Abstract
Telomere, the end of linear chromosome, is protected by DNA-protein complexes. These complexes cap the linear chromosome and play an important role in the maintenance of genomic stability. TRF1/PIN2, a double-stranded DNA binding protein is known to regulate telomere length by not only protecting telomere but also blocking the access of telomerase to telomere in cis. To better understand the mechanism through which TRF1/PIN2 regulates telomere length, we performed the yeast two-hybrid screening and identified the transcriptional activator c-Myc as a TRF1/PIN2-binding protein. The c-Myc-TRF1/PIN2 interaction was observed both in vitro and in vivo. This interaction is mediated by the basic helix-loop-helix (bHLH) domain of c-Myc. Importantly, overexpression of this TRF1/PIN2-interacting domain of c-Myc leads to telomere elongation in vivo. Together, these results suggest that c-Myc may be involved in the regulation of telomere length through its direct binding with TRF1/PIN2.
Keywords: TRF1/PIN2, Myc, Telomere
Telomere consists of several thousand copies of repetitive DNA sequence (TTAGGG in vertebrates) and locates at the ends of linear chromosome. Telomere is normally protected by nucleoprotein-DNA complexes, which contain proteins including TRF1, TRF2, TIN2, PTOP, RAP1, and POT1 [1]. These nucleoprotein-DNA complexes form a structure called “T loop”, which caps and protects linear chromosome ends [2]. This capping or “T loop” structure allows telomere ends to be distinguished from DNA double stranded breaks. It is generally believed that this capping structure protects telomere ends from nucleolytic degradation and DNA repair process, and thus prevents chromosomes to form end to end fusions [3, 4].
TRF1 or PIN2 [5] (derived by alternative splicing) is one of these telomere-binding proteins. TRF1 binds double-stranded telomeric DNA and is known to associate with proteins including TIN2 [6] and tankyrase [7]. Through its ability to interact with telomeric DNA and other cellular proteins, TRF1 plays important roles in the regulation of telomere length and telomere end protection. Loss of TRF1 in mice leads to embryonic lethality and telomere end fusions [8, 9].
In order to better understand the role of TRF1 in telomere length regulation and telomere maintenance, we performed yeast two-hybrid screening and identified c-Myc onco-protein as a new TRF1 binding protein. Our results demonstrate that c-Myc binds directly to TRF1 and participates in telomere maintenance.
MATERIALS AND METHODS
Plasmids
DNA fragment containing TRF1/PIN2 coding sequence was generated by PCR and cloned into pGBK-T7 (Clontech, Palo Alto, CA) for yeast two-hybrid screening. The mammalian expression plasmids encoding Myc-tagged full-length and the deletion mutants of human TRF1/PIN2 and c-Myc were also generated by PCR and cloned into pcDNA3 βm-1 (Pharmacia, Piscataway, NJ). The S-Flag-tagged mammalian expression plasmids encoding wild-type, N-terminal and C-terminal deletion mutants of c-Myc (pIRES-SF-Myc, pIRES-SF-Myc-N and pIRES-SF-Myc-C respectively) and TRF1/PIN2 (pIRES-SF-TRF1/PIN2) were constructed by inserting the corresponding PCR fragments into the EcoRI (5') and Hind III (3') restriction sites of pIRES-S-Flag vector (Invitrogen, San Diego, CA). The GST-fusion constructs of wild-type c-Myc and TRF1/PIN2 were similarly generated by PCR and subcloned into pGEX-4T-1 vector (Pharmacia, Piscataway, NJ).
Yeast two-hybrid screening
Full-length human TRF1/PIN2 was used as a bait in yeast two-hybrid screenings. In this study, we used two cDNA libraries, the human testis library and 293T cDNA library generated in the pACT2 vector (Clontech, Palo Alto, CA). The bait and the library DNAs were co-transformed into AH109 yeast strain using the lithium acetate method as previously described [10]. The transformants were selected for growth on the Leu−, Trp−, His− and Ade− solid media containing 30 mM 3-aminotriazole (3-AT). The β-galactosidase assay was performed by 4 incubating freeze-fractured colonies on nitrocellulose in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.03 mM β-mercaptoethanol and 2.5 M X-gal) at 30°C for 30 min.
Transient transfection
Cells (3 × 106 cells) were plated on 100 mm plates and transfected with various plasmids using Fugene6 reagent according to manufacture’s instruction (Roche. Inc). The total amount of DNA transfected was normalized using pcDNA3.1 plasmid as carrier DNA.
GST pull-down assay
The GST fusion proteins containing full-length c-Myc or TRF1/PIN2 were expressed in Escherichia coli and purified as previously described [11]. In vitro transcription and translation experiments were carried out using the TNT-T7 Coupled Reticulocyte Lysate System (Promega, Medison, WI) to obtain [35S]-methionine-labeled wild-type or deletion mutants of c-Myc and TRF1/PIN2. For GST pull-down assay, 2 μg of GST-TRF1/PIN2 fusion proteins was immobilized on the glutathione-Sepharose 4B beads and then incubated with the [35S]-labeled c-Myc proteins in NETN buffer (0.5% Nonidet P-40, 20 mM Tris [pH 8.0], 50 mM NaCl, 50 mM NaF, 100 μM Na3VO4, 1 mM DTT and 50 μg/ml PMSF) for 2 hrs at 4°C. After washing with NETN buffer, the samples were analyzed by SDS-PAGE and autoradiography.
Antibodies, immunoprecipitation and immunoblotting
Anti-Myc and anti-Flag antibodies were obtained from commercial sources (Sigma, Inc). Rabbit polyclonal anti-TRF1/PIN2 antibodies were raised against GST-TRF1/PIN2 fusion protein. For immunoprecipitation, 293T cells (3 × 106 cells) grown in 100-mm plates were transfected with 4.5 μg of each expression plasmid as indicated in figure legends. 48 hours after transfection, cells were lysed in NETN buffer for 20 min on ice. Crude lysates were cleared by centrifugation at 14,000 rpm at 4 °C for 5 minutes. Supernatants were collected and then incubated with protein A- or G-agarose-conjugated primary antibodies. The immunocomplexes were washed three times with NETN buffer and subjected to SDS-PAGE. Western blotting was performed using antibodies as indicated in the figure legends.
Construction of stable cell lines
HT1080 cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained in RPMI supplemented with 10% FBS and antibiotics. To construct cell lines stably expressing various proteins, HT10180 cells were transfected with plasmids encoding deletion mutants of c-Myc (pIRES-SF-Myc-N or pIRES-SF-Myc-C) or control pIRES vector. 48 hours after transfection, cells were split at the 1:10 ratio and cultured in the medium containing 400 μg/ml G418 for 3 weeks. The individual G418-resistant colonies were isolated and the expression of various exogenous proteins was confirmed by Western blotting.
Telomere length assay and telomeric repeat amplification protocol (TRAP) assay
HT1080 cells stably expressing N-terminal or C-terminal fragments of c-Myc were passaged and harvested at various population doubling time points. Genomic DNA samples were prepared using DNeasy Tissue DNA purification Kit (Qiagen) and then digested with restriction enzymes HinF1 and RSA1 (New England Biolabs). Telomere length assays were performed using the 32P-labeled oligo probe (TTAGGG)4. The quantification of telomere length was carried out using TELORUN. Telomerase activity was determined by the telomeric repeat amplification protocol (TRAP) assay using the TRAP-eze kit obtained from Intergen, Inc.
RESULTS AND DISCUSSION
Identification of c-Myc as a TRF1/PIN2-binding protein
To identify new binding partners of TRF1/PIN2, the full-length PIN2 was used as bait for yeast two-hybrid screening using a human testis library and a 293T cDNA library. Among 2 x 106 transformants, 10 positive clones with the highest β-galactosidase activity were isolated from each cDNA library. Sequencing analysis revealed that three clones from 293T library and one clone from human testis library all encoded the C-terminal portion of c-Myc proto-oncogene (Fig. 1), suggesting that c-Myc may be a potential TRF1/PIN2 binding protein.
Fig. 1.

Yeast two-hybrid screening identified c-Myc as a TRF1/PIN2 binding protein. The scheme of the domain structure of c-Myc protein is presented (TAD, transcription activation domain; bHLH, basic helix-loop-helix domain; Z, leucine zipper). The lines below indicate various partial cDNAs obtained from the positive prey vectors.
c-Myc is a transcriptional activator important for cell growth and cell division [12, 13]. Deregulated c-Myc overexpression triggers the G0-to-G1 transition and leads to uncontrolled cell proliferation and eventually cancer development [14]. In agreement with a positive role of c-Myc in cell proliferation, experiments in mice and cell lines showed that loss of c-Myc expression results in decreased cell growth and proliferation [15–17]. An interaction between TRF1 and c-Myc suggests that c-Myc may bind directly with TRF1 and regulates TRF1 function at telomeres.
TRF1/PIN2 interacts with c-Myc
To confirm the interaction between PIN2 and c-Myc in human cells, we transfected 293T cells with plasmids encoding S-Flag-tagged TRF1/PIN2 and Myc-tagged Myc (pMyc-Myc). Cell lysates were subjected to immunoprecipitation using anti-Flag antibody. Subsequent Western blotting using anti-Myc antibody revealed that c-Myc interacts with TRF1/PIN2 in 293T cells (Fig. 2A). In addition, the reverse co-immunoprecipitation experiments also showed an interaction between TRF1/PIN2 and c-Myc (Fig. 2B).
Fig. 2.
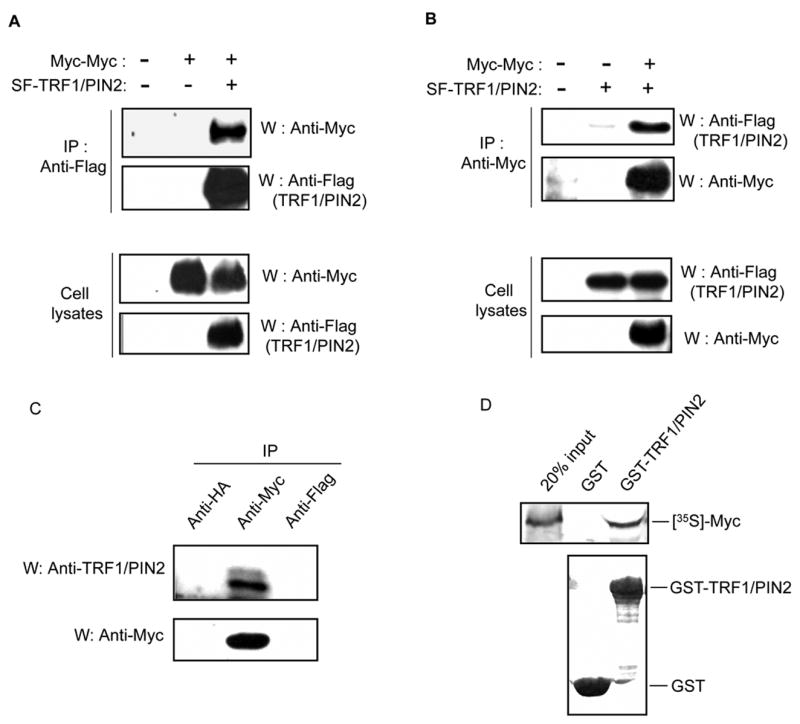
c-Myc binds TRF1/PIN2 both in vitro and in vivo. (A, B). Association of c-Myc with TRF1/PIN2 in 293T cells. A, 293T cells (3 × 106/100 mm dish) were transfected with 4.5 μg of plasmid encoding Myc-tagged c-Myc with or without plasmid encoding S-Flag-tagged TRF1/PIN2 as indicated. 48 hours after transfection, cells were collected and cell lysates were subjected to immunoprecipitation (IP) with anti-Flag antibody and immunoblotting (W) with anti-Myc antibody (first panel). The level of the immunoprecipitated Flag-TRF1/PIN2 was analyzed by Western blot using anti-Flag antibody (second panel). The amounts of c-Myc and TRF1/PIN2 in each lysate were shown respectively in the third and fourth panels. B, 293T cells were transfected with plasmids as indicated. Experiments were performed similar to that described above in A. C, Interaction of endogenous TRF1/PIN2 with c-Myc in the cell. Cell lysates were subjected to immunoprecipitation using anti-Myc, anti-HA or anti-Flag antibodies and immunoblotted with anti-TRF1/PIN2 antibody (top panel). The levels of the immunoprecipitated endogenous c-Myc were analyzed by anti-Myc Western blot (bottom panel). D, GST pull-down assay confirmed the interaction between TRF1/PIN2 and c-Myc. c-Myc proteins were labeled with [35S]-methionine by in vitro transcription and translation reactions and incubated with 2 μg of GST or GST-TRF1/PIN2 and glutathione beads for 2 hours at 4 °C. After extensive washing, the bound proteins were eluted, separated by SDS-PAGE and detected by autoradiograpy (upper panel). The bottom panel shows the equal amount of added GST and GST-TRF1/PIN2 proteins in these experiments.
To determine whether the interaction between TRF1/PIN2 and c-Myc occurs in a more physiologically relevant context, we examined the association of endogenous proteins in 293T cells. As shown in Fig. 2C, we co-immunoprecipitated endogenous TRF1/PIN2 with c-Myc protein. The specificity of this interaction was confirmed since TRF1/TIN2 did not precipitate TRF1/PIN2 with control anti-HA or anti-Flag antibodies (Fig. 2C).
To further explore whether these two proteins interact directly, we performed GST pull-down assay using control GST protein or GST-TRF1/PIN2 fusion protein and the [35S]-labeled c-Myc proteins produced by in vitro transcription and translation. Result showed that c-Myc specifically bound to GST-TRF1/PIN2 but not GST alone (Fig. 2D). In addition, we generated several deletion mutants of c-Myc to identify the region of c-Myc that interacts with TRF1/PIN2 (Fig. 3A). As shown in Fig. 3B, we found that B/HLH/Z domain of c-Myc is important for its interaction with TRF1/PIN2.
Fig. 3.
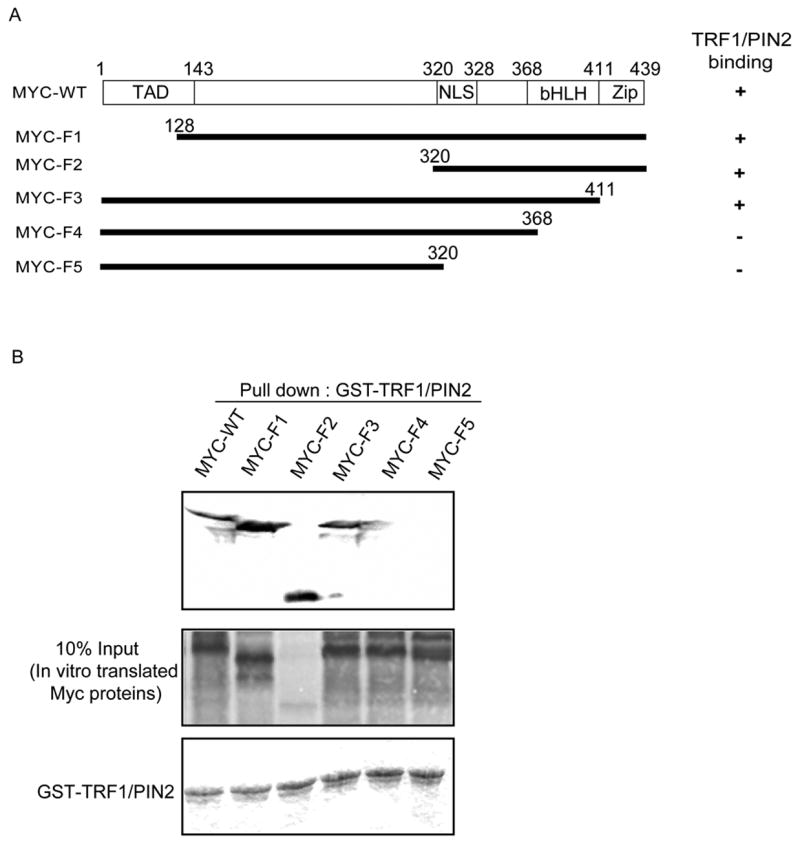
c-Myc bHLH region is important for its binding with TRF1/PIN2. A, Summary of the pull down experiments. B, wild-type or deletion mutants of c-Myc were labeled with [35S]-methionine by in vitro transcription and translation reactions and incubated with 2 μg of GST-TRF1/PIN2 for 2 hours at 4 °C. Washing and detection of Myc proteins were performed as described above in Fig. 2 (upper panel). The amounts of wild-type or mutants of c-Myc and GST-TRF1/PIN2 used in these experiments are shown in the lower panels.
Overexpression of C-terminal region of c-Myc affects telomere length in vivo
We have thus far shown that c-Myc is able to bind TRF1/PIN2 through its C-terminal region. Because TRF1/PIN2 participates in telomere regulation, we wanted to explore whether the interaction between c-Myc and TRF1/PIN2 is involved in telomere maintenance. Overexpression of c-Myc is known to regulate telomere length through its ability to function as a transcription activator that upregulates the expression of telomerase catalytic subunit. In order to circumvent this indirect role of c-Myc in the regulation of telomere length, we decided to establish stable HT1080 cell lines expressing only N-terminal or C-terminal region of c-Myc (Fig. 4A). Genomic DNA samples were prepared from these cells at different passages and their average telomere lengths were determined by the telomere restriction fragment assay. As shown in Fig. 4B and C, cells expressing C-terminal region of c-Myc consistently had longer telomeres at late passages when compared to control vector transfected control cells or cells expressing N-terminal region of c-Myc. As expected, there is no significant difference in telomerase activity in these stable cell lines as determined by TRAP assays (Fig. 4D). These data indicate that C-terminal region of c-Myc and its interaction with TRF1/PIN2 may be important for the c-Myc-mediated increase of telomere length.
Fig. 4.
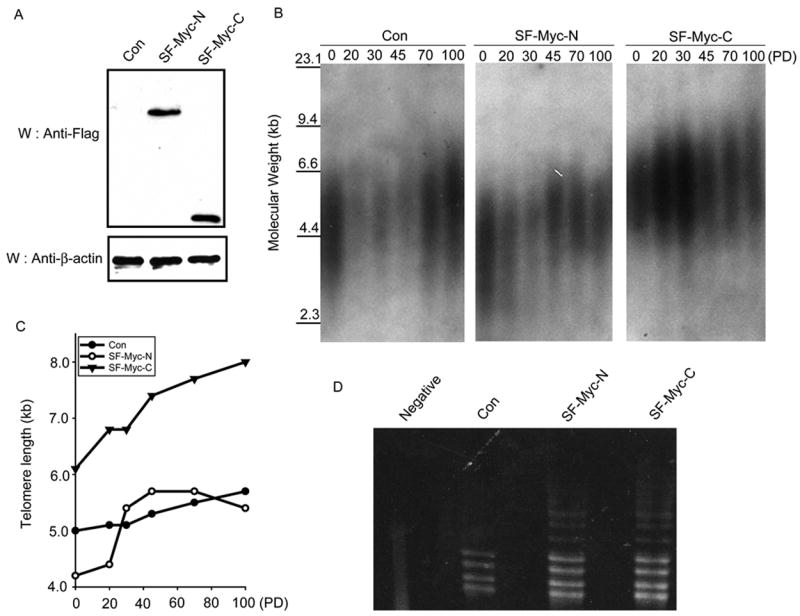
Expression of C-terminal region of c-Myc leads to an increase of telomere length. A, Establishment of HT1080 cell lines stably expressing c-Myc deletion mutants. Anti-Flag antibody was used to detect the expression levels of these c-Myc mutants. Anti-β-actin Western blot was used as a loading control. B, Telomere restriction fragment length analysis was performed using vector transfected control cell line and cell lines stably expressing c-Myc deletion mutants. These cell lines were collected at the indicated generations or population doublings. Extracted genomic DNA was digested with HinF1 and RSA1, separated on a 0.6% agarose gel and transferred to nylon-cellulose membranes. Southern blotting was performed using [32P]-ATP-labeled-(TTAGGG)4 probe. C, The mean size of telomere length was estimated using ImageQuant software and Telorun. Kb, kilobase; PD, population doublings. D, Telomerase activity in these stable cell lines was determined by TRAP assay. Heat-inactivated lysate prepared from cells stably expressing SF-Myc-C was included as a negative control (Negative).
Many studies have shown previously that overexpression of c-Myc leads to an increase of telomere length, probably via its role in the activation of telomerase transcription [18–20]. In this study, we observed an increase of telomere length in cells expressing only the C-terminal region of c-Myc, which does not contain the N-terminal transcriptional activation domain and should be transcriptional inactive. Therefore, these results indicate that the increase of telomere length observed in previous studies may not just depend on the activation of telomerase transcription by c-Myc. Indeed, recent studies suggest that c-Myc may have functions independent of its transcriptional activation activity. c-Myc can bind directly to MCMs and regulates DNA replication [21]. It is possible that a direct physical interaction between c-Myc and TRF1/PIN2 also contribute to the regulation of telomere length in vivo.
TRF1/PIN2 normally protects telomere and inhibits telomere replication. TRF1/PIN2 can hinder telomere C-strand DNA synthesis in vitro. Replication fork stalling at telomeric repeats occurred when TRF1/PIN2 proteins were added to in vitro and in vivo systems [22, 23]. Because of a positive role of c-Myc in the regulation of telomere length, we propose that c-Myc may block the inhibitory effect of TRF1 on telomere replication through a direct protein-protein interaction, and thus lead to an increase of telomere length when it is overexpressed. Future studies will reveal exactly how the C-terminal region of c-Myc counteracts the inhibitory activity of TRF1/PIN2 and participates in the regulation of telomere length in vivo.
Acknowledgments
We wish to thank other members of the Chen laboratory for their assistance and value discussions. We also thank Dr. K. P. Lu for providing us TRF1/PIN2 expression plasmid. This work was supported in part by grants from the National Institutes of Health (to J.C.). J.C is a recipient of the Era of Hope Scholars award from Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 3.Cervantes RB, Lundblad V. Mechanisms of chromosome-end protection. Curr Opin Cell Biol. 2002;14:351–356. doi: 10.1016/s0955-0674(02)00325-3. [DOI] [PubMed] [Google Scholar]
- 4.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21:532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 5.Shen M, Haggblom C, Vogt M, Hunter T, Lu KP. Characterization and cell cycle regulation of the related human telomeric proteins Pin2 and TRF1 suggest a role in mitosis. Proc Natl Acad Sci U S A. 1997;94:13618–13623. doi: 10.1073/pnas.94.25.13618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282:1484–1487. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 8.Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- 9.Karlseder J, Kachatrian L, Takai H, Mercer K, Hingorani S, Jacks T, de Lange T. Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol Cell Biol. 2003;23:6533–6541. doi: 10.1128/MCB.23.18.6533-6541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofer B, Backhaus S, Timmis KN. The biphenyl/polychlorinated biphenyl-degradation locus (bph) of Pseudomonas sp. LB400 encodes four additional metabolic enzymes. Gene. 1994;144:9–16. doi: 10.1016/0378-1119(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 12.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–699. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- 14.Lutz W, Leon J, Eilers M. Contributions of Myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- 15.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 16.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, Bishop JM. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- 18.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 19.Greenberg RA, O'Hagan RC, Deng H, Xiao Q, Hann SR, Adams RR, Lichtsteiner S, Chin L, Morin GB, DePinho RA. Telomerase reverse transcriptase gene is a direct target of c-Myc but is not functionally equivalent in cellular transformation. Oncogene. 1999;18:1219–1226. doi: 10.1038/sj.onc.1202669. [DOI] [PubMed] [Google Scholar]
- 20.Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer cells. Cancer Res. 1999;59:551–557. [PubMed] [Google Scholar]
- 21.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007 doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 22.Smucker EJ, Turchi JJ. TRF1 inhibits telomere C-strand DNA synthesis in vitro. Biochemistry. 2001;40:2426–2432. doi: 10.1021/bi001871o. [DOI] [PubMed] [Google Scholar]
- 23.Ohki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]


