Abstract
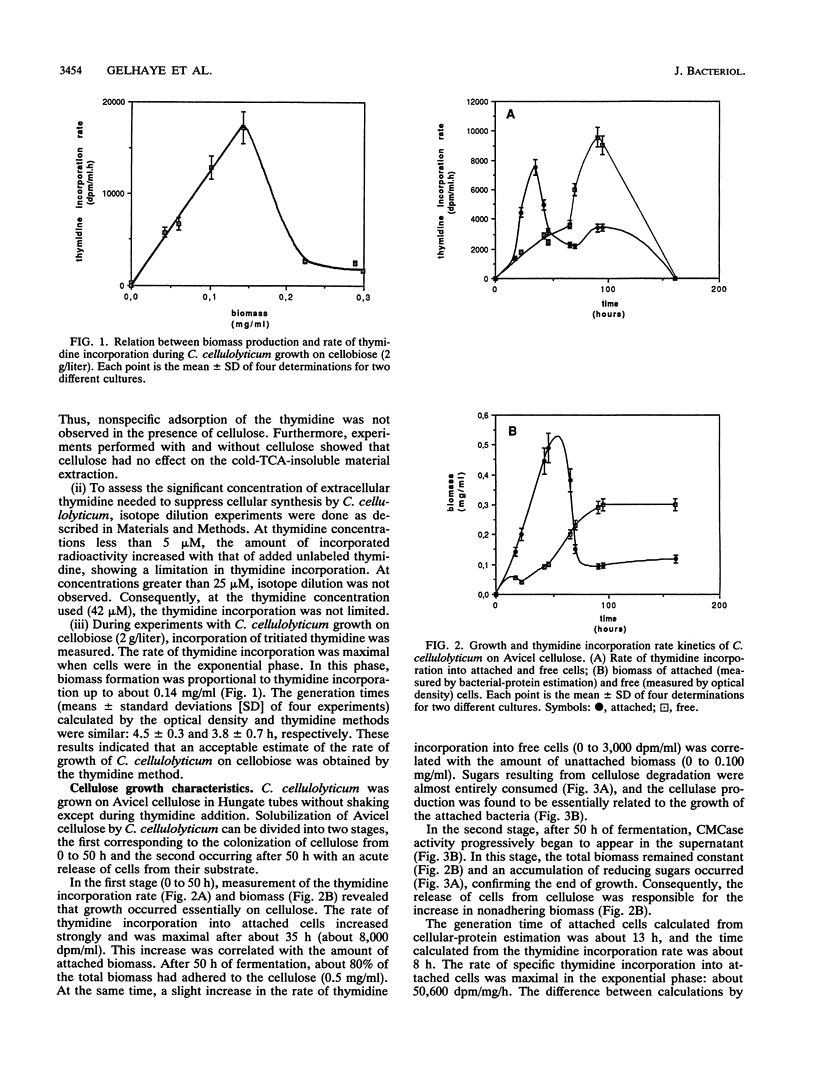
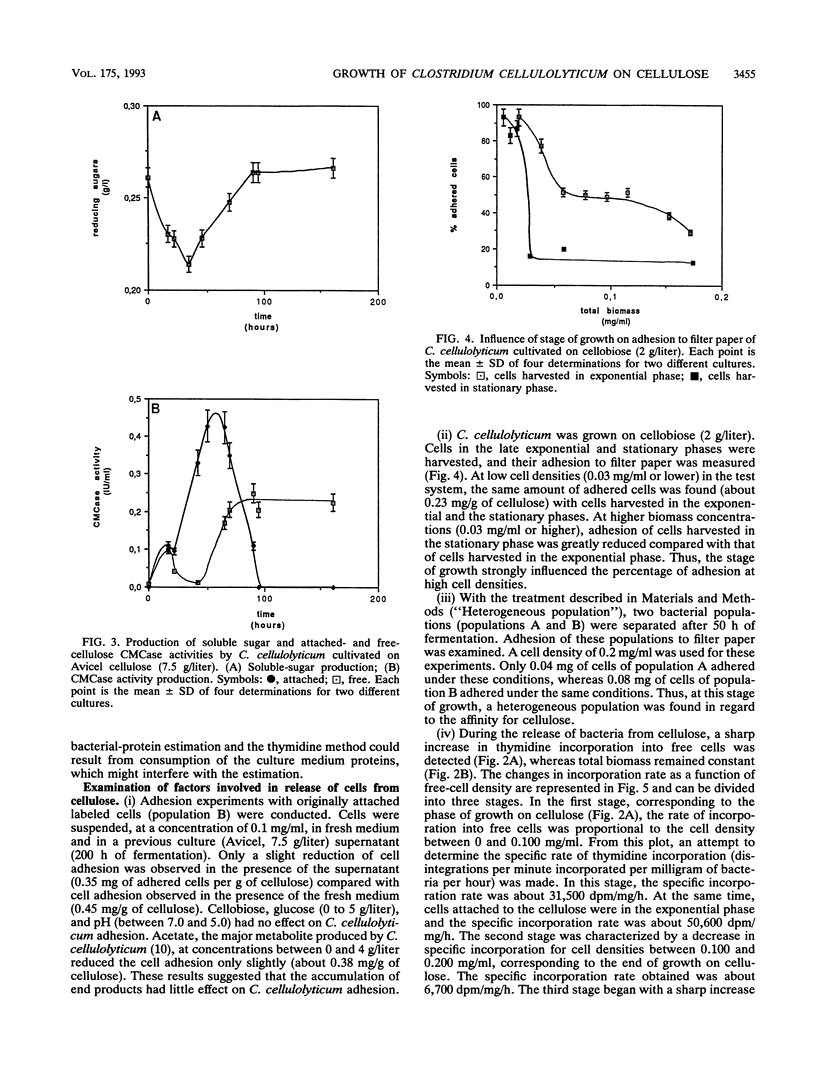
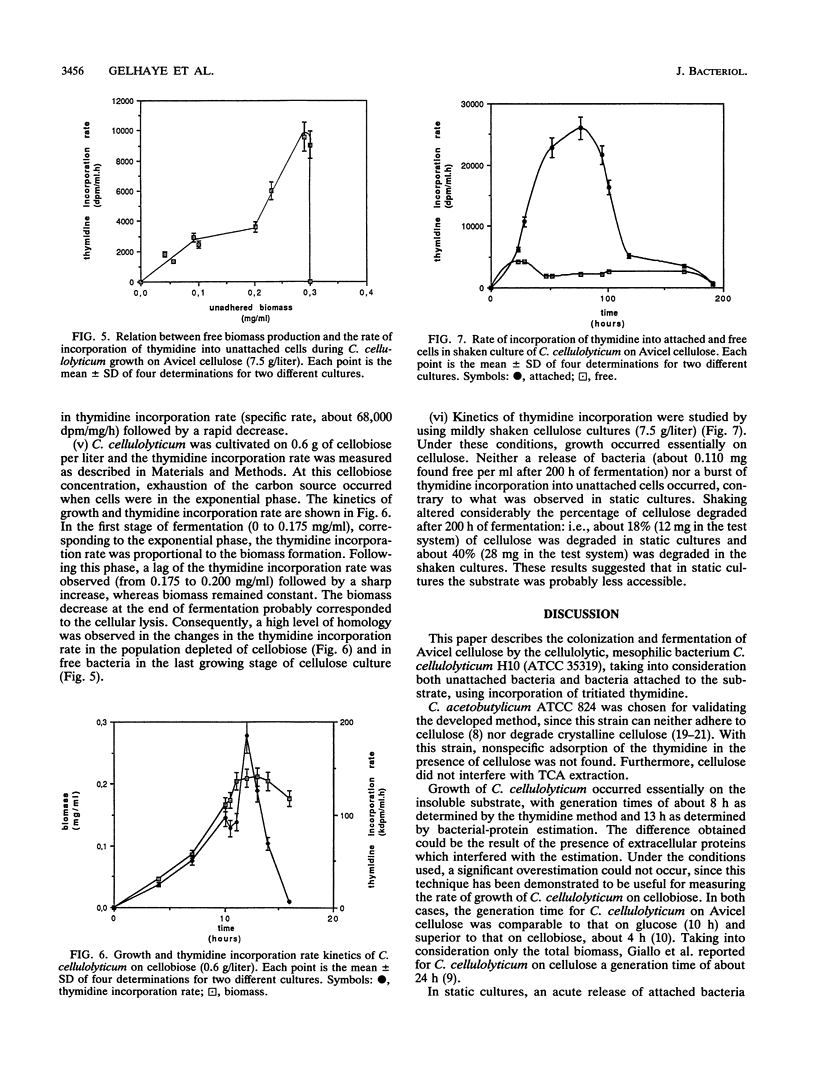
The rate of tritiated-thymidine incorporation into DNA was used to estimate Clostridium cellulolyticum H10 growth rates on Avicel cellulose, taking into consideration both the unattached cells and the cells adhered to the substrate. The generation time on cellobiose calculated from the data on cell density (4.5 h) agreed well with the generation time calculated by tritiated-thymidine incorporation (3.8 h). Growth on Avicel cellulose occurred when bacteria were adhered to their substrate; 80% of the biomass was detected on the cellulose. Taking into consideration attached and free bacteria, the generation time as determined by thymidine incorporation was about 8 h, whereas by bacterial-protein estimation it was about 13 h. In addition to the growth rate of the bacteria on the cellulose, the release of adhered cells constituted an important factor in the efficiency of the cellulolysis. The stage of growth influenced adhesion of C. cellulolyticum; maximum adhesion was found during the exponential phase. Under the conditions used, the end of growth was characterized by an acute release of biomass and cellulase activity from the cellulose. An exhaustion of the accessible cellulose could be responsible for this release.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bayer E. A., Kenig R., Lamed R. Adherence of Clostridium thermocellum to cellulose. J Bacteriol. 1983 Nov;156(2):818–827. doi: 10.1128/jb.156.2.818-827.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E. A., Setter E., Lamed R. Organization and distribution of the cellulosome in Clostridium thermocellum. J Bacteriol. 1985 Aug;163(2):552–559. doi: 10.1128/jb.163.2.552-559.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensadoun A., Weinstein D. Assay of proteins in the presence of interfering materials. Anal Biochem. 1976 Jan;70(1):241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- Bhat S., Wallace R. J., Orskov E. R. Adhesion of cellulolytic ruminal bacteria to barley straw. Appl Environ Microbiol. 1990 Sep;56(9):2698–2703. doi: 10.1128/aem.56.9.2698-2703.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavedon K., Leschine S. B., Canale-Parola E. Cellulase system of a free-living, mesophilic clostridium (strain C7). J Bacteriol. 1990 Aug;172(8):4222–4230. doi: 10.1128/jb.172.8.4222-4230.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure E., Belaich A., Bagnara C., Gaudin C., Belaich J. P. Sequence analysis of the Clostridium cellulolyticum endoglucanase-A-encoding gene, celCCA. Gene. 1989 Dec 7;84(1):39–46. doi: 10.1016/0378-1119(89)90137-6. [DOI] [PubMed] [Google Scholar]
- Giallo J., Gaudin C., Belaich J. P. Metabolism and Solubilization of Cellulose by Clostridium cellulolyticum H10. Appl Environ Microbiol. 1985 May;49(5):1216–1221. doi: 10.1128/aem.49.5.1216-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giallo J., Gaudin C., Belaich J. P., Petitdemange E., Caillet-Mangin F. Metabolism of glucose and cellobiose by cellulolytic mesophilic Clostridium sp. strain H10. Appl Environ Microbiol. 1983 Mar;45(3):843–849. doi: 10.1128/aem.45.3.843-849.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam S. H., Greene R. V., Griffin H. L. Adhesive properties of a symbiotic bacterium from a wood-boring marine shipworm. Appl Environ Microbiol. 1990 May;56(5):1317–1322. doi: 10.1128/aem.56.5.1317-1322.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey W. H., Paul J. H. Activity of an Attached and Free-Living Vibrio sp. as Measured by Thymidine Incorporation, p-Iodonitrotetrazolium Reduction, and ATP/DNA Ratios. Appl Environ Microbiol. 1986 Jan;51(1):150–156. doi: 10.1128/aem.51.1.150-156.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl D. M. Selected nucleic Acid precursors in studies of aquatic microbial ecology. Appl Environ Microbiol. 1982 Oct;44(4):891–902. doi: 10.1128/aem.44.4.891-902.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo H., Cheng K. J., Costerton J. W. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria from cellulose fibers. Can J Microbiol. 1987 Mar;33(3):267–272. doi: 10.1139/m87-045. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lamed R., Naimark J., Morgenstern E., Bayer E. A. Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987 Aug;169(8):3792–3800. doi: 10.1128/jb.169.8.3792-3800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamed R., Setter E., Bayer E. A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983 Nov;156(2):828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham M. J., Brooker B. E., Pettipher G. L., Harris P. J. Ruminococcus flavefaciens Cell Coat and Adhesion to Cotton Cellulose and to Cell Walls in Leaves of Perennial Ryegrass (Lolium perenne). Appl Environ Microbiol. 1978 Jan;35(1):156–165. doi: 10.1128/aem.35.1.156-165.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W., Gibbins L. N. Xylanolytic Activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985 Oct;50(4):1068–1076. doi: 10.1128/aem.50.4.1068-1076.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W. Isolation and Some Properties of a beta-d-Xylosidase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1987 Apr;53(4):651–654. doi: 10.1128/aem.53.4.651-654.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. F., Forsberg C. W., Rattray J. B. Purification and Characterization of Two Endoxylanases from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1987 Apr;53(4):644–650. doi: 10.1128/aem.53.4.644-650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell C. R., Konopka A. Thymidine incorporation by free-living and particle-bound bacteria in a eutrophic dimictic lake. Appl Environ Microbiol. 1985 Mar;49(3):501–504. doi: 10.1128/aem.49.3.501-504.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morag E., Halevy I., Bayer E. A., Lamed R. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol. 1991 Jul;173(13):4155–4162. doi: 10.1128/jb.173.13.4155-4162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P. C., Moriarty D. J. Validity of the tritiated thymidine method for estimating bacterial growth rates: measurement of isotope dilution during DNA synthesis. Appl Environ Microbiol. 1984 Dec;48(6):1076–1083. doi: 10.1128/aem.48.6.1076-1083.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoseyov O., Doi R. H. Essential 170-kDa subunit for degradation of crystalline cellulose by Clostridium cellulovorans cellulase. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2192–2195. doi: 10.1073/pnas.87.6.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner U., Egli T. Dynamics of microbial growth and cell composition in batch culture. FEMS Microbiol Rev. 1990 Mar;6(1):19–43. doi: 10.1111/j.1574-6968.1990.tb04084.x. [DOI] [PubMed] [Google Scholar]


