Abstract
Aging is associated with a decline of immune competence and an increase in markers of inflammation. There is considerable evidence that inflammatory processes play a role in aging and the determination of lifespan. Hypopituitary Ames dwarf mice have extended longevity and exhibit many symptoms of delayed aging, although various aspects of immune function are suppressed in the mutants. In the present study, the expression of genes related to immunity and inflammation was compared in peripheral blood leukocytes (PBL) from Ames dwarf and normal mice using Affymetrix Gene Chip arrays. Among the more than 3000 probe sets that were differentially expressed, 273 were identified as being associated with immunity and/or inflammation. Pathway analysis revealed interactions among 91 of these probe sets, centered on casp3, bcl2, il4, prkca, mapk14, and TGFβ1. Ames dwarf mice had reduced leukocyte expression of casp3 and TGFβ and increased expression of Bcl2. Alterations in the expression of these genes suggest likely functional changes in apoptosis, B and T cell homeostasis, prostaglandin synthesis, humoral immunity, chemokine activity, complement activation, hemostasis and wound healing pathways. Collectively, these results suggest that activation of both anti-inflammatory pathways and an anti-clotting mechanism combined with reduced turnover of leukocytes may contribute to delayed aging and extended longevity of Ames dwarf mice. We are also aware that alterations in gene expression in PBLs can be due to different composition of PBL populations when comparing Ames dwarf to WT animals, and it will be interesting to investigate these genes in particular PBL populations in the future. However, whole leucocytes population represents the function of immune system in these organisms.
Keywords: Ames dwarf, aging, PBL, inflammation
INTRODUCTION
The age-related immune decline, also termed immunosenescence, involves a reduced response of the adaptive immune system and a hyperactivity of the innate immune system resulting in chronic inflammation, higher susceptibility of the elderly to viral and bacterial infections, high incidence of cancer, and increased morbidity and mortality (DeVeale et al., 2004; Effros, 2001; Solana and Pawelec, 1998; Ginaldi et al., 1999; Finch and Crimmins, 2004, Pawelec et al., 1998). The deterioration of the adaptive immunity is caused by a shift in leukocytes population types, including a reduced proportion of naïve T cells relative to their memory counterparts (Chakravarti and Abraham, 1999) and a decline in lymphocyte functions (Solana and Pawelec, 1998). Aging is correlated with diminished antigen recognition, hyporesponsiveness to cell receptor stimulation, defects in the early events of signal transduction, and dysregulation of cytokine production and lymphocyte proliferative response to mitogenic stimulation (Chakravarti and Abraham, 1999; McGlauchlen and Vogel, 2003; Szakal et al., 2002; Miller, 2000). Aging also decreases the functions of the cells of the innate or ‘‘natural’’ immunity (Pawelec et al., 1998). Following lipopolysaccharide activation, aged monocytes show decreased cytotoxicity against tumor cells, reduced interleukin 1 and a decreased release of reactive oxygen and nitrogen intermediates (McLachlan et al., 1995 A & B). Diminished release of such compounds reflects a lowered capacity of macrophages to kill micro-organisms and lyse tumor cells. Aging significantly impairs chemotactic, phagocytic and killing capacities of polymorphonuclear cells in humans and mice (Polignano et al., 1994, Yoshino et al., 1992; Lloberas and Celada, 2002). Polymorphonuclear cells protect against invading micro-organisms and are the first, among the inflammatory cells, to be recruited to the inflammatory site. The chronic inflammation associated with aging may reflect an impairment of both innate and adaptive immunity, given the interdependence of the two immune systems. Inflammation has been implicated in the pathogenesis of atherosclerosis, and rigorous control of inflammatory reactions is thought to decrease the incidence of cardiovascular disease (Ross, 1993; Ridker et al., 1997). Blood elevation of inflammatory markers such as serum amyloid A, C-reactive protein, IL-6 and fibrinogen are risk factors for heart attack and stroke (Danesh et al., 2004, Pearson et al., 2003). It has been proposed that the historical increase in life expectancy of humans can be partially explained by a decreased exposure to chronic inflammation (Finch and Crimmins, 2004; Crimmins and Finch, 2006).
Prop1 and Pit1 mutations affecting development of the pituitary gland of mice produce long-lived dwarfs with deficiencies in serum growth hormone (GH), thyroid stimulating hormone and prolactin, as well as IGF-I, which is secreted by the liver in response to pituitary growth hormone (Sornson et al., 1996). There is considerable evidence that the GH/IGF-I deficiency is responsible for the positive longevity effects of the dwarfing mutations (Bartke, 2005, 2006). Calorie restriction, another intervention that consistently extends lifespan, may do so by altering the insulin/IGF-1 pathway, since it reduces the level of circulating insulin, GH and IGF-1 (Longo and Finch, 2003). Failure of CR to further increase longevity and to enhance insulin sensitivity in mice with targeted disruption of the GH receptor implies that somatotropic signaling mediates the effects of CR on lifespan (Bonkowski et al., 2006). The extended lifespan of dwarf and calorie-restricted mice has caused them to be extensively used as models of decelerated aging to identify the molecular mechanisms of the anti-aging effects.
In addition to extending longevity, both dwarfism and calorie restriction impede a whole host of age-induced pathologies including the development of neoplastic diseases, collagen cross-linking and deterioration of the cognitive function (Brown-Borg et al., 1996, Flurkey et al., 2001, 2002; Ikeno et al., 2003; Silberberg, 1972; Longo and Finch, 2003; Weindruch and Sohal, 1997). However, regarding immunity and inflammations pertinence to aging, only calorie restriction has been extensively investigated and repeatedly shown to potently enhance the immune functions and ameliorate inflammation (Pahlavani, 2000, 2004; Fernandes et al., 1997, Spaulding et al., 1997). In contrast, there are fewer reports concerning the effects of dwarfism on the immune system and inflammation, and their findings are often contradictory. Some studies suggested that dwarfism compromises the immune system development (Murphy et al., 1992; Dumont et al., 1979; Duquesnoy, 1972; Fabris et al., 1971; Baroni et al., 1969). Other studies reported that the immune competence of dwarf mice did not differ from their littermate controls (Schneider, 1976; Dumont et al., 1979; Cross et al., 1992).
Dwarf mice were shown to exhibit decreased peripheral blood cell and a deficiency of B cell progenitor populations in the bone marrow (Murphy et al., 1992), decreased splenic T and B lymphocytes (Dumont et al., 1979) and decreased thymocytes (Duquesnoy, 1972). Functional analysis showed that both cell-mediated immunity and humoral immunity were compromised (Fabris et al., 1971; Baroni et al., 1969) and restored with administration of either GH or a combination of GH and thyroxine (Fabris et al., 1971). In contrast to these findings, other studies of dwarf mice showed that the T-cell-dependent zones in the peripheral lymphoid tissues were not deficient in lymphocytes, and the thymuses were reported to have a normal cellular composition (Schneider, 1976). Dwarf mice were reported to have competent humoral and cell-mediated immunity with normal antibody production, mitogen-induced proliferation of both T and B cells and delayed-type hypersensitivity to a contact sensitizing agent (Schneider, 1976; Dumont et al., 1979). Another study concluded that the development of immunity in dwarf mice lags behind their normal littermates and is influenced by the age at which the mice are weaned, as well as the time between weaning and immunologic assessment (Cross et al., 1992). However, a fully functional immune system will develop if dwarf mice are allowed to mature to 32 days of age.
In regard to immunosenescence, dwarfism was shown to prevent age-dependent splenomegaly and splenic T cell proliferation impairment, and to diminish osteoarthritis of the knee joint (Silberberg, 1972; Flurkey, 1990). Changes in age-sensitive traits of connective tissue and immune system status were found to be decelerated in dwarf mice (Flurkey et al, 2001). Specifically, aged dwarf mice were shown to have youthful levels of CD4M and CD8M cells which express the CD44 surface marker typical of the memory T cell population. The proportions of mitogen-stimulated T cells that can proliferate and differentiate into cytotoxic effectors also were higher in older dwarf mice.
To address the dearth of information and the discrepancies in the reports concerning the effects of dwarfism on immunity and inflammation, we analyzed the gene expression profiles of the circulating leukocytes of Ames dwarf mice using Affymetrix oligonucleotide microarrays containing probes for more than 45,000 transcription units. The homozygosity for the Ames dwarf mutation extends longevity by more than 50% in both males and females (Brown-Borg et al., 1996). We have previously shown that the liver gene expression profiles of the Ames dwarf mice reflect increased insulin, glucagon and catecholamine sensitivity, gluconeogenesis, protein turnover, lipid β-oxidation, apoptosis, and xenobiotic and oxidant metabolism, as well as decreased cell proliferation, lipid and cholesterol synthesis, and chaperone expression (Tsuchiya et al., 2004). Also, using liver gene expression profiling, Ames dwarf mutation was shown to partially delay or decelerate age-related changes (Amador-Noguez et al., 2004). The findings of another liver gene expression profiling study of Ames dwarf mice were consistent with a reduced innate immune response (Amador-Noguez et al., 2005). Activation of the innate immune system can contribute to inflammation and the pathogenesis of autoimmune diseases.
In this study, we analyzed the gene expression profiles of leukocytes with focus on the genes and pathways related to immunity and inflammation. Circulating blood leukocytes are the main cells that provide immunity, mediate stress and inflammation, and produce cytokines, chemokines and growth factors that exert potent physiological and pathological actions on peripheral tissues. Furthermore, circulating leukocytes have the ability to mirror the body’s tissues and organs in health and disease. Dissecting leukocyte gene expression patterns of the long-lived Ames dwarf mice will advance our understanding of the effects of dwarfism on the immune functions. This molecular characterization should help in formulating hypotheses and developing future experiments to identify potential new immune mechanisms by which “longevity genes” postpone diseases and decelerate aging. Ultimately these studies may lead to the design and discovery of possible therapeutic interventions to enhance immune function in the elderly.
MATERIALS AND METHODS
Mice
Male and female mice of the Ames stock were bred and housed at Southern Illinois University. Ames dwarf (df/df) and normal (+/+ or +/df) mice were produced by crosses between df/+ parents or between fertile df/df males and df/+ females. Details of the animal husbandry were described previously (Masternak, 2004). Seven-month old 10 female Ames dwarf mice and 10 of their normal littermates were used in the study.
Leukocyte RNA isolation
Blood was drawn from mice in the presence of EDTA using the heart puncture method. Blood samples were immediately processed with the QIAamp® RNA Blood Mini Kit (Qiagen) according to the manufacturer’s instructions. Briefly, 2.5 ml of erythrocyte lysis buffer were added to 0.5 ml of blood to selectively lyse erythrocytes. After incubation on ice, the leukocyte-rich fraction was collected by centrifugation, resuspended in buffer RLT (Qiagen), and homogenized by centrifugation through a QIAshredder™ spin column. The homogenized lysate was mixed with ethanol and applied to the QIAamp spin column to bind the RNA to the silica-gel membrane. After washing, total RNA was eluted in 30 μl of RNase-free water.
Globin Reduction
Frequently, the erythrocyte lysis is incomplete resulting in high amounts of globin mRNA. To remove any globin mRNA from residual erythrocytes, the isolated total RNA was further processed with the GLOBINclear™–Mouse/Rat kit (Ambion). Briefly, 1 μg of total RNA was mixed with a biotinylated Capture Oligo Mix to hybridize the globin mRNA species with the biotinylated oligonucleotides that were subsequently captured on streptavidin Magnetic Beads. Total RNA, depleted from the globin mRNA, was then eluted after pulling the beads with a magnet. Purity and integrity of the obtained RNA were confirmed by the NanoDrop® ND-1000 UV-Vis Spectrophotometer (Wilmington, DE) which measures the 260/280 ratio, and by capillary electrophoresis (Agilent 2100 Bioanalyser, Agilent Inc), which measures the 28S/18S rRNA ratio and the RNA integrity number (RIN).
cRNA synthesis
Following globin reduction the total RNA obtained was amplified using the Affymetrix Two-Cycle Eukaryotic Target Labeling kit. The two-cycle labeling procedure includes two successive rounds of T7-based in vitro transcription incorporating biotin rNTPs in the second round reaction. Briefly, 100 ng total RNA was used in the first-round synthesis of double-stranded cDNA, followed by amplification with MEGAscript T7 kit (Ambion) using an unlabeled ribonucleotide mix. The unlabeled cRNA was reverse transcribed to generate the second-round double-stranded cDNA template followed by amplification with biotinylated nucleotide analog/ribonucleotide mix in the second in vitro transcription reaction. The quality of the resulting biotin-labeled cRNA was confirmed with the Agilent® 2100 bioanalyzer before any further processing.
Array hybridization
An aliquot (20 μg) of cRNA was fragmented by heat and ion-mediated hydrolysis at 94°C for 35 minutes. Fragmented cRNA was hybridized to the Affymetrix GeneChip® Mouse Genome 430 2.0, which is a single array with over 45,000 probe sets representing more than 34,000 well-substantiated mouse genes. One array was used for fragmented cRNA prepared from each of 20 mice, 10 mice for each experimental group (wild type and dwarf). The hybridization, washing, staining and scanning of the arrays were carried out at the Stanford Protein and Nucleic Acid Biotechnology Facility. After scanning of the chips, the raw probe intensities were stored in electronic files (in .DAT and .CEL formats) using the Affymetrix Microarray Suite 5.0 software.
Probe set expression measurement, normalization and filtering
The array images were inspected for the presence of experimental artifacts and the quality of the raw data was confirmed by measuring a set of QC metrics that are based on methods implemented in the Bioconductor (www.Bioconductor.org) packages ‘affy,’ ‘simpleaffy,’ ‘AffyQCReport,’ and ‘affyPLM’ (Gautier, et al., 2004; Wilson and Miller, 2005). The .CEL files from control (n=10) and Ames dwarf (n=10) groups were simultaneously normalized and expression values computed with the RMA (Robust Multichip Average) method using the Bioconductor package ‘affy’. The data were further filtered to exclude probe data sets that were “Absent” across all 20 arrays according to the MAS 5.0 detection algorithm. This filter produced 28,192 probe sets from the original 45,000 probe sets in the Mouse 430 2.0 array.
Statistical analysis of differential gene expression
All statistical analyses were performed using the BioConductor packages in R (http://www.r-project.org/). To determine the differentially expressed genes, we compared the data from Ames dwarf to normal mice using two separate statistical procedures, SAM (Significance Analysis of Microarrays) and LIMMA (Linear Models for MicroArray data; Smyth et al., 2005; Tusher et al., 2001).
For SAM analysis, the significance cutoff was set at a median false discovery rate < 5%, and the delta value was chosen so that the number of falsely discovered genes is less than one. For LIMMA analysis, we used a false discovery rate adjusted p- value < 5%.
PathwayArchitect analysis of molecular interactions between the differentially expressed genes related to immunity and/or inflammation
The genes found differentially expressed with SAM and LIMMA were searched to identify genes with association to immunity and inflammation using the NetAffx Analysis Center (Liu et al., 2003; http://www.affymetrix.com). The obtained list of genes associated with immunity and inflammation was imported into PathwayArchitect (version 1.0.4; http://stratagene.com). This pathway analysis tool surveys the PubMed literature and retrieves information on each gene relating to its transcriptional regulation, binding partners and any other gene that modifies or interacts with it. The collected information is used to build a graphical network of connections between the genes, illustrating all the extracted types of interaction.
Gene ontology analysis of gene expression using GenMAPP/MAPPfinder
Microarray data were analyzed with GenMAPP/MAPPFinder using MAPPs (MicroArray Pathway Profiles) generated from the Gene Ontology (GO) database and MAPPs specifically designed for GenMAPP (also called local MAPPs; Doniger et al., 2003). Gene expression data were imported into GenMAPP to produce a GenMAPP Expression Data set file. MAPPFinder was used to assign the differentially expressed genes to the GO biological process, cellular component and molecular function terms and calculate z-score as well as a non-parametric permutation test p-value. A positive z-score indicates that there are more genes meeting the criterion in a GO Term than would be expected by random chance. A z-score ≥ 2 is considered a statistically significant association between the differentially regulated genes and their corresponding GO terms. The non-parametric permutation test of the data addresses the multiple testing concerns that occur in GO analysis when calculating z-scores. In addition to a z-score ≥ 2, a GO Term is considered significant if the permutation p-value ≤ 0.05. The up-regulated or down-regulated GO terms were calculated separately in the MAPPFinder analysis. The MAPPFinder results were manually filtered to remove redundant GO Terms and to include only pathways with a number of significantly regulated genes greater than 3 and fewer than 100. GO Terms with very small or very large number of genes are unlikely to be informative because they may be too specific or too general. We focused on the biological processes category of GO and selected GO Terms that represent immunity and inflammation. GenMAPP was used to visualize gene expression data on maps representing biological pathways and groups of genes (Dahlquist et al., 2002).
Gene Ontology analysis using Global Testing
Gene ontology was searched for all pathways associated with immunity and inflammation using the browsing tool AmiGO (http://www.godatabase.org/cgi-bin/amigo/go.cgi). The retrieved GO pathways associated with immunity and inflammation were used to analyze the microarray data from control and Ames dwarf using the R-package “globaltest” (Goeman et al., 2004). The globaltest uses an empirical Bayesian generalized linear model to generate a p-value for testing whether or not some pre-specified groups of genes are differentially expressed. The algorithm applies a goodness-of-fit test on this model by combining the influence of the separate genes into a single test score for the group of genes. The p-values were adjusted for multiple hypotheses testing with Bonferroni correction to control the false discovery rate (FDR). We chose a value < 0.05 as a threshold of significance of the FDR-adjusted p-value. The pathways were further assessed with the “geneplot” function of the globaltest to determine the influence of individual genes on the outcome of the globaltest results.
RESULTS
We used Affymetrix microarrays to assess the effects of a dwarfing mutation that extends life span and postpones diseases on the expression of genes related to immunity and inflammation in circulating blood leukocytes. The procedure of RNA extraction from blood included a combination of erythrocyte lysis and a globin reduction to ensure a complete depletion of globin mRNA species and thus reliable microarray data. The obtained gene expression profiles were subjected to pathway analysis to identify potential immunity and inflammation pathways and biological processes regulated in circulating blood leukocytes of Ames dwarfs. Recently, tools for pathway and Gene Ontology mapping have been developed to translate the information from the individual gene level into functional information with biological processes and regulatory networks pathways (Curtis et al., 2005). The pathway analysis approach helps to overcome the limited sensitivity of microarray expression profiling by detecting more subtle changes in expression than the statistical methods commonly used to generate the gene lists.
Improvement of the target cRNA quality by erythrocyte lysis and globin mRNA reduction
The method used to prepare RNA from circulating blood cells has been shown to significantly impact the quality of microarray data (Feezor et al., 2004, Debey et al., 2004). Utilization of total RNA isolated from whole blood can produce poor quality expression data because the presence of high levels of globin mRNA causes nonspecific binding to other sequences, resulting in higher variability and decreased sensitivity, especially for relatively low expressed genes from leukocytes (Debey et al., 2004). To ensure enhanced detection sensitivity for mRNAs from leukocytes and to reduce the variability observed in microarray data, we depleted globin mRNA from the blood samples in two steps. First, we lysed the erythrocytes, which are the source of globin transcripts when whole blood is used to isolate total RNA. Second, to further deplete any residual globin mRNA derived from erythrocytes carried over from the lysis step, we performed a globin reduction using specific oligonucleotides designed to capture and remove the globin transcript. This combination of erythrocyte lysis with globin reduction to isolate total RNA isolation from leukocytes yielded RNA with integrity and purity high enough to be successfully used in microarray experiments to generate reliable gene expression data (Fig. 1A). As shown in Fig. 1B, the cRNA electropherogram of the sample extracted from whole blood without erythrocyte lysis or globin reduction shows a predominant peak (approximately 700 bases) that represents amplified globin mRNA. This peak was attenuated in the sample extracted with erythrocyte lysis, reflecting a significant decrease in the globin mRNA transcripts. Combination of both erythrocyte lysis and globin reduction produced a smooth curve with no peak, indicating the successful removal of globin mRNA transcripts from the amplification reaction. The gel representations also show that erythrocyte lysis attenuates the globin mRNA band and erythrocyte lysis combined with globin reduction increased the intensity and broadened the cRNA smear indicating that non-globin messages have been recovered.
Fig. 1.
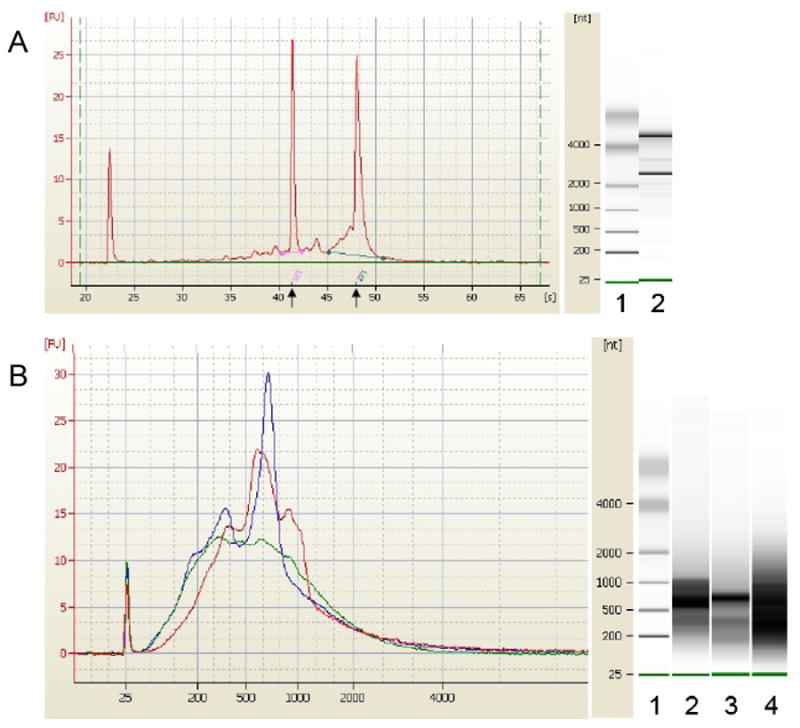
(A) Profile of total RNA extracted with the erythrocyte lysis method followed by the globin reduction procedure. RNA samples were run on the Agilent® 2100 bioanalyzer to obtain electropherogram (left) and virtual gel image (right). The profile is consistent with adequate RNA quality for microarray analysis. Arrows indicate 18S and 28S rRNA positions. The leftmost peak is the marker. The RNA Integrity Nunber (RIN) and the 28S/18S ratio were 9.1 and 1.9 respectively. Lane 1: markers; lane 2: Total RNA extracted from whole blood with erythrocyte lysis followed by globin reduction.
(B) Profile of cRNA samples. 100 ng of total RNA was amplified using the Affymetrix Two-Cycle Eukaryotic Target Labeling kit to generate biotinylated cRNA. Equal amounts of cRNA (450 ng) were run on the Agilent® 2100 bioanalyzer to obtain electropherograms (left) and virtual gel images (right). Red electropherogram represents cRNA from whole blood with no erythrocyte lysis or globin reduction; blue, erythrocyte lysis but no globin reduction; green, erythrocyte lysis followed by globin reduction. Lane 1 of the gel image represents markers; lane 2: cRNA of sample extracted from whole blood with no erythrocyte lysis or globin reduction; lane 3: erythrocyte lysis but no globin reduction; lane 4: erythrocyte lysis followed by globin reduction.
Identification of differentially regulated genes in circulating blood leukocytes of dwarf mice
Two independent statistical methods (SAM and LIMMA) were used to compare the microarray data from Ames dwarf mice with the normal phenotype to determine the gene expression profile specific to the dwarf phenotype. Both SAM and LIMMA are algorithms designed for the statistical analysis of gene expression microarrays and they are currently the most widely used tests for the assessment of microarray differential gene expression. To increase statistical confidence in the results, only genes found significantly changed by both statistical methods were considered differentially expressed in Ames dwarf mice.
In our experience with Affymetrix arrays over the years we found an increasing and significant correlation between array and real-time PCR data (Dhahbi et al. 2006, Dhahbi et al. 2004, Tsuchiya et al 2004). Moreover, the precision and sensitivity in detecting mRNA signals and the accuracy in detecting differential gene expression are increasing by the continuous improvement of the Affymetrix technology and the development of new statistical methods (Sandberg et al. 2007, Chen et al. 2006, Irizarry et al.2006). In addition to the credibility of its data, the Affymetrix platform has proved to be very stable and reliable which makes any further confirmation of its findings by another method such as real time-PCR redundant (de Reynies et al. 2006, Dalma-Weiszhausz et al. 2006).
At a 1.2-fold change cut-off, 3077 probe sets showed significantly different levels of expression in Ames dwarfs compared with controls. We found 1757 of the probe sets were down-regulated, and the remaining 1320 probe sets were up-regulated. We arbitrarily chose 1.2-fold as a cutoff for significance. Application of arbitrary thresholds may fail to detect true changes in gene expression. Genes with large fold changes can be highly variable among samples and thus have no biological significance. On the other hand, genes with small fold changes can be biologically important if the changes in expression of these genes are highly significant statistically because they are measured with high precision as a result of replication.
The 3077 differentially expressed probe sets were searched to identify 273 probes associated with immunity and inflammation using the NetAffx Analysis Center (http://www.affymetrix.com). The complete list of the 273 differentially expressed probe sets associated with immunity and inflammation genes is available as supplementary material accompanying the paper (Table 1).
Table 1.
Immunity and inflammation gene ontology terms and contributed pathways (also called local MAPPS) that showed the most significantly changed gene expression in the leukocytes of dwarf mice. MAPPFinder was used to identify the differentially expressed GO Terms and local MAPPs based on the z-score (>= 2) and permutation p-value (< 0.05). Only GO Terms and local MAPPS with a number of significantly regulated genes more than 3 and fewer than 100 genes were listed in Table.
| GO Term/Local MAPP Name | GOID/Local MAPP1 | Genes Measured2 | Genes Changed3 | Z-score4 | Permutation p-value5 |
|---|---|---|---|---|---|
| Wound healing | GO:0042060 | 41 | 14 | 5.6 | < 0.001 |
| Hemostasis | GO:0007599 | 32 | 12 | 5.6 | < 0.001 |
| Myeloid cell differentiation | GO:0030099 | 51 | 9 | 2.1 | 0.042 |
| Immune cell migration | GO:0050900 | 16 | 5 | 3.1 | 0.013 |
| Regulation of endocytosis | GO:0030100 | 23 | 6 | 2.8 | 0.008 |
| Fatty acid biosynthesis | GO:0006633 | 39 | 8 | 2.5 | 0.017 |
| Mm_Cytokines_and_Inflammatory_Response_Biocarta | Local MAPP | 15 | 6 | 3.8 | 0.004 |
| Mm_Prostaglandin_synthesis_regulation | Local MAPP | 25 | 8 | 3.6 | 0.003 |
| Mm_Eicosanoid_Synthesis | Local MAPP | 17 | 5 | 2.6 | 0.023 |
GO identity number or contributed pathway (local MAPP).
Number of genes measured in the mouse 430 2.0 chip.
Number of genes that meet the criteria for a significant change in expression.
Z-scores are calculated by subtracting the number of genes expected to be randomly changed in a GO term or local MAPP from the observed number of changed genes in that GO term or local MAPP. The obtained value is divided by the standard deviation of the observed number of genes under a hypergeometric distribution.
To calculate the permutation p-value, a non parametric statistic based on 2000 permutations of the data was used. The gene associations were randomized for each sample to generate a distribution of Z scores, which are then used to assign the permutation p-values.
Characterization of biological interactions between the differentially regulated genes related to immunity and inflammation
Upon determination of differentially expressed genes, a list of 273 probe sets associated with immunity and inflammation was generated and subjected to pathway analysis with the PathwayArchitect software. Of the 273 probe sets, PathwayArchitect database established direct interactions between 91 probe sets to produce a gene regulation network centered around 6 main genes, namely casp3, bcl2, il4, prkca, mapk14, and TGFβ1 (Fig. 2). To simplify this complex network for interpretation and discussion purposes, we extracted the subsets of genes that interact with the 6 main genes as illustrated in Fig. 3, S1–S6. These Figures are accessible in supplementary information online as clickable html web files with hyperlinks to information about the genes and the nature of their interactions. Notably, dwarfism decreased the expression of Caspase-3 while it increased the expression of bcl2. Dwarfism also decreased the expression of TGFβ1.
Fig. 2.
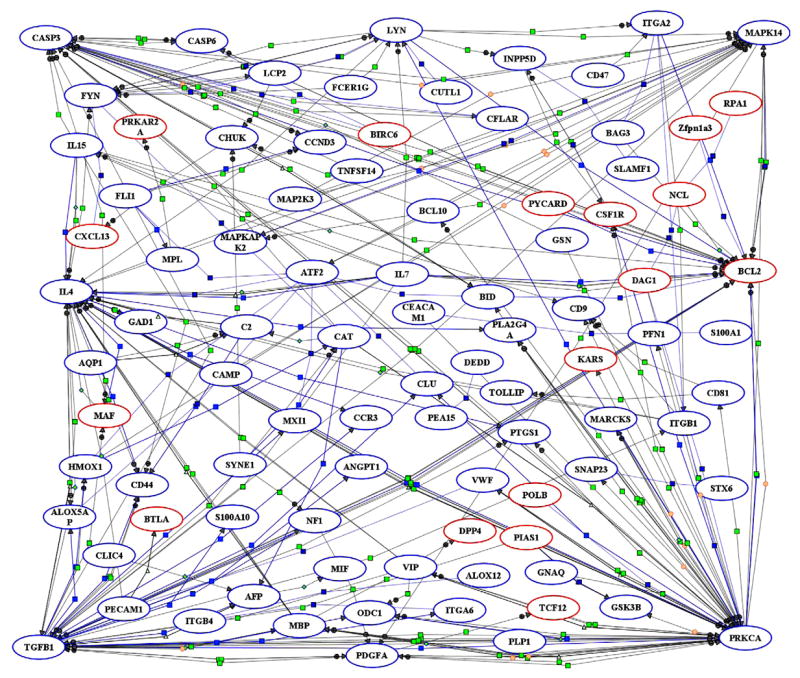
Schematic representation of putative signaling pathways identified in circulating blood leukocytes of dwarf mice. Microarray data were imported into PathwayArchitect and all known biological relationships between the differentially expressed genes were graphically identified as a complex network of genes associated with immunity and inflammation. Red ovals indicate the genes identified as significantly up-regulated. Down-regulated genes are shown as blue ovals. Lines represent functional associations between the genes. Light blue small squares refer to expression; dark blue squares, binding; neon green squares, regulation; light green triangles, transport; green diamond, metabolism; orange circle, protein modification; and green circle, promoter binding. Interactive graphical version of this network is accessible online in supplementary material with hyperlinks to information about the genes and their interactions.
Fig. 3.
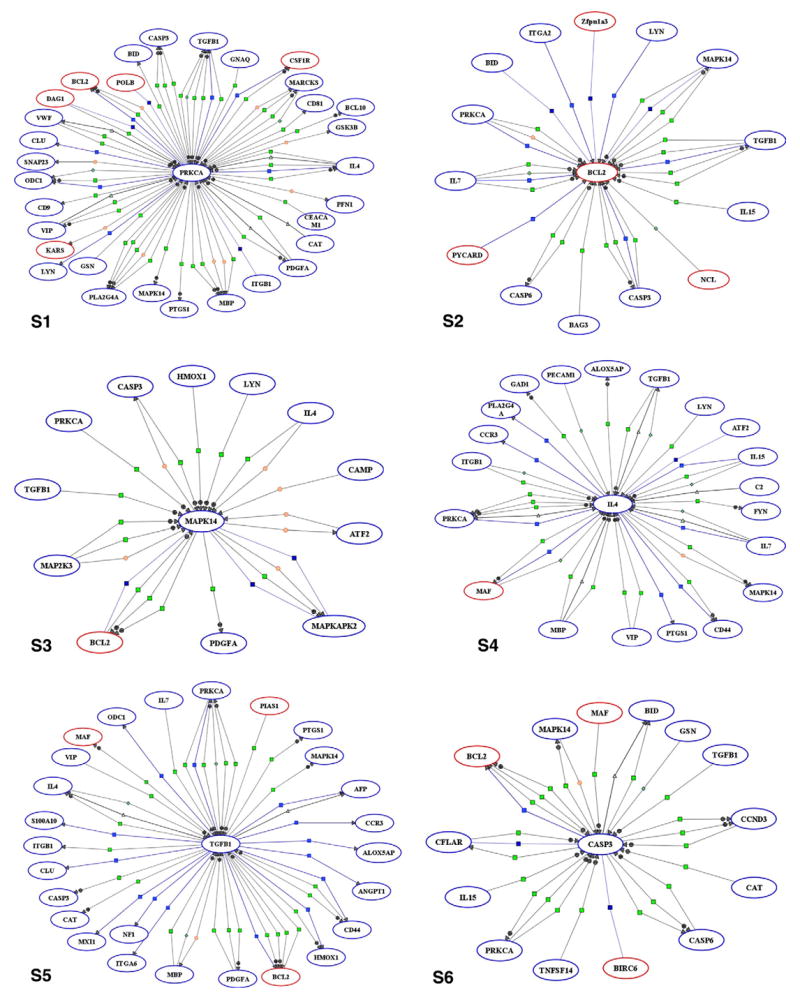
Simplified networks of immunity- and inflammation-related genes interacting with casp3, bcl2, il4, prkca, mapk14, and TGFβ1 in circulating blood leukocytes of dwarf mice. Red ovals indicate the genes identified as significantly up-regulated. Down-regulated genes are shown as blue ovals. Lines represent functional associations between the genes. Light blue small squares refer to expression; dark blue squares, binding; neon green squares, regulation; light green triangles, transport; green diamond, metabolism; orange circle, protein modification; and green circle, promoter binding.
MAPPFinder/GenMAPP analysis reveals altered immunity and inflammation pathways in circulating blood leukocytes of dwarf mice
As noted above, only a portion of the differentially expressed probe sets associated with immunity and inflammation (91 out of 273) were recognized by the PathwayArchitect database as having biological interactions. Thus, the biological significance of a large number of differentially expressed genes remains undetermined. To expand the pathway analysis to more differentially expressed genes and complement the findings of PathwayArchitect analysis, we performed a gene ontology analysis using MAPPFinder/GenMAPP, which is another pathway mapping tool that identifies overrepresented biological themes within gene expression data. Of the 3077 differentially expressed probe sets in Ames dwarf animals, 1686 probe sets were linked to GO terms by MAPPFinder. The output of MAPPFinder was screened to select only statistically significant GO terms and local MAPPS that represent immunity and inflammation pathways. As shown in Table 1, dwarfism altered the expression of 6 GO terms and 3 local MAPPs. The genes assigned to these pathways are represented in Figs. 4–6. The GO pathways most significantly affected in Ames dwarf mice were wound healing and hemostasis (Table 1; z-score = 5.6 and Permute-p < 0.001).
Figs. 4–6.
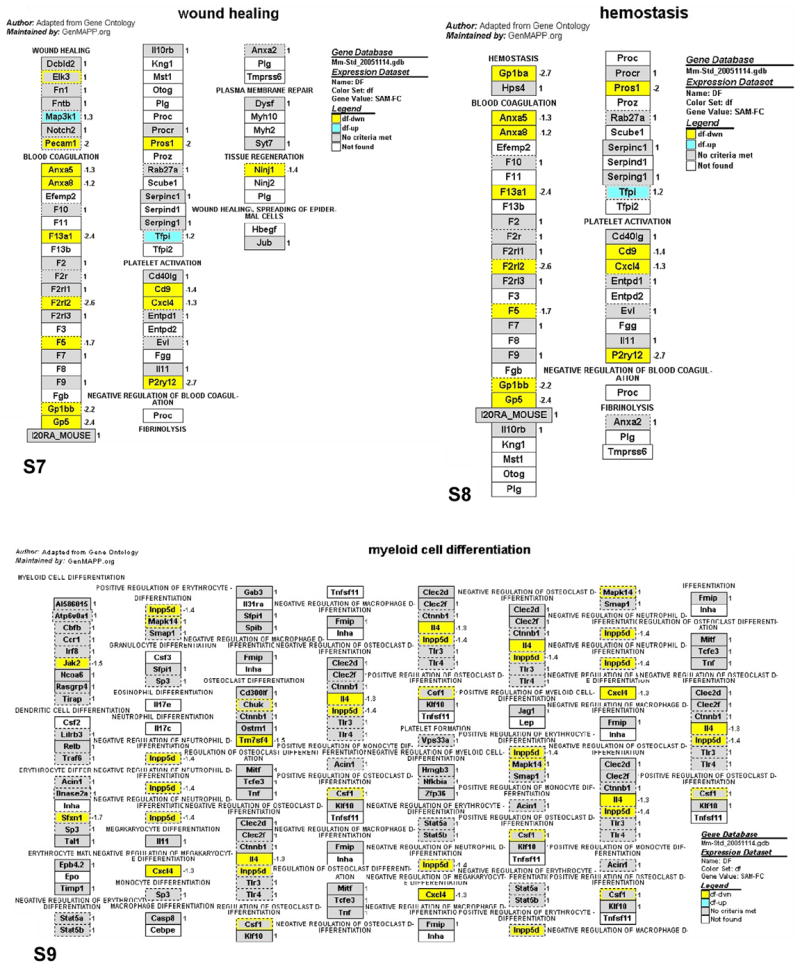
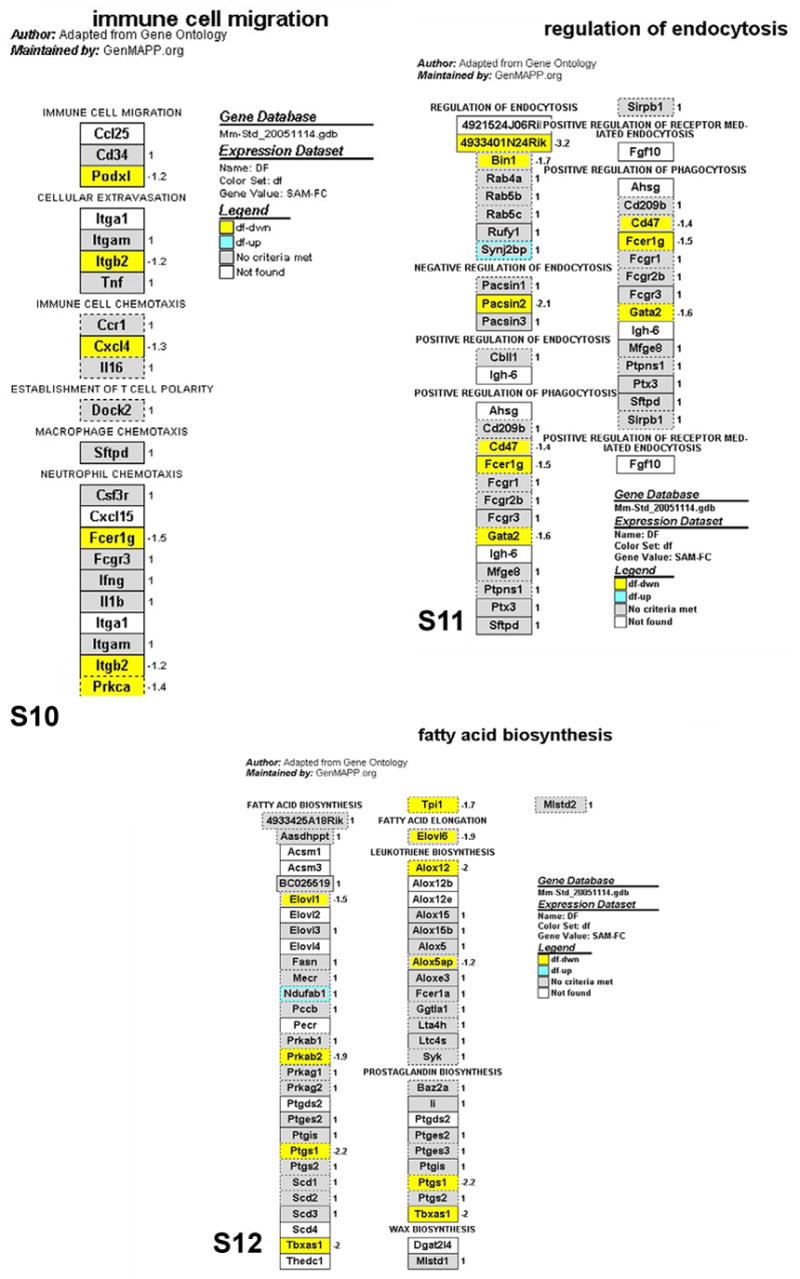
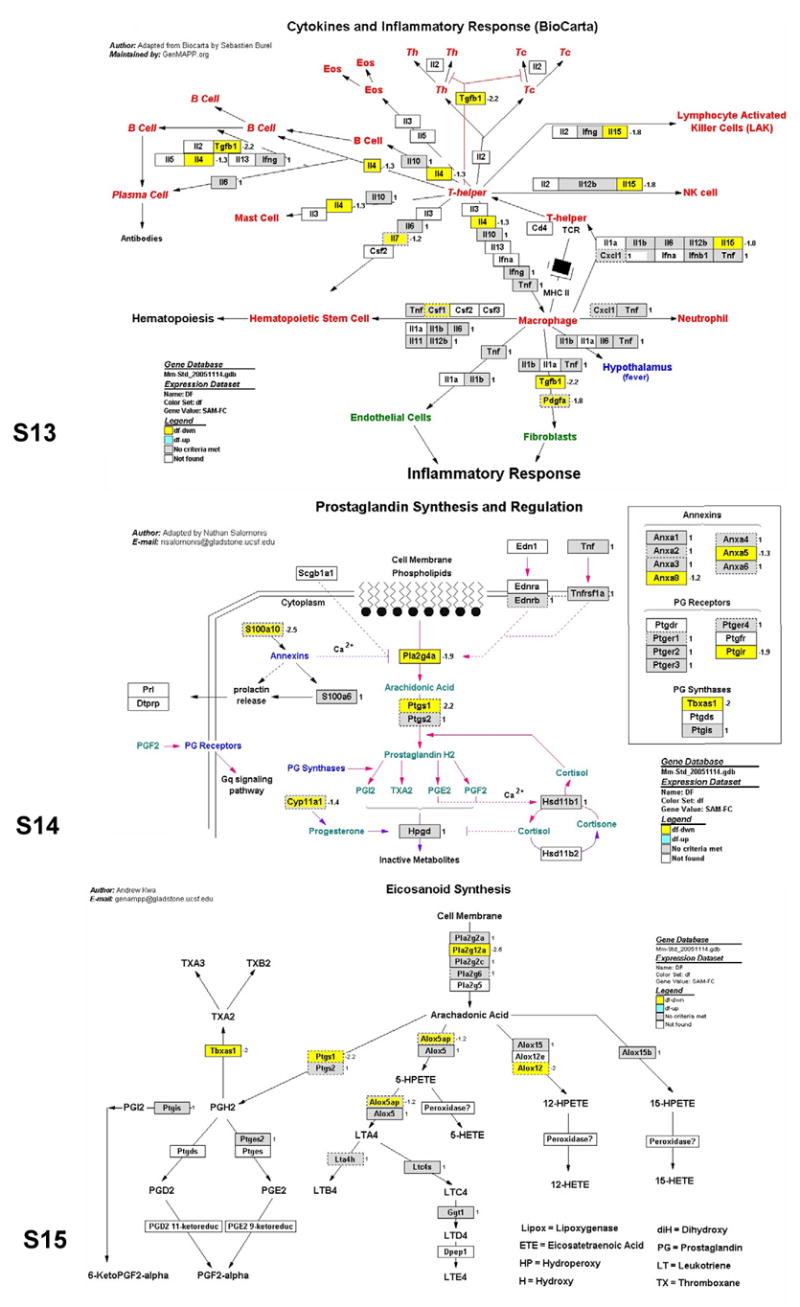
GenMAPP pathways integrating the gene expression data from circulating blood leukocytes in Ames dwarf mice. Yellow, blue and gray boxes indicate significant decrease, increase and no significant change of mRNA levels in the Ames dwarf samples, respectively. White boxes indicate that the gene was not present on the list of genes imported into GenMAPP. Boxes with multiple colors indicate that there was more than one location for the gene existing on the array. The inner color represents the result for the first value determined for the gene and the outer color represents the subsequent value. The gene expression fold change in the leukocytes of Ames dwarf compared to wild-type mice is indicated by numbers to the right of the gene boxes. Gene annotations are available on the GenMAPP interactive version of these pathways online.
Other immunity and inflammation molecular pathways altered in circulating blood leukocytes of dwarf mice
As with PathwayArchitect analysis, MAPPFinder analysis also did not encompass all the differentially expressed genes. Among the 3077 differentially expressed probe sets in Ames dwarf animals, MAPPFinder linked only 1686 probe sets to a GO Term. In order to achieve a more inclusive pathway analysis, the microarray data were analyzed with a third pathway analysis tool, the “globaltest” from the R-package (Goeman et al., 2004). In this approach we started by searching the GO database for all terms associated with immunity and inflammation. The retrieved immunity and inflammation-related GO Terms were then subjected to Global Test analysis to identify the pathways with increased proportions of differentially expressed genes. An advantage of Global Test compared to other methods is its ability to identify pathways with genes that have small changes in expression. As expected, Global Test analysis revealed many more immunity and inflammation pathways altered by dwarfism (Table 2). To determine the genes that contribute to the expression changes of the altered pathways, we used “Geneplot” which is a function that assesses the influence of individual genes in a given pathway produced by Global Test analysis (Table 3). A pathway can be significantly altered because a few genes are highly differentially expressed or because most genes are slightly differentially expressed. This can reflect significant biological differences by distinguishing the key player genes of the pathway. Identifying which genes contribute more to the expression changes can be of great help in guiding further investigation of the pathways.
Table 2.
Gene Ontology inflammation and immunity pathways responsive to dwarfism revealed by Global Test.
| Pathway Name | GO ID | Category1 | Number of genes in pathway2 | Number of genes measured3 | Number of genes affected4 | p-Value5 |
|---|---|---|---|---|---|---|
| B cell homeostasis | GO:0001782 | P | 21 | 21 | 5 | 8.2E-06 |
| T cell homeostasis | GO:0043029 | P | 5 | 5 | 2 | 8.4E-06 |
| Cellular extravasation | GO:0045123 | P | 2 | 2 | 2 | 9.8E-03 |
| Complement activation | GO:0006956 | P | 30 | 30 | 9 | 1.7E-02 |
| Humoral defense mechanism (sensu Vertebrata) | GO:0016064 | P | 42 | 41 | 15 | 2.3E-03 |
| Humoral immune response | GO:0006959 | P | 21 | 21 | 8 | 4.0E-03 |
| Negative regulation of immune response | GO:0050777 | P | 7 | 7 | 4 | 9.4E-03 |
| Phagocytosis, engulfment | GO:0006911 | P | 30 | 30 | 8 | 3.6E-02 |
| Positive regulation of immune response | GO:0050778 | P | 17 | 16 | 4 | 1.6E-02 |
| Regulation of immune response | GO:0050776 | P | 26 | 25 | 9 | 2.2E-02 |
| Wound healing | GO:0042060 | P | 13 | 13 | 5 | 3.9E-03 |
| Chemokine activity | GO:0008009 | F | 54 | 54 | 11 | 1.3E-03 |
| Antigen binding | GO:0003823 | F | 25 | 25 | 5 | 1.9E-02 |
| MHC class I protein complex | GO:0042613 | C | 6 | 6 | 5 | 1.8E-02 |
| MHC protein complex | GO:0042612 | C | 24 | 24 | 4 | 7.7E-03 |
The three highest-level branches in the GO tree: biological processes (P), molecular functions (F), and cellular components (C).
Number of genes within each functional gene category.
Number of genes measured in the array.
Genes differentially expressed with z-score 2.0. The identities of these genes are listed in Table 3.
The P-values were adjusted for multiple hypotheses testing with Bonferroni to control the false discovery rate (FDR).
Table 3.
Influential genes with significant expression changes contributing to the GO pathways listed in Table 2.
| Probe Set ID | Gene Title | Gene Symbol | Influence1 | Z-score2 | Gene expression change |
|---|---|---|---|---|---|
| Humoral defense mechanism (sensu Vertebrata) (GO:0016064) | |||||
| 1418021_at | Complement component 4 (within H-2S) | C4 | 1528 | 3.1 | INCREASE |
| 1455332_x_at | Fc receptor, IgG, low affinity IIb | Fcgr2b | 191 | 4.2 | INCREASE |
| 1459872_x_at | Histocompatibility 2, class II, locus DMa | H2-DMa | 237 | 3.9 | INCREASE |
| 1427351_s_at | Immunoglobulin heavy chain 6 (heavy chain of IgM) | Igh-6 | 306 | 5.3 | INCREASE |
| 1418972_at | B-cell leukemia/lymphoma 10 | Bcl10 | 193 | 7.9 | DECREASE |
| 1418340_at | Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1g | 197 | 5.8 | DECREASE |
| 1424195_a_at | Inositol polyphosphate-5-phosphatase D | Inpp5d | 116 | 7.0 | DECREASE |
| 1422847_a_at | Protein kinase C, delta | Prkcd | 72 | 4.1 | DECREASE |
| Humoral immune response (GO:0006959) | |||||
| 1424305_at | Immunoglobulin joining chain | Igj | 134 | 3.1 | INCREASE |
| 1424931_s_at | Immunoglobulin lambda chain, variable 1 | Igl-V1 | 334 | 3.5 | INCREASE |
| 1420176_x_at | Immunoglobulin lambda-like polypeptide 1 | Igll1 | 25 | 3.0 | INCREASE |
| 1448733_at | Polycomb group ring finger 4 | Pcgf4 | 178 | 4.8 | INCREASE |
| 1449393_at | SH2 domain protein 1A | Sh2d1a | 101 | 3.5 | INCREASE |
| 1448508_at | Traf3 interacting protein 2 | Traf3ip2 | 89 | 3.3 | INCREASE |
| 1419607_at | Tumor necrosis factor | Tnf | 20 | 3.9 | DECREASE |
| Regulation of immune response (GO:0050776) | |||||
| 1435996_at | Caspase recruitment domain family, member 11 | Card11 | 196 | 3.5 | INCREASE |
| 1449130_at | CD1d1 antigen | Cd1d1 | 236 | 4.5 | INCREASE |
| 1439221_s_at | CD40 antigen | Cd40 | 268 | 3.6 | INCREASE |
| 1453389_a_at | Adaptor protein with pleckstrin homology and src | MGI:1345171 | 7 | 2.2 | DECREASE |
| 1418340_at | Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1g | 197 | 5.8 | DECREASE |
| 1449864_at | Interleukin 4 | Il4 | 65 | 7.1 | DECREASE |
| 1445251_at | Tumor necrosis factor (ligand) superfamily, member 13b | Tnfsf13b | 22 | 2.6 | DECREASE |
| Negative regulation of immune response (GO:0050777) | |||||
| 1455332_x_at | Fc receptor, IgG, low affinity IIb | Fcgr2b | 191 | 4.2 | INCREASE |
| 1424195_a_at | Inositol polyphosphate-5-phosphatase D | Inpp5d | 116 | 7.0 | DECREASE |
| Positive regulation of immune response (GO:0050778) | |||||
| 1459872_x_at | Histocompatibility 2, class II, locus DMa | H2-DMa | 237 | 3.9 | INCREASE |
| 1418340_at | Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1g | 197 | 5.8 | DECREASE |
| 1418219_at | Interleukin 15 | Il15 | 354 | 5.5 | DECREASE |
| B cell homeostasis (GO:0001782 ) | |||||
| 1426165_a_at | Caspase 3 | Casp3 | 1442 | 9.9 | DECREASE |
| 1421209_s_at | Inhibitor of kappaB kinase gamma | Ikbkg | 21 | 2.6 | DECREASE |
| 1445251_at | Tumor necrosis factor (ligand) superfamily, member 13b | Tnfsf13b | 22 | 2.6 | DECREASE |
| T cell homeostasis (GO:0001782 ) | |||||
| 1426165_a_at | Caspase 3 | Casp3 | 1442 | 9.9 | DECREASE |
| Cellular extravasation (GO:0045123) | |||||
| 1419607_at | Tumor necrosis factor | Tnf | 20 | 3.9 | DECREASE |
| 1422046_at | Integrin alpha M | Itgam | 190 | 3.6 | DECREASE |
| Complement activation (GO:0006956) | |||||
| 1455821_x_at | Complement component 1, q subcomponent binding protein | C1qbp | 329 | 8.4 | INCREASE |
| 1449401_at | Complement component 1, q subcomponent, gamma polypeptide | C1qg | 1206 | 2.1 | INCREASE |
| 1437726_x_at | Complement component 1, q subcomponent, beta polypeptide | C1qb | 2352 | 3.6 | INCREASE |
| 1418021_at | Complement component 4 (within H-2S) | C4 | 1528 | 3.1 | INCREASE |
| 1417381_at | Complement component 1, q subcomponent, alpha polypeptide | C1qa | 1758 | 4.0 | INCREASE |
| 1441912_x_at | Complement component 2 (within H-2S) | C2 | 96 | 5.1 | DECREASE |
| Phagocytosis, engulfment (GO:0006911) | |||||
| 1420911_a_at | Milk fat globule-EGF factor 8 protein | Mfge8 | 43 | 3.9 | INCREASE |
| 1421840_at | ATP-binding cassette, sub-family A (ABC1), member 1 | Abca1 | 140 | 4.7 | INCREASE |
| 1455332_x_at | Fc receptor, IgG, low affinity IIb | Fcgr2b | 191 | 4.2 | INCREASE |
| 1416986_a_at | Protein tyrosine phosphatase, non-receptor type substrate 1 | Ptpns1 | 46 | 3.4 | DECREASE |
| 1418340_at | Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1g | 197 | 5.8 | DECREASE |
| 1432092_a_at | GULP, engulfment adaptor PTB domain containing 1 | Gulp1 | 21 | 3.0 | DECREASE |
| Wound healing (GO:0042060) | |||||
| 1424850_at | Mitogen activated protein kinase kinase kinase 1 | Map3k1 | 86 | 5.7 | INCREASE |
| 1417662_at | ELK3, member of ETS oncogene family | Elk3 | 26 | 5.5 | DECREASE |
| 1421287_a_at | Platelet/endothelial cell adhesion molecule 1 | Pecam1 | 481 | 6.7 | DECREASE |
| Chemokine activity (GO:0008009) | |||||
| 1448823_at | Chemokine (C-X-C motif) ligand 12 | Cxcl12 | 119 | 3.4 | INCREASE |
| 1417851_at | Chemokine (C-X-C motif) ligand 13 | Cxcl13 | 3260 | 5.9 | INCREASE |
| 1421688_a_at | Chemokine (C-C motif) ligand 1 | Ccl1 | 27 | 5.1 | DECREASE |
| 1420380_at | Chemokine (C-C motif) ligand 2 | Ccl2 | 22 | 8.2 | DECREASE |
| 1417925_at | Chemokine (C-C motif) ligand 22 | Ccl22 | 59 | 7.4 | DECREASE |
| 1421578_at | Chemokine (C-C motif) ligand 4 | Ccl4 | 52 | 4.1 | DECREASE |
| 1415804_at | Chemokine (C-X3-C motif) ligand 1 | Cx3cl1 | 14 | 3.7 | DECREASE |
| 1419697_at | Chemokine (C-X-C motif) ligand 11 | Cxcl11 | 52 | 2.5 | DECREASE |
| 1448995_at | Chemokine (C-X-C motif) ligand 4 | Cxcl4 | 62 | 8.5 | DECREASE |
| 1418480_at | Chemokine (C-X-C motif) ligand 7 | Cxcl7 | 82 | 7.8 | DECREASE |
| Antigen binding (GO:0003823) | |||||
| 1420176_x_at | Immunoglobulin lambda-like polypeptide 1 | Igll1 | 25 | 3.0 | INCREASE |
| 1424305_at | Immunoglobulin joining chain | Igj | 134 | 3.1 | INCREASE |
| 1424931_s_at | Immunoglobulin lambda chain, variable 1 | Igl-V1 | 334 | 3.5 | INCREASE |
| 1427351_s_at | Immunoglobulin heavy chain 6 (heavy chain of IgM) | Igh-6 | 306 | 5.3 | INCREASE |
| MHC class I protein complex (GO:0042613) | |||||
| 1443783_x_at | Histocompatibility 2, class II antigen A, alpha, mRNA | H2-Aa | 275 | 3.0 | DECREASE |
| 1451721_a_at | Histocompatibility 2, class II antigen A, beta 1 | H2-Ab1 | 183 | 2.5 | DECREASE |
| MHC protein complex (GO:0042612) | |||||
| 1418536_at | Similar to MHC Q8/9d surface antigen | LOC386462 | 379 | 4.4 | INCREASE |
| 1425336_x_at | Histocompatibility 2, K1, K region | H2-K1 | 310 | 5.1 | INCREASE |
| 1426324_at | Histocompatibility 2, D region locus 1 | H2-D1 | 76 | 2.3 | INCREASE |
| 1450702_at | Hemochromatosis | Hfe | 75 | 4.6 | DECREASE |
Indicates the influence of the gene on the test result of the whole pathway.
Represents the global test statistic for the single gene pathway containing only that gene.
As shown in Table 2, B and T cell homeostasis were among the pathways significantly affected in Ames dwarf mice (p-value < 0.00001). Geneplot analysis showed that caspase 3 was the most influential gene in B and T cell homeostasis pathways (Table 3; z-score = 9.9, Influence = 1442). This is in line with SAM and LIMMA analyses, which found that caspase 3 mRNA expressions were decreased 3.1-fold in Ames dwarf mice (Table S1). Global Testing also revealed that the Ames dwarf mutation significantly altered the humoral defense mechanism pathway (p-value = 0.002). The most influential genes responsible for the alteration of this pathway by dwarfism are complement component 4 (Table 3; z-score = 3.1, Influence = 1528) followed by immunoglobulin heavy chain 6 (z-score = 5.3, Influence = 306). However, the expression of both of these genes was not significantly changed with SAM and LIMMA analyses. This is an example of how Global Testing is able to unravel subtle gene expression changes that remain undetectable with microarray traditional methods aimed at identifying the statistically significant genes. Dwarfism altered the expression of other important pathways related to immunity and inflammation including chemokine activity, complement activation and wound healing pathways (Tables 2 & 3).
DISCUSSION
In the present study, the gene expression profiles of circulating blood leukocytes in Ames dwarf mice and their normal littermates were analyzed. Ames dwarf mice are homozygous for a spontaneous autosomal recessive loss-of-function mutation of the prophet of pituitary factor 1 gene (Prop1df). These mutants are remarkably long-lived and healthier than normal animals, with an increased “health span” and reduced incidence of age-related disease (Bartke and Brown-Borg, 2004). Interestingly, Ames dwarfs share many characteristics with animals subjected to calorie restriction (CR). However, it is known that CR enhances immune function and ameliorates inflammation, while the immune system status of Ames dwarf mice is not well established (Pahlavani, 2000, 2004; Fernandes et al., 1997, Spaulding et al., 1997). To evaluate the effects of Prop1df mutation on immunity and inflammation, we analyzed expression of 34,000 genes. The list of the genes that were differentially expressed in normal and dwarf mice included 273 probe sets involved in immunity and inflammation. Analysis of direct interactions between 91 probe sets based on the Pathway Architect data base identified 6 main genes: casp3, bcl2, IL4, mapk14, TGFβ1 and prkca.
Two of these genes, casp3 and bcl2 play important roles in apoptosis of lymphocytes. Caspase 3 plays a central role in the execution-phase of apoptosis, and is also essential for the regulation of B and T cell homeostasis. Bcl2 blocks the apoptotic death of some cells including lymphocytes. Decrease of the expression of pro-apoptotic casp3 and a coinciding increase of anti-apoptotic Bcl2 gene expression suggest decreased apoptosis in Ames dwarf’s peripheral blood leukocytes (PBLs) when compared to normal controls. Dwarfism also decreased the expression of TGFβ1, a cytokine that controls cell proliferation, differentiation, can stimulate apoptosis, and plays a key role in inflammation and defense responses. Down-regulation of this gene could suggest decreased apoptosis and, more importantly, suppressed cell proliferation. These effects would be consistent with decreased peripheral blood cell population and B cell deficiency in Ames dwarf mice (Murphy et al., 1992). Finding of decreased TGFβ1 and a correlated reduction in peripheral blood cell population fits nicely with decreased mRNA expression of IL-4, a pleiotropic cytokine produced by activated T cells. IL4 is also known as a B cell stimulatory factor. The MAPK14 is known as a regulator of cellular processes such as proliferation, differentiation, transcription regulation and development. The inactivation of MAPK14 promotes activation of Bcl2, which additionally supports decreased apoptosis in circulating blood cells of Ames dwarf mice. The last of the 6 genes on this list, prkca was also decreased in Ames dwarf mice. This gene plays an important role in regulating cell-cycle control and apoptosis by activation of Bcl2.
In conclusion, the up-regulation of Bcl2 expression and opposite changes at the mRNA levels of casp3, IL4, TGFβ1, mapk14 and prkca in Ames dwarf mice in comparison to their control littermates suggest reduced apoptosis and decreased proliferation of PBLs. The suggested decrease of apoptosis in dwarf PBLs contrasts with the results reported by Kennedy et al. (2003), which indicated increased apoptosis in the liver of these long-lived mutants. The authors suggested that increased apoptosis in some organs, such as the liver, can be beneficial for increased longevity by possibly higher efficiency of eliminating damaged or harmful neoplastic cells. However, there is still wide debate about the role of apoptosis in aging and longevity. Moreover, it is known that in different organs apoptosis can have different roles, which additionally complicates the relationship of apoptosis and aging (Pollack and Leewenburgh, 2001).
On the basis of the present findings, it is possible to speculate that compared to normal animals Ames dwarf mice have a more advanced cellular repair system and a better PBLs selection system during development. Reduced apoptosis and cell proliferation may serve to extend maintenance of an efficient immune system in keeping with the increased lifespan of these mutants.
Genes involved in wound healing and hemostasis showed greatest differences in expression in dwarf as compared to normal mice. Based on the studies of FXIII (F13a1)-deficient mice (Lauer et al., 2002, Inbal et al., 2005) and the function of other affected genes (Figures 7 and 8), the down-regulation of these genes suggests that Ames dwarf mice could have impaired wound healing and delayed arrest of bleeding. However, this defective repair system could be a trade-off for other aspect of health and/or a potentially longer lifespan. The data obtained in Par3 (F2rl2) knockout mice indicated that the deficiency of these genes was protective against ferric chloride-induced thrombosis of mesenteric arterioles and against thromboplastin-induced pulmonary embolism (Weiss et al., 2002). Also factor V Leiden (FVL or F5) is known as a beneficial factor in hemophilia (Schlachterman et al., 2005), which suggests that a low level of this molecule could prevent blood from clotting. However, from the other perspective, the expression of Gp1bb and Gp5 is also down-regulated, which can balance the risk for extensive bleeding in mutant animals. These genes are known to be elevated specifically in patients subjected to aspirin treatments that can increase bleeding (Aktas et al., 2005). In summary, some of the identified changes in gene expression in long-lived Ames dwarf mice could protect from cardiovascular disease by preventing blood clotting and promoting better circulation.
There were also multiple alterations of the myeloid cell differentiation pathway genes in Ames dwarf mice in comparison to their normal siblings. This suggests that the process of myeloid progenitor cell differentiation into erythrocyte, megakaryocyte, platelet, basophil, eosinophil, neutrophil, monocyte or macrophage cells is strongly affected by Prop1df mutation. More interestingly, dwarfism down-regulated pathways related to fatty acid biosynthesis, including the synthesis of two major inflammatory mediators, prostaglandins and leukotrienes. Other inflammatory factors such as the COX gene are suppressed in Ames dwarf mice, leading to decreased prostaglandin production, and contribute toward maintaining a healthier and longer life in the animal. Aspirin, ibuprofen and other anti-inflammatory drugs target these same genes. It is known that, when therapeutically used, aspirin can be protective from cancer by decreasing the level of prostaglandin synthesis, which nicely correlates with significantly decreased or delayed risk of cancer in these long-lived mutant mice (Ikeno et al., 2003). Also, decrease of the Alox12 gene that regulates leukotriene biosynthesis was shown to reduce oxidant stress (Li et al., 2005), which correlates with the finding that fibroblasts derived from Ames dwarf mice are more resistant to oxidative stress than fibroblasts derived from normal animals (Salmon et al., 2005). Many of the genes involved in the cytokine and inflammatory pathway were altered indicating that the Ames dwarf mutation exerts significant effects on immunity and inflammation.
SUMMARY
Analysis of peripheral blood leukocyte gene expression profiles indicated that many of the genes that regulate immunity responses such as apoptosis, wound healing, hemostasis and fatty acids biosynthesis were suppressed by Prop1df (Ames dwarf) mutation. These results indicate that Ames dwarfism affects the pathways related to immunity and inflammation. Suppression of blood coagulation could prevent cardiovascular diseases, while decreased synthesis of prostaglandin could act as an anti-inflammatory factor and, more importantly, protect from certain cancers. However, alterations in gene expression detected in the present study could also suggest that the suppression of immunity and inflammation represent an adaptation of the immune system that is maintained longer in long-lived Ames dwarf mice than in control animals with normal lifespan.
It is also important to point out that for more specific conclusions additional studies are necessary. It will be important to investigate observed alterations in particular PBLs populations to determine if observed changes are due to different composition of PBL populations in Ames dwarf mice vs. WT animals, or due to the genetic differences. It will be also important to perform functional studies to confirm the association between expression of genes related to a particular pathway and the function in these animals.
Supplementary Material
Acknowledgments
This project was supported by BioMarker Pharmaceuticals Inc. and NIA U19 AG023122. The authors would like to thank Steve Sandstrom for helping with the editing of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aktas B, Pozgajova M, Bergmeier W, Sunnarborg S, Offermanns S, Lee D, Wagner DD, Nieswandt B. Aspirin induces platelet receptor shedding via adam17 (tace) J Biol Chem. 2005;280:39716–39722. doi: 10.1074/jbc.M507762200. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Yagi K, Venable S, Darlington G. Gene expression profile of long-lived ames dwarf mice and little mice. Aging Cell. 2004;3:423–441. doi: 10.1111/j.1474-9728.2004.00125.x. [DOI] [PubMed] [Google Scholar]
- Amador-Noguez D, Zimmerman J, Venable S, Darlington G. Gender-specific alterations in gene expression and loss of liver sexual dimorphism in the long-lived ames dwarf mice. Biochem Biophys Res Commun. 2005;332:1086–1100. doi: 10.1016/j.bbrc.2005.05.063. [DOI] [PubMed] [Google Scholar]
- Baroni CD, Fabris N, Bertoli G. Effects of hormones on development and function of lymphoid tissues. Synergistic action of thyroxin and somatotropic hormone in pituitary dwarf mice. Immunology. 1969;17:303–314. [PMC free article] [PubMed] [Google Scholar]
- Bartke A. Minireview: Role of the growth hormone/insulin-like growth factor system in mammalian aging. Endocrinology. 2005;146:3718–3723. doi: 10.1210/en.2005-0411. [DOI] [PubMed] [Google Scholar]
- Bartke A. Long-lived klotho mice: New insights into the roles of igf-1 and insulin in aging. Trends Endocrinol Metab. 2006;17:33–35. doi: 10.1016/j.tem.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Curr Top Dev Biol. 2004;63:189–225. doi: 10.1016/S0070-2153(04)63006-7. [DOI] [PubMed] [Google Scholar]
- Bonkowski MS, Rocha JS, Masternak MM, Al Regaiey KA, Bartke A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci U S A. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384:33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- Chakravarti B, Abraham GN. Aging and t-cell-mediated immunity. Mech Ageing Dev. 1999;108:183–206. doi: 10.1016/s0047-6374(99)00009-3. [DOI] [PubMed] [Google Scholar]
- Chen Z, McGee M, Liu Q, Scheuermann RH. A distribution free summarization method for Affymetrix GeneChip arrays. Bioinformatics. 2007;1;23(3):321–7. doi: 10.1093/bioinformatics/btl609. [DOI] [PubMed] [Google Scholar]
- Crimmins EM, Finch CE. Infection, inflammation, height, and longevity. Proc Natl Acad Sci U S A. 2006;103:498–503. doi: 10.1073/pnas.0501470103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross RJ, Bryson JS, Roszman TL. Immunologic disparity in the hypopituitary dwarf mouse. J Immunol. 1992;148:1347–1352. [PubMed] [Google Scholar]
- Curtis RK, Oresic M, Vidal-Puig A. Pathways to the analysis of microarray data. Trends Biotechnol. 2005;23:429–435. doi: 10.1016/j.tibtech.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. Genmapp, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31:19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Dalma-Weiszhausz DD, Warrington J, Tanimoto EY, Miyada CG. The affymetrix GeneChip platform: an overview. Methods Enzymol. 2006;410:3–28. doi: 10.1016/S0076-6879(06)10001-4. Review. [DOI] [PubMed] [Google Scholar]
- Danesh J, Wheeler JG, Hirschfield GM, Eda S, Eiriksdottir G, Rumley A, Lowe GD, Pepys MB, Gudnason V. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- Debey S, Schoenbeck U, Hellmich M, Gathof BS, Pillai R, Zander T, Schultze JL. Comparison of different isolation techniques prior gene expression profiling of blood derived cells: Impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J. 2004;4:193–207. doi: 10.1038/sj.tpj.6500240. [DOI] [PubMed] [Google Scholar]
- de Reynies Reynies A, Geromin D, Cayuela JM, Petel F, Dessen P, Sigaux F, Rickman DS. Comparison of the latest commercial short and long oligonucleotide microarray technologies. BMC Genomics. 2006;15(7):51. doi: 10.1186/1471-2164-7-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeale B, Brummel T, Seroude L. Immunity and aging: The enemy within? Aging Cell. 2004;3:195–208. doi: 10.1111/j.1474-9728.2004.00106.x. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM, Tsuchiya T, Kim HJ, Mote PL, Spindler SR. Gene expression and physiologic responses of the heart to the initiation and withdrawal of caloric restriction. J Gerontol A Biol Sci Med Sci. 2006 Mar;61(3):218–31. doi: 10.1093/gerona/61.3.218. [DOI] [PubMed] [Google Scholar]
- Dhahbi JM, Kim HJ, Mote PL, Beaver RJ, Spindler SR. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc Natl Acad Sci U S A. 2004 Apr 13;101(15):5524–9. doi: 10.1073/pnas.0305300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. Mappfinder: Using gene ontology and genmapp to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont F, Robert F, Bischoff P. T and b lymphocytes in pituitary dwarf snell-bagg mice. Immunology. 1979;38:23–31. [PMC free article] [PubMed] [Google Scholar]
- Duquesnoy RJ. Immunodeficiency of the thymus-dependent system of the ames dwarf mouse. J Immunol. 1972;108:1578–1590. [PubMed] [Google Scholar]
- Effros RB. Ageing and the immune system. Novartis Found Symp. 2001;235:130–139. doi: 10.1002/0470868694.ch12. discussion 139–145, 146–139. [DOI] [PubMed] [Google Scholar]
- Fabris N, Pierpaoli W, Sorkin E. Hormones and the immunological capacity.Iv Restorative effects of developmental hormones or of lymphocytes on the immunodeficiency syndrome of the dwarf mouse. Clin Exp Immunol. 1971;9:227–240. [PMC free article] [PubMed] [Google Scholar]
- Feezor RJ, Baker HV, Mindrinos M, Hayden D, Tannahill CL, Brownstein BH, Fay A, MacMillan S, Laramie J, Xiao W, Moldawer LL, Cobb JP, Laudanski K, Miller-Graziano CL, Maier RV, Schoenfeld D, Davis RW, Tompkins RG. Whole blood and leukocyte rna isolation for gene expression analyses. Physiol Genomics. 2004;19:247–254. doi: 10.1152/physiolgenomics.00020.2004. [DOI] [PubMed] [Google Scholar]
- Fernandes G, Venkatraman JT, Turturro A, Attwood VG, Hart RW. Effect of food restriction on life span and immune functions in long-lived fischer-344 x brown norway f1 rats. J Clin Immunol. 1997;17:85–95. doi: 10.1023/a:1027344730553. [DOI] [PubMed] [Google Scholar]
- Finch CE, Crimmins EM. Inflammatory exposure and historical changes in human life-spans. Science. 2004;305:1736–1739. doi: 10.1126/science.1092556. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Harrison DE. In: Genetic Effects on Aging II. Harrison DE, editor. Telford; Caldwell, NJ: 1990. pp. 437–456. [Google Scholar]
- Flurkey K, Papaconstantinou J, Harrison DE. The snell dwarf mutation pit1 (dw) can increase life span in mice. Mech Ageing Dev. 2002;123:121–130. doi: 10.1016/s0047-6374(01)00339-6. [DOI] [PubMed] [Google Scholar]
- Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci U S A. 2001;98:6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy--analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Ginaldi L, De Martinis M, D’Ostilio A, Marini L, Loreto MF, Martorelli V, Quaglino D. The immune system in the elderly: Ii. Specific cellular immunity. Immunol Res. 1999;20:109–115. doi: 10.1007/BF02786467. [DOI] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A global test for groups of genes: Testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- Ikeno Y, Bronson RT, Hubbard GB, Lee S, Bartke A. Delayed occurrence of fatal neoplastic diseases in ames dwarf mice: Correlation to extended longevity. J Gerontol A Biol Sci Med Sci. 2003;58:291–296. doi: 10.1093/gerona/58.4.b291. [DOI] [PubMed] [Google Scholar]
- Inbal A, Lubetsky A, Krapp T, Castel D, Shaish A, Dickneitte G, Modis L, Muszbek L, Inbal A. Impaired wound healing in factor xiii deficient mice. Thromb Haemost. 2005;94:432–437. doi: 10.1160/TH05-04-0291. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Wu Z, Jaffee HA. Comparison of Affymetrix GeneChip expression measures. Bioinformatics. 2006;1;22(7):789–94. doi: 10.1093/bioinformatics/btk046. [DOI] [PubMed] [Google Scholar]
- Kennedy MA, Rakoczy SG, Brown-Borg HM. Long-living ames dwarf mouse hepatocytes readily undergo apoptosis. Exp Gerontol. 2003;38:997–1008. doi: 10.1016/s0531-5565(03)00164-5. [DOI] [PubMed] [Google Scholar]
- Lauer P, Metzner HJ, Zettlmeissl G, Li M, Smith AG, Lathe R, Dickneite G. Targeted inactivation of the mouse locus encoding coagulation factor xiii-a: Hemostatic abnormalities in mutant mice and characterization of the coagulation deficit. Thromb Haemost. 2002;88:967–974. [PubMed] [Google Scholar]
- Li SL, Dwarakanath RS, Cai Q, Lanting L, Natarajan R. Effects of silencing leukocyte-type 12/15-lipoxygenase using short interfering rnas. J Lipid Res. 2005;46:220–229. doi: 10.1194/jlr.M400328-JLR200. [DOI] [PubMed] [Google Scholar]
- Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. Netaffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloberas J, Celada A. Effect of aging on macrophage function. Exp Gerontol. 2002;37:1325–1331. doi: 10.1016/s0531-5565(02)00125-0. [DOI] [PubMed] [Google Scholar]
- Longo VD, Finch CE. Evolutionary medicine: From dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- Masternak MM, Al-Regaiey K, Bonkowski MS, Panici J, Sun L, Wang J, Przybylski GK, Bartke A. Divergent effects of caloric restriction on gene expression in normal and long-lived mice. J Gerontol A Biol Sci Med Sci. 2004;59:784–788. doi: 10.1093/gerona/59.8.b784. [DOI] [PubMed] [Google Scholar]
- McGlauchlen KS, Vogel LA. Ineffective humoral immunity in the elderly. Microbes Infect. 2003;5:1279–1284. doi: 10.1016/j.micinf.2003.09.001. [DOI] [PubMed] [Google Scholar]
- McLachlan JA, Serkin CD, Morrey KM, Bakouche O. Antitumoral properties of aged human monocytes. J Immunol. 1995;154:832–843. [PubMed] [Google Scholar]
- McLachlan JA, Serkin CD, Morrey-Clark KM, Bakouche O. Immunological functions of aged human monocytes. Pathobiology. 1995;63:148–159. doi: 10.1159/000163946. [DOI] [PubMed] [Google Scholar]
- Miller RA. Effect of aging on t lymphocyte activation. Vaccine. 2000;18:1654–1660. doi: 10.1016/s0264-410x(99)00502-2. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Durum SK, Anver MR, Longo DL. Immunologic and hematologic effects of neuroendocrine hormones. Studies on dw/j dwarf mice. J Immunol. 1992;148:3799–3805. [PubMed] [Google Scholar]
- Pahlavani MA. Caloric restriction and immunosenescence: A current perspective. Front Biosci. 2000;5:D580–587. doi: 10.2741/pahlavani. [DOI] [PubMed] [Google Scholar]
- Pahlavani MA. Influence of caloric restriction on aging immune system. J Nutr Health Aging. 2004;8:38–47. [PubMed] [Google Scholar]
- Pawelec G, Solana R, Remarque E, Mariani E. Impact of aging on innate immunity. J Leukoc Biol. 1998;64:703–712. doi: 10.1002/jlb.64.6.703. [DOI] [PubMed] [Google Scholar]
- Pearson TA, Mensah GA, Alexander RW, Anderson JL, Cannon RO, 3rd, Criqui M, Fadl YY, Fortmann SP, Hong Y, Myers GL, Rifai N, Smith SC, Jr, Taubert K, Tracy RP, Vinicor F. Markers of inflammation and cardiovascular disease: Application to clinical and public health practice: A statement for healthcare professionals from the centers for disease control and prevention and the american heart association. Circulation. 2003;107:499–511. doi: 10.1161/01.cir.0000052939.59093.45. [DOI] [PubMed] [Google Scholar]
- Polignano A, Tortorella C, Venezia A, Jirillo E, Antonaci S. Age-associated changes of neutrophil responsiveness in a human healthy elderly population. Cytobios. 1994;80:145–153. [PubMed] [Google Scholar]
- Pollack M, Leeuwenburgh C. Apoptosis and aging: Role of the mitochondria. J Gerontol A Biol Sci Med Sci. 2001;56:B475–482. doi: 10.1093/gerona/56.11.b475. [DOI] [PubMed] [Google Scholar]
- Ridker PM, Cushman M, Stampfer MJ, Tracy RP, Hennekens CH. Inflammation, aspirin, and the risk of cardiovascular disease in apparently healthy men. N Engl J Med. 1997;336:973–979. doi: 10.1056/NEJM199704033361401. [DOI] [PubMed] [Google Scholar]
- Ross R. The pathogenesis of atherosclerosis: A perspective for the 1990s. Nature. 1993;362:801–809. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- Salmon AB, Murakami S, Bartke A, Kopchick J, Yasumura K, Miller RA. Fibroblast cell lines from young adult mice of long-lived mutant strains are resistant to multiple forms of stress. Am J Physiol Endocrinol Metab. 2005;289:E23–29. doi: 10.1152/ajpendo.00575.2004. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Larsson O. Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics. 2007;8;8(1):48. doi: 10.1186/1471-2105-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlachterman A, Schuettrumpf J, Liu JH, Furlan Freguia C, Toso R, Poncz M, Camire RM, Arruda VR. Factor v leiden improves in vivo hemostasis in murine hemophilia models. J Thromb Haemost. 2005;3:2730–2737. doi: 10.1111/j.1538-7836.2005.01639.x. [DOI] [PubMed] [Google Scholar]
- Schneider GB. Immunological competence in snell-bagg pituitary dwarf mice: Response to the contact-sensitizing agent oxazolone. Am J Anat. 1976;145:371–393. doi: 10.1002/aja.1001450306. [DOI] [PubMed] [Google Scholar]
- Silberberg R. Articular aging and osteoarthrosis in dwarf mice. Pathol Microbiol (Basel) 1972;38:417–430. doi: 10.1159/000162458. [DOI] [PubMed] [Google Scholar]
- Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Solana R, Pawelec G. Molecular and cellular basis of immunosenescence. Mech Ageing Dev. 1998;102:115–129. doi: 10.1016/s0047-6374(98)00029-3. [DOI] [PubMed] [Google Scholar]
- Sornson MW, Wu W, Dasen JS, Flynn SE, Norman DJ, O’Connell SM, Gukovsky I, Carriere C, Ryan AK, Miller AP, Zuo L, Gleiberman AS, Andersen B, Beamer WG, Rosenfeld MG. Pituitary lineage determination by the prophet of pit-1 homeodomain factor defective in ames dwarfism. Nature. 1996;384:327–333. doi: 10.1038/384327a0. [DOI] [PubMed] [Google Scholar]
- Spaulding CC, Walford RL, Effros RB. The accumulation of non-replicative, non-functional, senescent t cells with age is avoided in calorically restricted mice by an enhancement of t cell apoptosis. Mech Ageing Dev. 1997;93:25–33. doi: 10.1016/s0047-6374(96)01808-8. [DOI] [PubMed] [Google Scholar]
- Szakal AK, Aydar Y, Balogh P, Tew JG. Molecular interactions of fdcs with b cells in aging. Semin Immunol. 2002;14:267–274. doi: 10.1016/s1044-5323(02)00059-3. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T, Dhahbi JM, Cui X, Mote PL, Bartke A, Spindler SR. Additive regulation of hepatic gene expression by dwarfism and caloric restriction. Physiol Genomics. 2004;17:307–315. doi: 10.1152/physiolgenomics.00039.2004. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch R, Sohal RS. Seminars in medicine of the beth israel deaconess medical center. Caloric intake and aging. N Engl J Med. 1997;337:986–994. doi: 10.1056/NEJM199710023371407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EJ, Hamilton JR, Lease KE, Coughlin SR. Protection against thrombosis in mice lacking par3. Blood. 2002;100:3240–3244. doi: 10.1182/blood-2002-05-1470. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ. Simpleaffy: A bioconductor package for affymetrix quality control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- Yoshino T, Tamura M, Kawabe M, Nomura H, Imai N, Ono M. Effects of recombinant human granulocyte colony-stimulating factor on neutrophil functions in aged animals. Br J Haematol. 1992;82:664–670. doi: 10.1111/j.1365-2141.1992.tb06941.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


