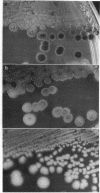
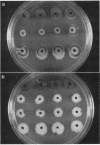
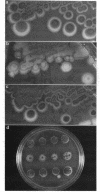
Abstract
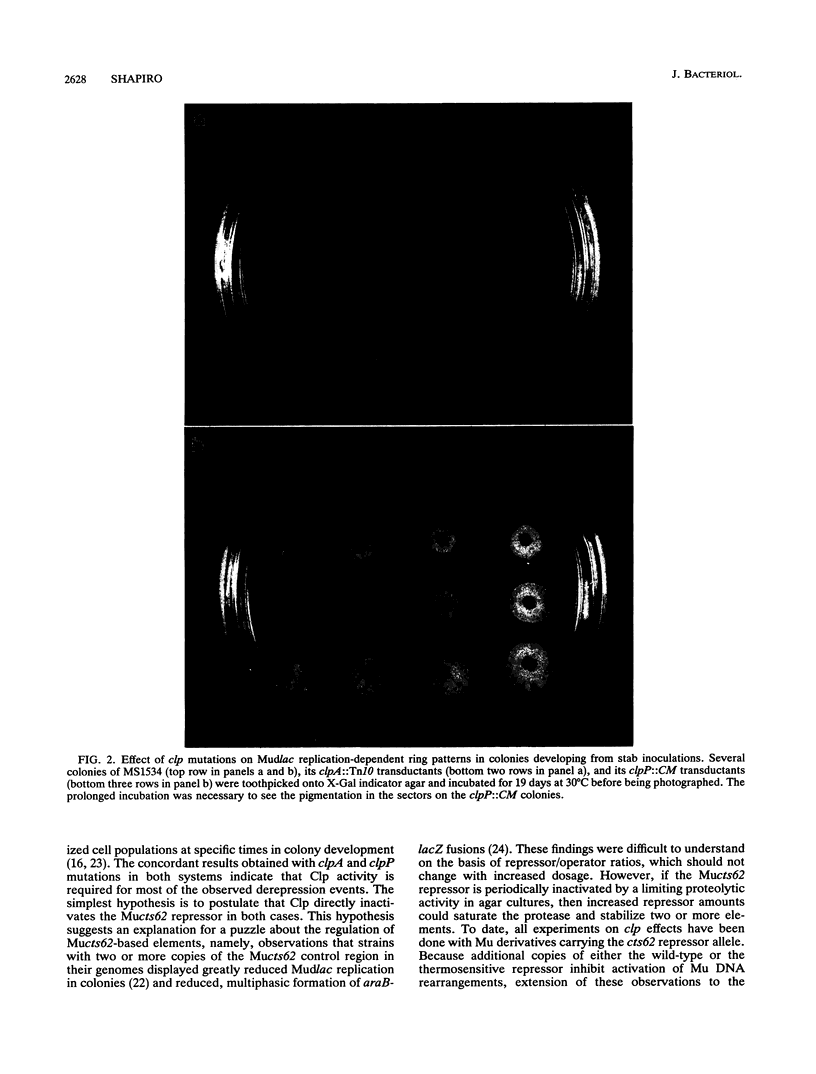
Bacteriophage Mu, one of the best-characterized mobile genetic elements, can be used effectively to answer fundamental questions about the regulation of biochemical machinery for DNA rearrangement. Previous studies of Mu virulence have implicated the Clp protease in repressor inactivation (V. Geuskens, A. Mhammedi-Alaoui, L. Desmet, and A. Toussaint, EMBO J. 13:5121-5127, 1992). These studies were extended by analyzing the phenotypic consequences of clp alleles in two Escherichia coli systems: (i) the periodic replication of Mudlac transposons in colonies and (ii) the action of a Mu prophage in forming araB-lacZ coding sequence fusions. The clpP::CM mutation, which removes the proteolytic subunit of Clp protease, caused a drastic reduction in Mu activity in both systems. The clpA::Tn10 mutation, which removes a regulatory subunit of Clp protease, altered the timing of Mu activity in both systems. A clpA deletion reduced the extent of Mudlac replication in colonies. These results point to temporal changes in Clp proteolysis of the Mucts62 repressor as a key molecular event in the regulation of one class of genomic change in E. coli.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cairns J., Overbaugh J., Miller S. The origin of mutants. Nature. 1988 Sep 8;335(6186):142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuskens V., Mhammedi-Alaoui A., Desmet L., Toussaint A. Virulence in bacteriophage Mu: a case of trans-dominant proteolysis by the Escherichia coli Clp serine protease. EMBO J. 1992 Dec;11(13):5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Squires C., Pichersky E., Carrington M., Hobbs M., Mattick J. S., Dalrymple B., Kuramitsu H., Shiroza T., Foster T. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci U S A. 1990 May;87(9):3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. G. Adaptive evolution that requires multiple spontaneous mutations. I. Mutations involving an insertion sequence. Genetics. 1988 Dec;120(4):887–897. doi: 10.1093/genetics/120.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y., Gottesman S., Pumphrey J., Rudikoff S., Clark W. P., Maurizi M. R. The two-component, ATP-dependent Clp protease of Escherichia coli. Purification, cloning, and mutational analysis of the ATP-binding component. J Biol Chem. 1988 Oct 15;263(29):15226–15236. [PubMed] [Google Scholar]
- Maurizi M. R., Clark W. P., Katayama Y., Rudikoff S., Pumphrey J., Bowers B., Gottesman S. Sequence and structure of Clp P, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J Biol Chem. 1990 Jul 25;265(21):12536–12545. [PubMed] [Google Scholar]
- Mittler J. E., Lenski R. E. New data on excisions of Mu from E. coli MCS2 cast doubt on directed mutation hypothesis. Nature. 1990 Mar 8;344(6262):173–175. doi: 10.1038/344173a0. [DOI] [PubMed] [Google Scholar]
- Parsell D. A., Sanchez Y., Stitzel J. D., Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991 Sep 19;353(6341):270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., Higgins N. P. Differential activity of a transposable element in Escherichia coli colonies. J Bacteriol. 1989 Nov;171(11):5975–5986. doi: 10.1128/jb.171.11.5975-5986.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A., Higgins N. P. Variation of beta-galactosidase expression from Mudlac elements during the development of Escherichia coli colonies. Ann Inst Pasteur Microbiol. 1988 Jan-Feb;139(1):79–103. doi: 10.1016/0769-2609(88)90098-1. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A., Leach D. Action of a transposable element in coding sequence fusions. Genetics. 1990 Oct;126(2):293–299. doi: 10.1093/genetics/126.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Mechanisms of DNA reorganization in bacteria. Int Rev Cytol. 1985;93:25–56. doi: 10.1016/s0074-7696(08)61371-6. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Natural genetic engineering in evolution. Genetica. 1992;86(1-3):99–111. doi: 10.1007/BF00133714. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Observations on the formation of clones containing araB-lacZ cistron fusions. Mol Gen Genet. 1984;194(1-2):79–90. doi: 10.1007/BF00383501. [DOI] [PubMed] [Google Scholar]
- Shapiro J. A. Organization of developing Escherichia coli colonies viewed by scanning electron microscopy. J Bacteriol. 1987 Jan;169(1):142–156. doi: 10.1128/jb.169.1.142-156.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. The use of Mudlac transposons as tools for vital staining to visualize clonal and non-clonal patterns of organization in bacterial growth on agar surfaces. J Gen Microbiol. 1984 May;130(5):1169–1181. doi: 10.1099/00221287-130-5-1169. [DOI] [PubMed] [Google Scholar]