Abstract
The risk of rapid pest adaptation to an insecticide is highly dependent on the initial frequency of resistance alleles in field populations. Because we have lacked empirical estimates of these frequencies, population–genetic models of resistance evolution have relied on a wide range of theoretical estimates. The recent commercialization of genetically engineered cotton that constitutively produces an insecticidal protein derived from the biocontrol agent, Bacillus thuringiensis (Bt) has raised concern that we lack data needed to quantify the risk of insect pests such as Heliothis virescens rapidly adapting to this ecologically valuable class of toxins. By individually mating over 2,000 male H. virescens moths collected in four states to females of a Bt toxin-resistant laboratory strain, and screening F1 and F2 offspring for tolerance of the toxic protein, we were able to directly estimate the field frequency of alleles for resistance as 1.5 × 10−3. This high initial frequency underscores the need for caution in deploying transgenic cotton to control insect pests. Our single-pair mating technique greatly increases the efficiency of detecting recessive resistance alleles. Because alleles that decrease target site sensitivity to Bt toxins and other insecticides are often recessive, this technique could be useful in estimating resistance allele frequencies in other insects exposed to transgenic insecticidal crops or conventional insecticides.
Keywords: transgenic crops, insecticides, evolution
Insect resistance to pesticides is a major problem in modern agriculture. Over 500 arthropod species are resistant to at least one pesticide (1), and some major insect pests have evolved resistance to new insecticides within 1–3 years (2, 3). In 1996, genetically engineered cotton that produces an insecticidal protein originally found in the biocontrol agent, Bacillus thuringiensis (Bt), was planted on 1.8 million acres of farm land. Because this toxin is produced by the plants from seedling stage to harvest, there will be intense selection for resistant pest genotypes as the acreage planted to this transgenic cotton increases (4, 5). Concern over the risk of pest resistance to insecticidal cotton led the U.S. Environmental Protection Agency to offer only conditional registration for the marketing of varieties producing Bt toxin (6).
During the past 20 years, a number of population–genetic models have been used to gain insight into the process of resistance evolution, and to assess the risk of rapid pest adaptation in a given situation (7–11). Recently, these models have been modified to examine approaches for deploying insecticidal crops in ways that could delay pest adaptation (12–15). The utility of these models has been limited by the lack of accurate empirical data needed for setting genetic and ecological model parameters (16). One parameter, the initial frequency of resistance-conferring alleles, has never been systematically assessed before the widespread use of an insecticide (17, 18). Instead, genetic models have assumed initial allelic frequencies from 10−2 to 10−6 based on theoretical assumptions about the balance between mutation and selection (17). Because the rate at which resistance develops is tightly linked to initial allelic frequency (7–15), the ability to determine the risk of rapid resistance development could be substantially improved if empirical estimates of this parameter were available.
Here we present a direct estimate of the frequency of alleles in Heliothis virescens that confer resistance to the specific Bt toxin, CryIA(c), that is produced by commercial, transgenic cotton varieties. Because our estimate is based on insects collected prior to the first commercial planting of transgenic cotton it represents the initial conditions in the system. In the Midsouth region of the United States where most of this genetically engineered cotton will be grown, the major pest targeted for control by the Bt toxin is H. virescens. This lepidopteran species has evolved resistance to almost all registered conventional insecticides (19), so farmers have a strong incentive to use engineered plants that produce the Bt toxin.
Laboratory selection experiments have already shown that H. virescens can become resistant to CryIA(c) and other Bt-derived toxins. Two laboratory strains developed moderate levels of resistance (20, 21) and one strain, YHD2, selected in our laboratory, developed a very high level of resistance to CryIA(c) and related CryI toxins (22). The toxin concentration needed to kill 50% (LC50) of the larvae from the YHD2 strain was >2,000 times higher than the LC50 for susceptible H. virescens larvae (22), and recent experiments have shown that this strain can survive on cotton that produces Bt toxin (F.G., L. Carter, and L. Seltmann, unpublished data). Genetic crosses demonstrated that a major portion of the resistance in the YHD2 strain is encoded by a single gene (or a set of linked genes) with mostly recessive inheritance (22). Recent work using genetic markers indicates that this major resistance gene (BTR4) is located on H. virescens linkage group 9, and there is some evidence that YHD2 contains a minor resistance gene on linkage group 11 (23). Biochemical analyses indicate that resistance in the YHD2 strain is due to decreased toxin binding to the membrane of larval midgut cells which is the toxin’s site of action (24). Similar decreases in Bt toxin binding have also been found in Bt-resistant strains of other lepidopteran species (25, 26). In all cases where Bt resistance is associated with decreased toxin binding, inheritance is partially or completely recessive (22, 25, 26).
The selection experiment that resulted in the YHD2 strain was begun with 490 diploid eggs (i.e., 980 haploid genomes) collected from three adjacent North Carolina counties. At least 1 of these 980 haploid genomes must have carried the major resistance allele. Assuming that the eggs collected were a random sample of the H. virescens population (22), the initial frequency, p, of the Bt resistance allele can be estimated as 1/980 or ≈10−3. This tentative estimate of initial frequency could be an underestimate because there may have been more than one copy of the resistance allele in the sample, and it is also probable that not all 980 individuals contributed offspring to the F4 generation (the first generation selected for resistance). The 10−3 estimate also could be considered too high because a number of selection experiments for Bt resistance in H. virescens were conducted in our laboratory (21, 22) and in other laboratories (20) and only one led to the high level of resistance found in the YHD2 strain.
The goal of the work reported here was to develop an independent estimate of the frequency of major Bt resistance alleles by directly screening field populations of H. virescens prior to commercialization of cotton that produced Bt toxin.
MATERIALS AND METHODS
Methodology for Resistance Gene Screening.
Typical screening for resistance traits involves collecting eggs or young larvae and exposing them to the toxin in question. Such an approach could easily detect a major resistance gene if it occurs at a high frequency (≈10−3) and is expressed in heterozygotes. However, for recessive genes such as that in YHD2 (and most Bt resistance traits found in other Lepidoptera), only homozygous-resistant individuals would stand out in such screening (22). If the initial frequency (p) of a recessive resistance allele is 10−3, the expected frequency of homozygous resistant individuals (p2) is 10−6. Screening for such rare individuals is not feasible with H. virescens or most other Lepidoptera.
Because we already had a strain (YHD2) in which p was ≈1.0, another screening option was available for assessing the frequency of this recessive allele. Male moths captured in the field could be individually mated to females of the YHD2 strain that were homozygous for the recessive resistance allele (rr). If the male was homozygous susceptible (SS), then all of the offspring would be heterozygotes (rS) and would only have a low level of resistance. If a male was heterozygous for a resistance allele (r′) at the same locus as the YHD2 r allele, then 50% of the progeny would be expected to be heterozygotes (rS) and 50% resistant homozygotes (r′r). (The r′ allele could be genetically identical to the YHD2 r allele or it could differ genetically and also could confer a different level of Bt resistance.) This screening procedure also would detect major resistance alleles at other loci if they had dominant expression.
Resistance Bioassay.
A bioassay developed in our laboratory can differentiate rr from rS H. virescens larvae based on their growth rate when chronically exposed to a sublethal concentration of Bt toxin incorporated into an artificial diet (22). In this assay, individual neonate larvae are reared for 10–14 days in 30 ml cups containing ≈5 ml artificial diet with a Bt toxin concentration of 0.064 μg/ml. When offspring of a (rr × rS) cross were assayed by this procedure, there was a clear bimodality in larval weight, with ≈50% large and 50% small larvae (22).
To estimate the initial allelic frequency in field populations of H. virescens, we needed to screen ≈50 offspring from at least 1,000 single pair crosses so we modified our assay to make it more efficient by rearing 5 larvae per cup for only 7 days. Because larvae can interfere with each other’s growth and because there is some cannibalism when larvae reach third instar (27), this assay is less precise than the original assay, but it can differentiate between offspring of (rr × SS) and those from (rr × rS) crosses.
All of the fast-growing F1 offspring from an (rr × r′S) cross are expected to have the rr′ genotype. If a single gene is responsible for larval growth on toxin-containing diet, then the F2 offspring from crosses between fast-growing larvae from a single F1 family are all expected to be rr′ and grow rapidly on Bt-toxin-containing diet. We used growth assays of these F2 larvae to confirm F1 tests that indicated the presence of a resistance allele in a field-collected male.
Field Collections.
From July 9 to October 20, 1993, male moths trapped in cotton-producing areas of the Midsouth (Bolivar and Washington Counties of Mississippi, Franklin and Bossier Parishes of Louisiana, and Burleson County, Texas) were sent by overnight courier to North Carolina State University. Additionally, eggs were collected during July in the area of North Carolina where the YHD2 strain originated (22). Trapped males and males that developed from the egg collections were used in single-pair crosses to YHD2 females. Offspring from the crosses between wild males and YHD2 females were reared for 7 days on artificial diet containing 0.064 μg/ml CryIA(c) (28), as described above.
Eliminating False Positive Families.
An F1 family could be falsely identified as positive if, for some reason, a container used for mating contained one rr YHD2 adult and one rS YHD2 adult. Two specific laboratory procedures guarded against such an occurrence: (i) as larvae, all YHD2 individuals were exposed to concentrations of Bt toxin that were expected to kill 100% of heterozygotes (i.e., rS); and (ii) field-collected moths were transferred directly from shipping cartons to containers used for the single-pair matings. Another source of a false positive would be if a mistake was made in sexing YHD2 pupae and a rr male emerged among the female YHD2 adults and mated with one rr female before she was transferred to an individual mating container (see ref. 22). If the female subsequently mated with an SS wild male, it seems feasible that she could lay 50% rr and 50% rS eggs. Experimental work with H. virescens (29–31) indicates that this is unlikely because the second male either completely replaces the first male or fertilizes almost 0% of the female’s eggs.
As a final measure to confirm that we did not have a false positive family, moths from large larvae of the F2 generation were frozen and later subjected to electrophoretic screening to establish that they had at least one genetically stable electrophoretic allele that was not found in the YHD2 strain. A set of 14 allozymes previously established as inherited in Mendelian ratios (ref. 23, and D.S., unpublished data) was used to examine the F2 offspring.
RESULTS
Single Pair Mating of Field-Collected Males.
From a total of 2,289 single pair matings, 1,025 produced sufficient offspring for testing [Mississippi (MS), 124 males; Louisiana (LA), 399 males; Texas (TX), 422 males; North Carolina (NC), 80 males). An average of 46.9 larvae were assayed per family of offspring. For offspring of the first 262 successful crosses, larval instar was recorded for each survivor. In the later 763 crosses, when the overall growth of the offspring was slow, we recorded only whether any larvae had grown beyond second instar.
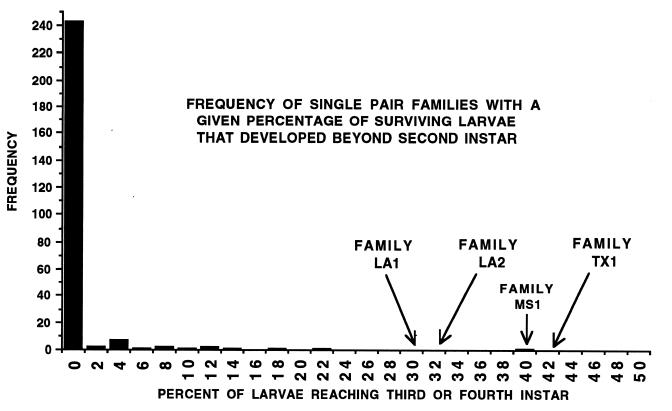
In 92.7% of the first 262 single-pair families assayed, 0% of the larvae reached third instar. Because instar data were recorded for every surviving larva in the first 262 families assayed, it was possible to calculate the percentage of larvae from each family that had at least reached third instar. A frequency histogram, Fig. 1, shows the number of families with a given percentage of third and fourth instars. In 11 of the 19 single pair families where more than 0% of the larvae had grown beyond second instar, only a single larva in that family had reached third instar. However, in one family of offspring (MS1), over 40% of the larvae developed beyond second instar.
Figure 1.
Histogram of the percentage of larvae from each of the first 262 successful single pair crosses that grew beyond second instar in 7 days of rearing on an artificial diet containing 0.064 μg/ml CryIA(c). Depicted only by arrows are the three families from the later 763 crosses with atypically high percentages of large larvae.
In assays of larvae from the later 763 crosses, 88.4% had no larvae reaching third instar, but in three single-pair families (LA1, LA2, and TX1) 30% or more of the offspring developed to third or fourth instar by day 7 (see arrows in Fig. 1). Although none of the four atypical families had the 50% large larvae expected from perfect Mendelian segregation, these families were clearly outliers compared with the other families and were considered the potential result of (rr × r′S) matings, or the mating of YHD2 with a wild male that was heterozygous for a dominant resistance gene.
Bioassays of F2 Offspring.
If these four outlier families of offspring were the result of (rr × r′S) matings, the genotype of the large larvae was expected to be r′r and that of the small larvae was expected to be rS. To test this prediction, all surviving larvae from three of these families (LA1, LA2, and TX1) were removed from the toxic diet, weighed, and then reared to pupation in individual cups of nontoxic diet (the MS1 larvae were discarded before this approach was instituted). When moths emerged, single-pair matings were set up between males and females (within a family) that had been among the 1/3 largest larvae after 7 days, and between males and females that had been among the 1/3 smallest larvae. Six of the large by large (lg × lg) matings and two of the small by small (sm × sm) matings were successful.
Offspring from these eight crosses were reared on diet containing 0.064 μg/ml CryIA(c), and the larvae were weighed after 10 or 14 days. If the wild male in the original single pair mating was r′S, then the large larvae in the F1 generation would be r′r and the small larvae would be rS. Offspring of the (lg × lg) crosses would therefore be expected to have a 1:2:1 mixture of rr:r′r:r′r′, so all larvae would be large. (Because the r′ allele from the wild male could confer a different level of resistance from that conferred by the r allele from the YHD2 female, additional variance in larval size might be seen.) Offspring of the (sm × sm) cross would be expected to have a 1:2:1 mixture of SS:rS:rr, resulting in 25% large individuals (or somewhat more if a significant proportion of SS larvae died). If our prediction was incorrect and a wild male had actually been SS with a large percentage of its F1 offspring developing beyond second instar due to nongenetic factors, then offspring of the (lg × lg) and (sm × sm) crosses would be expected to be similar and have a 1:2:1 mixture of SS:rS:rr.
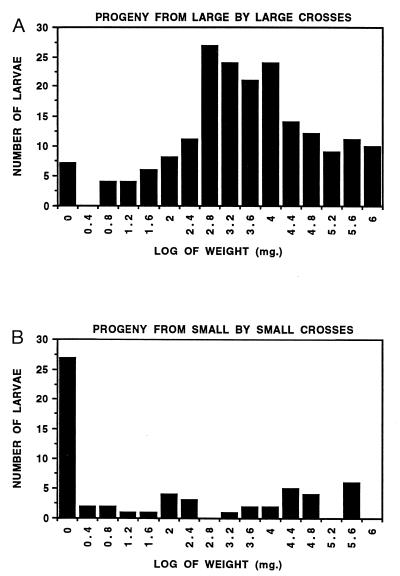
Results of the assays indicate that offspring from (lg × lg) crosses differed significantly from those derived from (sm × sm) crosses (Fig. 2), confirming the prediction that the wild males were not SS and that our original screening procedure had succeeded in identifying heterozygous males. Pooled offspring weights from all (lg × lg) crosses (Fig. 2A) were significantly greater than weights of offspring from all (sm × sm) crosses (Fig. 2B) based on Wilcoxon two-sample test (P < 0.001). In addition to statistical analysis of pooled data, weights of offspring from the one (sm × sm) and two (lg × lg) crosses from within the LA2 family and the one (sm × sm) and two (lg × lg) crosses from within the TX1 family were analyzed separately by Wilcoxon two-sample tests [the LA1 family only had successful (lg × lg) crosses]. In these analyses there was always a significant difference between the (sm × sm) cross and each of the two (lg × lg) crosses within a family (P < 0.005) but no difference between the two (lg × lg) crosses from either the LA2 or the TX1 family (P > 0.10).
Figure 2.
Histograms depicting the frequency of F2 larvae in given size classes (natural log) after rearing on 0.064 μg/ml CryIA(c). (A) F2 larvae resulting from all single pair crosses of F1 larvae that had grown well on Bt-containing diet. (B) F2 larvae resulting from all single pair crosses of F1 larvae that had grown poorly on Bt-containing diet.
Electrophoresis of F2 offspring demonstrated that families LA1 and LA2 had SoDH and HBDH alleles, respectively, that are not found in the YHD2 strain. This finding is further evidence that these families did not arise from mistaken mating of two YHD2 individuals.
DISCUSSION
We conclude that at least three males in a sample of 1,025 were heterozygous for a major resistance allele. Because the males are diploid, we estimate the frequency of resistant alleles in the population as 3 out of 2,050, or 1.5 × 10−3. We determined lower and upper 95% confidence intervals for the frequency of resistant alleles as 3.0 × 10−4 and 4.1 × 10−3, respectively, based on the Clopper–Pearson method (32). [We did not estimate frequencies for each state because of low sample size and the fact that U.S. H. virescens populations are considered homogeneous for genetic traits that have not been under strong selection pressure (33). We did not include the MS1 family in our analysis because we did not conduct an F2 test on this family. Had this family been included, our estimate of initial allelic frequency would have been 2.0 × 10−3.] These data support our preliminary estimate of 10−3 for the initial frequency of the YHD2 resistance allele (22).
Because high levels of resistance to Bt toxins and other insecticides are often encoded by recessive or partially recessive alleles (5, 17) they are difficult to detect using conventional resistance screening techniques. Our single-pair mating design offers an alternative approach when resistant laboratory strains are available. Currently such Bt-resistant strains are available for Plutella xylostella and Plodia interpunctella. Laboratory selection experiments could produce similar strains of other pests targeted for control with Bt or other toxins. These strains could be used not only for assessing the initial field frequency of resistance alleles, they also could be used for monitoring changes in allele frequencies as the toxins become deployed in transgenic plants or by conventional application.
Genetic models indicate that a recessive allele at the frequency detected here in H. virescens could lead to rapid evolution of resistant populations if Bt toxin-producing cotton is grown without adequate refuges for toxin-susceptible larvae (12–15). Although the hypothesis that the field-collected males carried a recessive resistance allele (r′) offers the best fit to our data, we cannot statistically rule out the possibility that one or more of the males carried a major dominant gene for resistance. If one or more of the 1,025 males carried a dominant resistance gene, the risk of resistance would be even higher (34).
In the H. virescens/cotton system, the Bt–cotton cultivars currently deployed produce concentrations of toxin that are high enough to kill 100% of heterozygous larvae with low levels of Bt toxin resistance (F.G., L. Carter, and L. Seltmann, unpublished data). Assuming that resistance alleles are at least partially recessive and occur at ≈10−3, and the Environmental Protection Agency mandate of a 4% refuge to maintain SS moths is followed, it should take at least 10 years before Bt resistance becomes a problem in H. virescens populations (12). However, currently deployed Bt cotton and corn cultivars are not as toxic to other pest species as they are to H. virescens. For example, fewer than 90% of natural, unselected larvae of the cotton bollworm (Helicoverpa zea) and the European corn borer (Ostrinia nubilalis) are killed by commercially available Bt crops (ref. 35; F.G., unpublished data). If we assume that there is a partially recessive Bt resistance gene in these pest species that also occurs at 10−3, genetic models (12) predict that populations could become resistant to the Bt crops in 3–4 years, even with a 4% refuge. Unless cultivars can be developed that are more toxic to these pests, much larger refuges will be needed if we are to sustain the utility of Bt toxin-expressing crops.
Acknowledgments
We thank L. Seltmann, A. Reynolds, and L. Li for technical help, and C. Brownie for statistical advice. This work was supported by the U.S. Department of Agriculture/National Research Initiative Competitive Grants Program and the National Science Foundation/North Carolina State University Center for Integrated Pest Management.
ABBREVIATION
- Bt
Bacillus thuringiensis
References
- 1.Georghiou G P, Lagunes-Tejeda A. The Occurrence of Resistance to Pesticides in Arthropods. Rome: Food and Agriculture Organization; 1991. [Google Scholar]
- 2.Metcalf R L. Pestic Sci. 1989;26:333–358. [Google Scholar]
- 3.Talekar N S, Shelton A M. Annu Rev Entomol. 1993;38:275–301. [Google Scholar]
- 4.Gould F. BioScience. 1988;38:26–33. [Google Scholar]
- 5.McGaughey W H, Whalon M E. Science. 1992;258:1451–1455. doi: 10.1126/science.258.5087.1451. [DOI] [PubMed] [Google Scholar]
- 6.Matten, S. R. & Lewis P. T. (1995) ISB News Rep. December, pp. 3–5.
- 7.Tabashnik B E. In: Pesticide Resistance in Arthropods. Roush R T, Tabashnik B E, editors. New York: Chapman & Hall; 1990. pp. 153–182. [Google Scholar]
- 8.Comins H N. J Theor Biol. 1977;64:177–197. doi: 10.1016/0022-5193(77)90119-9. [DOI] [PubMed] [Google Scholar]
- 9.Curtis C F, Cook L M, Wood R J. Ecol Entomol. 1978;3:273–287. [Google Scholar]
- 10.Roush R T. Pestic Sci. 1989;26:423–441. [Google Scholar]
- 11.Taylor C E, Georghiou G P. J Econ Entomol. 1979;72:105–109. [Google Scholar]
- 12.Gould F. Environ Entomol. 1986;15:11–23. [Google Scholar]
- 13.Mallet J, Porter P. Proc R Soc London Ser B. 1992;250:165–169. [Google Scholar]
- 14.Roush R T. Biocontrol Sci Techol. 1994;4:501–516. [Google Scholar]
- 15.Alstad D N, Andow D A. Science. 1995;268:1894–1896. doi: 10.1126/science.268.5219.1894. [DOI] [PubMed] [Google Scholar]
- 16.Taylor C E. In: Pest Resistance to Pesticides. Georghiou G P, Saito T, editors. New York: Plenum; 1983. pp. 163–173. [Google Scholar]
- 17.Roush R T, McKenzie J A. Annu Rev Entomol. 1987;32:361–380. doi: 10.1146/annurev.en.32.010187.002045. [DOI] [PubMed] [Google Scholar]
- 18.Roush R T. In: Advances in Insect Control: The Role of Transgenic Crops. Carozzi N, Koziel M, editors. London: Taylor & Francis; 1997. pp. 271–294. [Google Scholar]
- 19.Martin S H, Elzen G W, Graves J B, Micinski S, Leonard B R, Burris E. J Econ Entomol. 1995;88:505–511. [Google Scholar]
- 20.Sims S R, Stone T B. J Invertebr Pathol. 1991;57:206–210. [Google Scholar]
- 21.Gould F, Martinez-Ramirez A, Anderson A, Ferre J, Silva F J, Moar W J. Proc Natl Acad Sci USA. 1992;89:7986–7988. doi: 10.1073/pnas.89.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gould F, Anderson A, Reynolds A, Bumgarner L, Moar W J. J Econ Entomol. 1995;88:1545–1559. [Google Scholar]
- 23.Heckel D G, Gahan L C, Gould F, Anderson A. J Econ Entomol. 1997;90:75–86. [Google Scholar]
- 24.Lee M K, Rajamohan F, Gould F, Dean D H. Appl Environ Microbiol. 1995;61:3836–3842. doi: 10.1128/aem.61.11.3836-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Rie J, McGaughey W H, Johnson D E, Barnett D B, Van Mallaert H. Science. 1990;247:72–74. doi: 10.1126/science.2294593. [DOI] [PubMed] [Google Scholar]
- 26.Ferre J, Real M D, Van Rie J, Jansens S, Peferoen M. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould F, Holtzman G, Rabb R L, Smith M. Ann Entomol Soc Am. 1980;73:243–250. [Google Scholar]
- 28.Feitelson J S, Payne J, Kim L. Bio/Technology. 1992;10:271–275. [Google Scholar]
- 29.Flint H M, Kressin E L. J Econ Entomol. 1968;61:477–483. [Google Scholar]
- 30.Drummond B A. In: Sperm Competition and the Evolution of Animal Mating Systems. Smith R L, editor. New York: Academic; 1984. pp. 291–370. [Google Scholar]
- 31.Gwynne D T. In: Sperm Competition and the Evolution of Animal Mating Systems. Smith R L, editor. New York: Academic; 1984. pp. 117–149. [Google Scholar]
- 32.Santner T J, Duffy D E. The Statistical Analysis of Discrete Data. New York: Springer; 1989. pp. 33–34. [Google Scholar]
- 33.Korman A K, Mallet J, Goodenough J L, Graves J B, Hayes J L, Hendricks D E, Luttrell R, Pair S D, Wall M. Ann Entomol Soc Am. 1993;86:182–188. [Google Scholar]
- 34.Heckel D G. Biocontrol Sci Techol. 1994;4:405–418. [Google Scholar]
- 35.Lambert A L, Bradley J R, Van Duyn J W. Proc 1996 Beltwide Cotton Conf. 1996;1:931–932. [Google Scholar]