Abstract
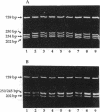
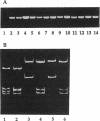
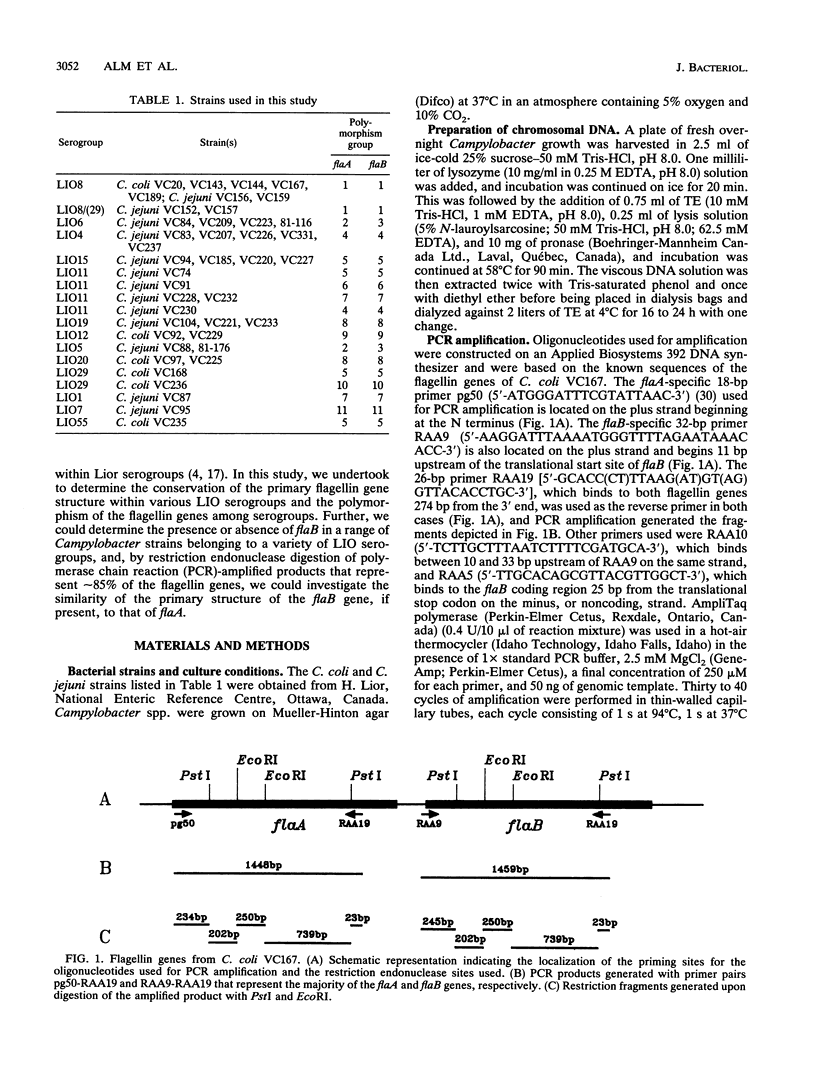
The complex flagellar filaments of the LIO8 serogroup member Campylobacter coli VC167 are composed of two highly related subunit proteins encoded by the flaA and flaB genes which share 92% identity. Using oligonucleotide primers based on the known DNA sequence of both the flaA and flaB genes from C. coli VC167 in the polymerase chain reaction, we have shown conservation of both fla genes among isolates within the LIO8 heat-labile serogroup by digestion of the amplified product with PstI and EcoRI restriction endonucleases. Amplification and subsequent restriction analysis of the flaA flagellin gene from Campylobacter isolates belonging to 13 different LIO serogroups further identified 10 unique polymorphic groups. Within most of the serogroups examined, isolates appeared to contain flaA genes with conserved primary structures. Only in serogroups LIO11 and LIO29 did independent isolates possess flagellin genes with different primary structures. Furthermore, by employing primers specific for the flaB gene of C. coli VC167, all serogroups examined contained a second fla gene corresponding to flaB. In all serogroups except the LIO5 and LIO6 isolates which were identical to each other, the polymorphic pattern of this flaB gene was identical to that of the corresponding flaA gene. These data indicate that the presence of a second highly homologous flagellin gene is widespread throughout Campylobacter isolates and that in most instances, the primary structure of the two fla genes is conserved within isolates belonging to the same heat-labile LIO serogroup. This may represent the presence of clonal evolutionary groups in Campylobacter spp.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Pluschke G. Clonal analysis of descent and virulence among selected Escherichia coli. Annu Rev Microbiol. 1986;40:185–210. doi: 10.1146/annurev.mi.40.100186.001153. [DOI] [PubMed] [Google Scholar]
- Aeschbacher M., Piffaretti J. C. Population genetics of human and animal enteric Campylobacter strains. Infect Immun. 1989 May;57(5):1432–1437. doi: 10.1128/iai.57.5.1432-1437.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A., Guerry P., Power M. E., Lior H., Trust T. J. Analysis of the role of flagella in the heat-labile Lior serotyping scheme of thermophilic Campylobacters by mutant allele exchange. J Clin Microbiol. 1991 Nov;29(11):2438–2445. doi: 10.1128/jcm.29.11.2438-2445.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm R. A., Guerry P., Power M. E., Trust T. J. Variation in antigenicity and molecular weight of Campylobacter coli VC167 flagellin in different genetic backgrounds. J Bacteriol. 1992 Jul;174(13):4230–4238. doi: 10.1128/jb.174.13.4230-4238.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P., Roth J. Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci U S A. 1981 May;78(5):3113–3117. doi: 10.1073/pnas.78.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P., Musser J. M., Helmuth R., Farmer J. J., 3rd, Frerichs W. M., Wachsmuth I. K., Ferris K., McWhorter A. C., Wells J. G., Cravioto A. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Mocca L. F., Frasch C. E., Frøholm L. O., Zollinger W. D., Selander R. K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987 Jun;169(6):2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov O. V., Kostyukova A. S., Pyatibratov M. G. Architectonics of a bacterial flagellin filament subunit. FEBS Lett. 1988 Dec 5;241(1-2):145–148. doi: 10.1016/0014-5793(88)81048-2. [DOI] [PubMed] [Google Scholar]
- Fischer S. H., Nachamkin I. Common and variable domains of the flagellin gene, flaA, in Campylobacter jejuni. Mol Microbiol. 1991 May;5(5):1151–1158. doi: 10.1111/j.1365-2958.1991.tb01888.x. [DOI] [PubMed] [Google Scholar]
- Gerl L., Sumper M. Halobacterial flagellins are encoded by a multigene family. Characterization of five flagellin genes. J Biol Chem. 1988 Sep 15;263(26):13246–13251. [PubMed] [Google Scholar]
- Guerry P., Alm R. A., Power M. E., Logan S. M., Trust T. J. Role of two flagellin genes in Campylobacter motility. J Bacteriol. 1991 Aug;173(15):4757–4764. doi: 10.1128/jb.173.15.4757-4764.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P., Logan S. M., Thornton S., Trust T. J. Genomic organization and expression of Campylobacter flagellin genes. J Bacteriol. 1990 Apr;172(4):1853–1860. doi: 10.1128/jb.172.4.1853-1860.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L. A., Logan S. M., Guerry P., Trust T. J. Antigenic variation of Campylobacter flagella. J Bacteriol. 1987 Nov;169(11):5066–5071. doi: 10.1128/jb.169.11.5066-5071.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T. Genetics of structure and function of bacterial flagella. Annu Rev Genet. 1977;11:161–182. doi: 10.1146/annurev.ge.11.120177.001113. [DOI] [PubMed] [Google Scholar]
- Joys T. M. Identification of an antibody binding site in the phase-1 flagellar protein of Salmonella typhimurium. Microbios. 1976;15(61-62):221–228. [PubMed] [Google Scholar]
- Joys T. M., Rankis V. The primary structure of the phase-1 flagellar protein of Salmonella typhimurium. I. The tryptic peptides. J Biol Chem. 1972 Aug 25;247(16):5180–5193. [PubMed] [Google Scholar]
- Joys T. M. The flagellar filament protein. Can J Microbiol. 1988 Apr;34(4):452–458. doi: 10.1139/m88-078. [DOI] [PubMed] [Google Scholar]
- Kalmokoff M. L., Jarrell K. F. Cloning and sequencing of a multigene family encoding the flagellins of Methanococcus voltae. J Bacteriol. 1991 Nov;173(22):7113–7125. doi: 10.1128/jb.173.22.7113-7125.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmokoff M. L., Jarrell K. F., Koval S. F. Isolation of flagella from the archaebacterium Methanococcus voltae by phase separation with Triton X-114. J Bacteriol. 1988 Apr;170(4):1752–1758. doi: 10.1128/jb.170.4.1752-1758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrzynska M., Betts J. D., Austin J. W., Trust T. J. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J Bacteriol. 1991 Feb;173(3):937–946. doi: 10.1128/jb.173.3.937-946.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwajima G. Flagellin domain that affects H antigenicity of Escherichia coli K-12. J Bacteriol. 1988 Jan;170(1):485–488. doi: 10.1128/jb.170.1.485-488.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lior H., Woodward D. L., Edgar J. A., Laroche L. J., Gill P. Serotyping of Campylobacter jejuni by slide agglutination based on heat-labile antigenic factors. J Clin Microbiol. 1982 May;15(5):761–768. doi: 10.1128/jcm.15.5.761-768.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuijten P. J., van Asten F. J., Gaastra W., van der Zeijst B. A. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J Biol Chem. 1990 Oct 15;265(29):17798–17804. [PubMed] [Google Scholar]
- Nuijten P. J., van der Zeijst B. A., Newell D. G. Localization of immunogenic regions on the flagellin proteins of Campylobacter jejuni 81116. Infect Immun. 1991 Mar;59(3):1100–1105. doi: 10.1128/iai.59.3.1100-1105.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olyhoek T., Crowe B. A., Achtman M. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev Infect Dis. 1987 Jul-Aug;9(4):665–692. doi: 10.1093/clinids/9.4.665. [DOI] [PubMed] [Google Scholar]
- Oyofo B. A., Thornton S. A., Burr D. H., Trust T. J., Pavlovskis O. R., Guerry P. Specific detection of Campylobacter jejuni and Campylobacter coli by using polymerase chain reaction. J Clin Microbiol. 1992 Oct;30(10):2613–2619. doi: 10.1128/jcm.30.10.2613-2619.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallesen L., Hindersson P. Cloning and sequencing of a Treponema pallidum gene encoding a 31.3-kilodalton endoflagellar subunit (FlaB2). Infect Immun. 1989 Jul;57(7):2166–2172. doi: 10.1128/iai.57.7.2166-2172.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parales J., Jr, Greenberg E. P. N-terminal amino acid sequences and amino acid compositions of the Spirochaeta aurantia flagellar filament polypeptides. J Bacteriol. 1991 Feb;173(3):1357–1359. doi: 10.1128/jb.173.3.1357-1359.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton C. M., Wachsmuth I. K., Evins G. M., Kiehlbauch J. A., Plikaytis B. D., Troup N., Tompkins L., Lior H. Evaluation of 10 methods to distinguish epidemic-associated Campylobacter strains. J Clin Microbiol. 1991 Apr;29(4):680–688. doi: 10.1128/jcm.29.4.680-688.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffaretti J. C., Kressebuch H., Aeschbacher M., Bille J., Bannerman E., Musser J. M., Selander R. K., Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A. 1989 May;86(10):3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleier E., Schmitt R. Identification and sequence analysis of two related flagellin genes in Rhizobium meliloti. J Bacteriol. 1989 Mar;171(3):1467–1475. doi: 10.1128/jb.171.3.1467-1475.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Beltran P., Smith N. H., Barker R. M., Crichton P. B., Old D. C., Musser J. M., Whittam T. S. Genetic population structure, clonal phylogeny, and pathogenicity of Salmonella paratyphi B. Infect Immun. 1990 Jun;58(6):1891–1901. doi: 10.1128/iai.58.6.1891-1901.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Beltran P., Selander R. K. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J Bacteriol. 1990 May;172(5):2209–2216. doi: 10.1128/jb.172.5.2209-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Selander R. K. Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):603–609. doi: 10.1128/jb.172.2.603-609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam G., Kalmokoff M. L., Jarrell K. F., Koval S. F., Beveridge T. J. Isolation, characterization, and cellular insertion of the flagella from two strains of the archaebacterium Methanospirillum hungatei. J Bacteriol. 1990 Jun;172(6):3221–3228. doi: 10.1128/jb.172.6.3221-3228.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow L. S., Rittenberg S. C. Waveform analysis and structure of flagella and basal complexes from Bdellovibrio bacteriovorus 109J. J Bacteriol. 1985 Sep;163(3):1038–1046. doi: 10.1128/jb.163.3.1038-1046.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton S. A., Logan S. M., Trust T. J., Guerry P. Polynucleotide sequence relationships among flagellin genes of Campylobacter jejuni and Campylobacter coli. Infect Immun. 1990 Aug;58(8):2686–2689. doi: 10.1128/iai.58.8.2686-2689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlsty T. D., Albertini A. M., Miller J. H. Gene amplification in the lac region of E. coli. Cell. 1984 May;37(1):217–224. doi: 10.1016/0092-8674(84)90317-9. [DOI] [PubMed] [Google Scholar]
- Trachtenberg S., DeRosier D. J. Three-dimensional reconstruction of the flagellar filament of Caulobacter crescentus. A flagellin lacking the outer domain and its amino acid sequence lacking an internal segment. J Mol Biol. 1988 Aug 20;202(4):787–808. doi: 10.1016/0022-2836(88)90559-1. [DOI] [PubMed] [Google Scholar]
- Vonderviszt F., Aizawa S., Namba K. Role of the disordered terminal regions of flagellin in filament formation and stability. J Mol Biol. 1991 Oct 20;221(4):1461–1474. doi: 10.1016/0022-2836(91)90946-4. [DOI] [PubMed] [Google Scholar]
- Vonderviszt F., Kanto S., Aizawa S., Namba K. Terminal regions of flagellin are disordered in solution. J Mol Biol. 1989 Sep 5;209(1):127–133. doi: 10.1016/0022-2836(89)90176-9. [DOI] [PubMed] [Google Scholar]
- Wachsmuth I. K., Kiehlbauch J. A., Bopp C. A., Cameron D. N., Strockbine N. A., Wells J. G., Blake P. A. The use of plasmid profiles and nucleic acid probes in epidemiologic investigations of foodborne, diarrheal diseases. Int J Food Microbiol. 1991 Jan;12(1):77–89. doi: 10.1016/0168-1605(91)90049-u. [DOI] [PubMed] [Google Scholar]
- Wei L. N., Joys T. M. Covalent structure of three phase-1 flagellar filament proteins of Salmonella. J Mol Biol. 1985 Dec 20;186(4):791–803. doi: 10.1016/0022-2836(85)90397-3. [DOI] [PubMed] [Google Scholar]
- Wei L. N., Joys T. M. The nucleotide sequence of the H-1r gene of Salmonella rubislaw. Nucleic Acids Res. 1986 Oct 24;14(20):8227–8227. doi: 10.1093/nar/14.20.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988 Sep;56(9):2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect Immun. 1988 Sep;56(9):2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]