Abstract
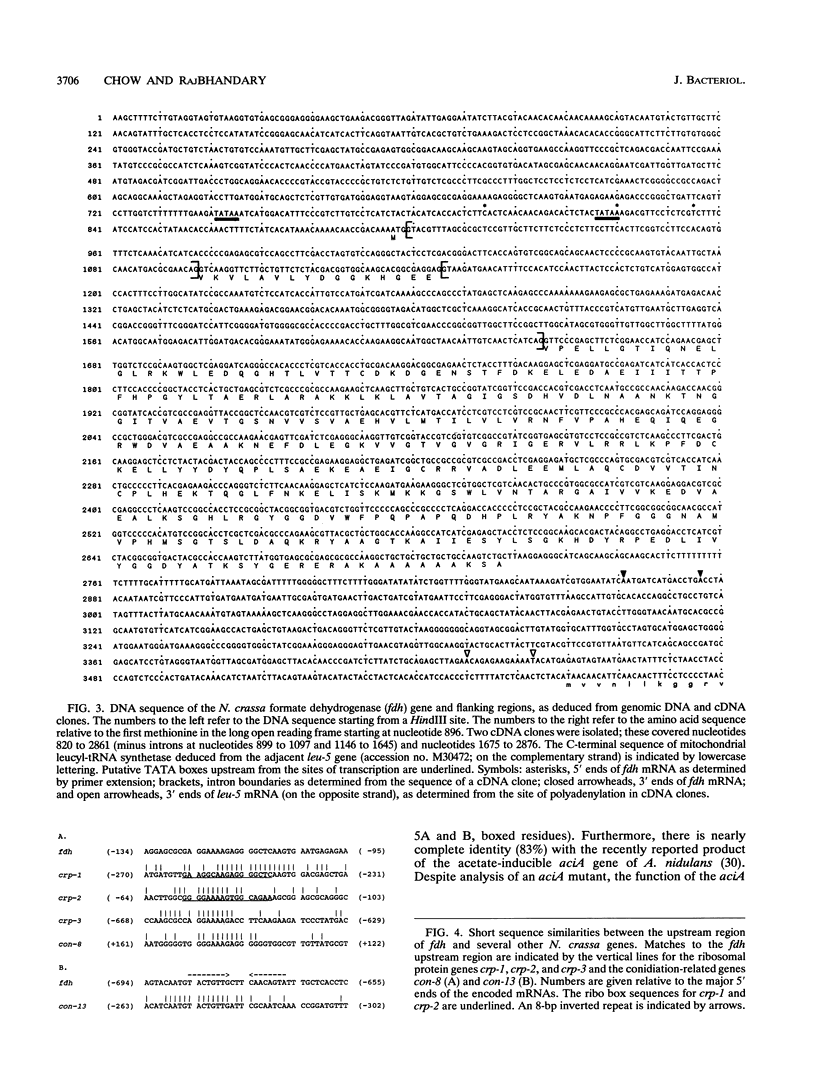
We have isolated and characterized a gene, fdh, from Neurospora crassa which is developmentally regulated and which produces formate dehydrogenase activity when expressed in Escherichia coli. The gene is closely linked (less than 0.6 kb apart) to the leu-5 gene encoding mitochondrial leucyl-tRNA synthetase; the two genes are transcribed convergently from opposite strands. The expression patterns of these genes differ: fdh mRNA is found only during conidiation and early germination and is not detectable during mycelial growth, while leu-5 mRNA appears during germination and mycelial growth. The structure of the fdh gene was determined from the sequence of cDNA and genomic DNA clones and from mRNA mapping studies. The gene encodes a 375-amino-acid-long protein with sequence similarity to NAD-dependent dehydrogenases of the E. coli 3-phosphoglycerate dehydrogenase (serA gene product) subfamily. In particular, there is striking sequence similarity (52% identity) to formate dehydrogenase from Pseudomonas sp. strain 101. All of the residues thought to interact with NAD in the crystal structure of the Pseudomonas enzyme are conserved in the N. crassa enzyme. We have further shown that expression of the N. crassa gene in E. coli leads to the production of formate dehydrogenase activity, indicating that the N. crassa gene specifies a functional polypeptide.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Applegarth D. A., Kirby L. Annotated bibliography: chorionic villus biopsy and diagnosis of inherited diseases using recombinant DNA analysis. Pediatr Pathol. 1984;2(4):501–502. doi: 10.3109/15513818409025897. [DOI] [PubMed] [Google Scholar]
- Axley M. J., Grahame D. A., Stadtman T. C. Escherichia coli formate-hydrogen lyase. Purification and properties of the selenium-dependent formate dehydrogenase component. J Biol Chem. 1990 Oct 25;265(30):18213–18218. [PubMed] [Google Scholar]
- Bell-Pedersen D., Dunlap J. C., Loros J. J. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 1992 Dec;6(12A):2382–2394. doi: 10.1101/gad.6.12a.2382. [DOI] [PubMed] [Google Scholar]
- Benarous R., Chow C. M., RajBhandary U. L. Cytoplasmic leucyl-tRNA synthetase of Neurospora crassa is not specified by the leu-5 locus. Genetics. 1988 Aug;119(4):805–814. doi: 10.1093/genetics/119.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):849–855. doi: 10.1128/mcb.5.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Protein changes during the asexual cycle of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):839–848. doi: 10.1128/mcb.5.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chow C. M., Metzenberg R. L., Rajbhandary U. L. Nuclear gene for mitochondrial leucyl-tRNA synthetase of Neurospora crassa: isolation, sequence, chromosomal mapping, and evidence that the leu-5 locus specifies structural information. Mol Cell Biol. 1989 Nov;9(11):4631–4644. doi: 10.1128/mcb.9.11.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C. M., Rajbhandary U. L. Regulation of the nuclear genes encoding the cytoplasmic and mitochondrial leucyl-tRNA synthetases of Neurospora crassa. Mol Cell Biol. 1989 Nov;9(11):4645–4652. doi: 10.1128/mcb.9.11.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant G. A. A new family of 2-hydroxyacid dehydrogenases. Biochem Biophys Res Commun. 1989 Dec 29;165(3):1371–1374. doi: 10.1016/0006-291x(89)92755-1. [DOI] [PubMed] [Google Scholar]
- Hager K. M., Yanofsky C. Genes expressed during conidiation in Neurospora crassa: molecular characterization of con-13. Gene. 1990 Dec 15;96(2):153–159. doi: 10.1016/0378-1119(90)90247-o. [DOI] [PubMed] [Google Scholar]
- Hao R., Schmit J. C. Purification and characterization of glutamate decarboxylase from Neurospora crassa conidia. J Biol Chem. 1991 Mar 15;266(8):5135–5139. [PubMed] [Google Scholar]
- Kinnaird J. H., Fincham J. R. The complete nucleotide sequence of the Neurospora crassa am (NADP-specific glutamate dehydrogenase) gene. Gene. 1983 Dec;26(2-3):253–260. doi: 10.1016/0378-1119(83)90195-6. [DOI] [PubMed] [Google Scholar]
- Kreader C. A., Heckman J. E. Isolation and characterization of a Neurospora crassa ribosomal protein gene homologous to CYH2 of yeast. Nucleic Acids Res. 1987 Nov 11;15(21):9027–9042. doi: 10.1093/nar/15.21.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamzin V. S., Aleshin A. E., Popov B. O., Arutiunian E. G. NAD-zavisimaia formiatdegidrogenaza metilotrofnykh bakterii Pseudomonas sp. 101. II. Prostranstvennaia struktura fermenta pri razreshenii 3,0 A. Bioorg Khim. 1990 Mar;16(3):336–344. [PubMed] [Google Scholar]
- Lamzin V. S., Aleshin A. E., Strokopytov B. V., Yukhnevich M. G., Popov V. O., Harutyunyan E. H., Wilson K. S. Crystal structure of NAD-dependent formate dehydrogenase. Eur J Biochem. 1992 Jun 1;206(2):441–452. doi: 10.1111/j.1432-1033.1992.tb16945.x. [DOI] [PubMed] [Google Scholar]
- Lauter F. R., Russo V. E., Yanofsky C. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 1992 Dec;6(12A):2373–2381. doi: 10.1101/gad.6.12a.2373. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbach M. J., Sachs M. S., Yanofsky C. The Neurospora crassa arg-2 locus. Structure and expression of the gene encoding the small subunit of arginine-specific carbamoyl phosphate synthetase. J Biol Chem. 1990 Jul 5;265(19):10981–10987. [PubMed] [Google Scholar]
- Roberts A. N., Berlin V., Hager K. M., Yanofsky C. Molecular analysis of a Neurospora crassa gene expressed during conidiation. Mol Cell Biol. 1988 Jun;8(6):2411–2418. doi: 10.1128/mcb.8.6.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. N., Yanofsky C. Genes expressed during conidiation in Neurospora crassa: characterization of con-8. Nucleic Acids Res. 1989 Jan 11;17(1):197–214. doi: 10.1093/nar/17.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M. S., Bertrand H., Metzenberg R. L., RajBhandary U. L. Cytochrome oxidase subunit V gene of Neurospora crassa: DNA sequences, chromosomal mapping, and evidence that the cya-4 locus specifies the structural gene for subunit V. Mol Cell Biol. 1989 Feb;9(2):566–577. doi: 10.1128/mcb.9.2.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M. S., Yanofsky C. Developmental expression of genes involved in conidiation and amino acid biosynthesis in Neurospora crassa. Dev Biol. 1991 Nov;148(1):117–128. doi: 10.1016/0012-1606(91)90322-t. [DOI] [PubMed] [Google Scholar]
- Saleeba J. A., Cobbett C. S., Hynes M. J. Characterization of the amdA-regulated aciA gene of Aspergillus nidulans. Mol Gen Genet. 1992 Nov;235(2-3):349–358. doi: 10.1007/BF00279380. [DOI] [PubMed] [Google Scholar]
- Sawers G., Heider J., Zehelein E., Böck A. Expression and operon structure of the sel genes of Escherichia coli and identification of a third selenium-containing formate dehydrogenase isoenzyme. J Bacteriol. 1991 Aug;173(16):4983–4993. doi: 10.1128/jb.173.16.4983-4993.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit J. C., Brody S. Biochemical genetics of Neurospora crassa conidial germination. Bacteriol Rev. 1976 Mar;40(1):1–41. doi: 10.1128/br.40.1.1-41.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. G., Tyler B. M. Coordinate expression of ribosomal protein genes in Neurospora crassa and identification of conserved upstream sequences. Nucleic Acids Res. 1991 Dec 11;19(23):6511–6517. doi: 10.1093/nar/19.23.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman T. C. Biosynthesis and function of selenocysteine-containing enzymes. J Biol Chem. 1991 Sep 5;266(25):16257–16260. [PubMed] [Google Scholar]
- Timberlake W. E., Marshall M. A. Genetic regulation of development in Aspergillus nidulans. Trends Genet. 1988 Jun;4(6):162–169. doi: 10.1016/0168-9525(88)90022-4. [DOI] [PubMed] [Google Scholar]
- Tyler B. M., Harrison K. A Neurospora crassa ribosomal protein gene, homologous to yeast CRY1, contains sequences potentially coordinating its transcription with rRNA genes. Nucleic Acids Res. 1990 Oct 11;18(19):5759–5765. doi: 10.1093/nar/18.19.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]



