Abstract
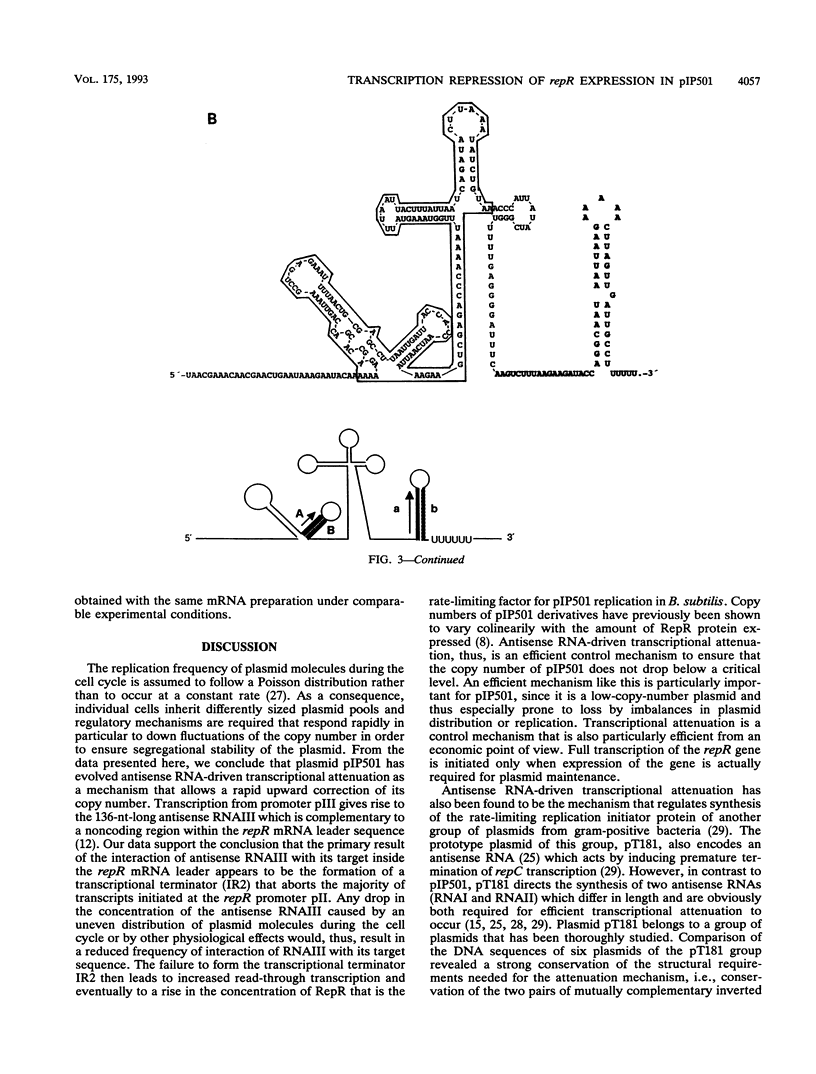
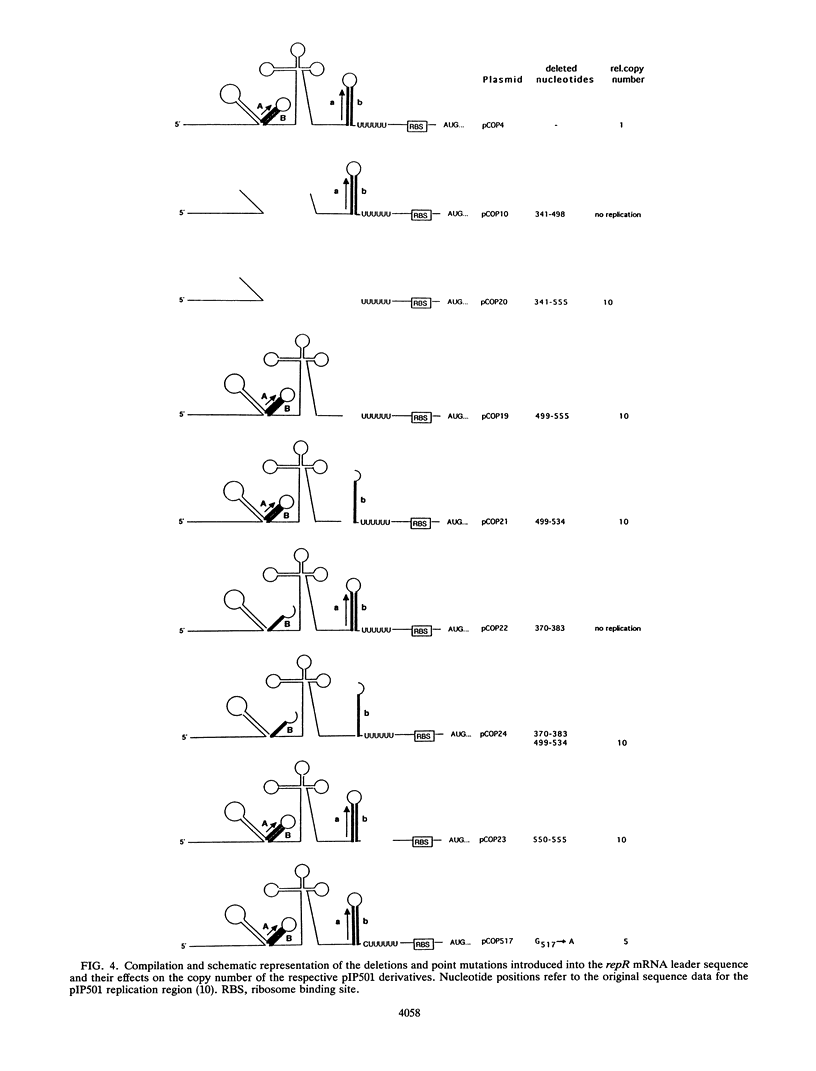
Expression of the rate-limiting initiator protein RepR of plasmid pIP501 is controlled by the antisense RNAIII. Mutational alteration of individual G residues within the single-stranded loops of RNAIII led to an increase in copy number. In contrast to the G-rich single-stranded loops, two smaller AT-rich loops of RNAIII were found to be dispensable for its inhibitory function. Reciprocal mutations in the same loop compensated for each other's effect, and a destabilization of the major stem structure of RNAIII also resulted in an increased copy number. These data were consistent with the idea that the interaction of RNAIII with its target starts with the formation of a kissing complex between the single-stranded loops of both molecules. The repR mRNA leader sequence, which includes the target of RNAIII, is able to assume two alternative structures due to the presence of two inverted repeats the individual sequences of which are mutually complementary. In the presence of the antisense RNAIII, one of these inverted repeats (IR2) is forced to fold into a transcriptional terminator structure that prevents transcription of the repR gene. In the absence of RNAIII, formation of the transcriptional terminator is prevented and expression of the essential repR gene can proceed normally. This antisense RNA-driven transcriptional attenuation mechanism was supported by extensive deletional analysis and direct evidence that IR2 functions as a transcriptional terminator.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barthelemy I., Salas M., Mellado R. P. In vivo transcription of bacteriophage phi 29 DNA: transcription initiation sites. J Virol. 1986 Dec;60(3):874–879. doi: 10.1128/jvi.60.3.874-879.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke D., Ferretti J. J. Physical mapping of plasmid pDB101: a potential vector plasmid for molecular cloning in streptococci. Plasmid. 1980 Sep;4(2):130–138. doi: 10.1016/0147-619x(80)90002-5. [DOI] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S., Ferretti J. J. Plasmid pGB301, a new multiple resistance streptococcal cloning vehicle and its use in cloning of a gentamicin/kanamycin resistance determinant. Mol Gen Genet. 1981;182(3):414–421. doi: 10.1007/BF00293929. [DOI] [PubMed] [Google Scholar]
- Behnke D., Gilmore M. S. Location of antibiotic resistance determinants, copy control, and replication functions on the double-selective streptococcal cloning vector pGB301. Mol Gen Genet. 1981;184(1):115–120. doi: 10.1007/BF00271206. [DOI] [PubMed] [Google Scholar]
- Behnke D., Malke H., Hartmann M., Walter F. Post-transformational rearrangement of an in vitro reconstructed group-A streptococcal erythromycin resistance plasmid. Plasmid. 1979 Oct;2(4):605–616. doi: 10.1016/0147-619x(79)90058-1. [DOI] [PubMed] [Google Scholar]
- Blomberg P., Nordström K., Wagner E. G. Replication control of plasmid R1: RepA synthesis is regulated by CopA RNA through inhibition of leader peptide translation. EMBO J. 1992 Jul;11(7):2675–2683. doi: 10.1002/j.1460-2075.1992.tb05333.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Behnke D., Alonso J. C. Molecular analysis of the replication region of the conjugative Streptococcus agalactiae plasmid pIP501 in Bacillus subtilis. Comparison with plasmids pAM beta 1 and pSM19035. Nucleic Acids Res. 1990 Aug 25;18(16):4783–4790. doi: 10.1093/nar/18.16.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Behnke D. Characterization of the minimal origin required for replication of the streptococcal plasmid pIP501 in Bacillus subtilis. Mol Microbiol. 1992 Dec;6(23):3501–3510. doi: 10.1111/j.1365-2958.1992.tb01785.x. [DOI] [PubMed] [Google Scholar]
- Brantl S., Behnke D. Copy number control of the streptococcal plasmid pIP501 occurs at three levels. Nucleic Acids Res. 1992 Feb 11;20(3):395–400. doi: 10.1093/nar/20.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Behnke D. The amount of RepR protein determines the copy number of plasmid pIP501 in Bacillus subtilis. J Bacteriol. 1992 Aug;174(16):5475–5478. doi: 10.1128/jb.174.16.5475-5478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Nowak A., Behnke D., Alonso J. C. Revision of the nucleotide sequence of the Streptococcus pyogenes plasmid pSM19035 repS gene. Nucleic Acids Res. 1989 Dec 11;17(23):10110–10110. doi: 10.1093/nar/17.23.10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brantl S., Nuez B., Behnke D. In vitro and in vivo analysis of transcription within the replication region of plasmid pIP501. Mol Gen Genet. 1992 Jul;234(1):105–112. doi: 10.1007/BF00272351. [DOI] [PubMed] [Google Scholar]
- Bruand C., Ehrlich S. D., Jannière L. Unidirectional theta replication of the structurally stable Enterococcus faecalis plasmid pAM beta 1. EMBO J. 1991 Aug;10(8):2171–2177. doi: 10.1002/j.1460-2075.1991.tb07752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buu-Hoï A., Bieth G., Horaud T. Broad host range of streptococcal macrolide resistance plasmids. Antimicrob Agents Chemother. 1984 Feb;25(2):289–291. doi: 10.1128/aac.25.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton S., Projan S. J., Highlander S. K., Moghazeh S. M., Novick R. P. Control of pT181 replication II. Mutational analysis. EMBO J. 1984 Oct;3(10):2407–2414. doi: 10.1002/j.1460-2075.1984.tb02147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Yagi Y., Dunny G. M., Schultz S. K. Characterization of three plasmid deoxyribonucleic acid molecules in a strain of Streptococcus faecalis: identification of a plasmid determining erythromycin resistance. J Bacteriol. 1974 Jan;117(1):283–289. doi: 10.1128/jb.117.1.283-289.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel H. W., Soedirman N., Rost J. A., van Leeuwen W. J., van Embden J. D. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J Bacteriol. 1980 May;142(2):407–413. doi: 10.1128/jb.142.2.407-413.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson E. M., Chace N. M., London S. B., London J. Transfer of plasmid-mediated antibiotic resistance from streptococci to lactobacilli. J Bacteriol. 1979 Jan;137(1):614–619. doi: 10.1128/jb.137.1.614-619.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. F., Kunka B. S. Plasmid transfer in Pediococcus spp.: intergeneric and intrageneric transfer of pIP501. Appl Environ Microbiol. 1983 Jul;46(1):81–89. doi: 10.1128/aem.46.1.81-89.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlander S. K., McKay L. L., Schachtele C. F. Molecular cloning of the lactose-metabolizing genes from Streptococcus lactis. Appl Environ Microbiol. 1984 Aug;48(2):347–351. doi: 10.1128/aem.48.2.347-351.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horodniceanu T., Bouanchaud D. H., Bieth G., Chabbert Y. A. R plasmids in Streptococcus agalactiae (group B). Antimicrob Agents Chemother. 1976 Nov;10(5):795–801. doi: 10.1128/aac.10.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Bruand C., Ehrlich S. D. Structurally stable Bacillus subtilis cloning vectors. Gene. 1990 Mar 1;87(1):53–61. doi: 10.1016/0378-1119(90)90495-d. [DOI] [PubMed] [Google Scholar]
- Kawamura F., Doi R. H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984 Oct;160(1):442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C. C., Novick R. P. Plasmid pT181 replication is regulated by two countertranscripts. Proc Natl Acad Sci U S A. 1985 Feb;82(3):638–642. doi: 10.1073/pnas.82.3.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Light J. Control of replication of bacterial plasmids: genetics, molecular biology, and physiology of the plasmid R1 system. Plasmid. 1984 Sep;12(2):71–90. doi: 10.1016/0147-619x(84)90054-4. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Projan S. J., Carleton S., Highlander S. K., Gruss A., Khan S. A., Iordanescu S. Control of pT181 replication I. The pT181 copy control function acts by inhibiting the synthesis of a replication protein. EMBO J. 1984 Oct;3(10):2399–2405. doi: 10.1002/j.1460-2075.1984.tb02146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Iordanescu S., Projan S. J., Kornblum J., Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989 Oct 20;59(2):395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: formation of an initial transient complex is rate-limiting for antisense RNA--target RNA pairing. EMBO J. 1990 Nov;9(11):3777–3785. doi: 10.1002/j.1460-2075.1990.tb07591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: kinetics of in vitro interaction between the antisense RNA, CopA, and its target, CopT. EMBO J. 1988 Oct;7(10):3279–3288. doi: 10.1002/j.1460-2075.1988.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson C., Wagner E. G., Nordström K. Control of replication of plasmid R1: structures and sequences of the antisense RNA, CopA, required for its binding to the target RNA, CopT. EMBO J. 1990 Nov;9(11):3767–3775. doi: 10.1002/j.1460-2075.1990.tb07590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich P. M., Haykinson MYa, Arutyunova L. S., Yomantas YuV, Stepanov A. I. The structure and source of plasmid DNA determine the cloning properties of vectors for Bacillus subtilis. Basic Life Sci. 1985;30:635–656. doi: 10.1007/978-1-4613-2447-8_44. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaberg D. R., Clewell D. B., Glatzer L. Conjugative transfer of R-plasmids from Streptococcus faecalis to Staphylococcus aureus. Antimicrob Agents Chemother. 1982 Aug;22(2):204–207. doi: 10.1128/aac.22.2.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinfield T. J., Oultram J. D., Thompson D. E., Brehm J. K., Minton N. P. Physical characterisation of the replication region of the Streptococcus faecalis plasmid pAM beta 1. Gene. 1990 Mar 1;87(1):79–90. [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication. Intermediates in the binding of RNA I and RNA II. J Mol Biol. 1990 Apr 20;212(4):683–694. doi: 10.1016/0022-2836(90)90230-j. [DOI] [PubMed] [Google Scholar]
- Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984 Oct;38(3):861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Som T. Control of ColE1 plasmid replication: enhancement of binding of RNA I to the primer transcript by the Rom protein. Cell. 1984 Oct;38(3):871–878. doi: 10.1016/0092-8674(84)90282-4. [DOI] [PubMed] [Google Scholar]
- Wagner E. G., Blomberg P., Nordström K. Replication control in plasmid R1: duplex formation between the antisense RNA, CopA, and its target, CopT, is not required for inhibition of RepA synthesis. EMBO J. 1992 Mar;11(3):1195–1203. doi: 10.1002/j.1460-2075.1992.tb05160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]