:1548-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pariente, N., A. Airaksinen, and E. Domingo. 2003. Mutagenesis versus inhibition in the efficiency of extinction of foot-and-mouth disease virus. J. Virol. 77:7131-7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pariente, N., S. Sierra, P. R. Lowenstein, and E. Domingo. 2001. Efficient virus extinction by combinations of a mutagen and antiviral inhibitors. J. Virol. 75:9723-9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pfeiffer, J. K., and K. Kirkegaard. 2003. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. USA 100:7289-7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilger, B. D., R. Perozzo, F. Alber, C. Wurth, G. Folkers, and L. Scapozza. 1999. Substrate diversity of herpes simplex virus thymidine kinase. Impact of the kinematics of the enzyme. J. Biol. Chem. 274:31967-31973. [DOI] [PubMed] [Google Scholar]
- 45.Pogolotti, A. L., Jr., and D. V. Santi. 1982. High-pressure liquid chromatography-ultraviolet analysis of intracellular nucleotides. Anal. Biochem. 126:335-345. [DOI] [PubMed] [Google Scholar]
- 46.Ruiz-Jarabo, C. M., C. Ly, E. Domingo, and J. C. de la Torre. 2003. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV). Virology 308:37-47. [DOI] [PubMed] [Google Scholar]
- 47.Schuster, P., and J. Swetina. 1988. Stationary mutant distributions and evolutionary optimization. Bull. Math. Biol. 50:635-660. [DOI] [PubMed] [Google Scholar]
- 48.Severson, W. E., C. S. Schmaljohn, A. Javadian, and C. B. Jonsson. 2003. Ribavirin causes error catastrophe during Hantaan virus replication. J. Virol. 77:481-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sierra, S., M. Davila, P. R. Lowenstein, and E. Domingo. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinhauer, D. A., E. Domingo, and J. J. Holland. 1992. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene 122:281-288. [DOI] [PubMed] [Google Scholar]
- 51.Stone, M. J., A. N. Nedderman, D. H. Williams, P. K. Lin, and D. M. Brown. 1991. Molecular basis for methoxyamine initiated mutagenesis. 1H nuclear magnetic resonance studies of base-modified oligodeoxynucleotides. J. Mol. Biol. 222:711-723. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, T., K. Moriyama, C. Otsuka, D. Loakes, and K. Negishi. 2006. Template properties of mutagenic cytosine analogues in reverse transcription. Nucleic Acids Res. 34:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Temiakov, D., M. Anikin, and W. T. McAllister. 2002. Characterization of T7 RNA polymerase transcription complexes assembled on nucleic acid scaffolds. J. Biol. Chem. 277:47035-47043. [DOI] [PubMed] [Google Scholar]
- 54.van Nimwegen, E., J. P. Crutchfield, and M. Huynen. 1999. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 96:9716-9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Rompay, A. R., M. Johansson, and A. Karlsson. 2003. Substrate specificity and phosphorylation of antiviral and anticancer nucleoside analogues by human deoxyribonucleoside kinases and ribonucleoside kinases. Pharmacol. Ther. 100:119-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vignuzzi, M., J. K. Stone, J. J. Arnold, C. E. Cameron, and R. Andino. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439:344-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner, C. R., V. V. Iyer, and E. J. McIntee. 2000. Pronucleotides: toward the in vivo delivery of antiviral and anticancer nucleotides. Med. Res. Rev. 20:417-451. [DOI] [PubMed] [Google Scholar]
- 58.Wilke, C. O. 2005. Quasispecies theory in the context of population genetics. BMC Evol. Biol. 5:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilke, C. O., J. L. Wang, C. Ofria, R. E. Lenski, and C. Adami. 2001. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412:331-333. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, S., R. Liu, B. M. Baroudy, B. A. Malcolm, and G. R. Reyes. 2003. The effect of ribavirin and IMPDH inhibitors on hepatitis C virus subgenomic replicon RNA. Virology 310:333-342. [DOI] [PubMed] [Google Scholar]